Abstract
Genetic anomalies are known to affect about 15% of infertile patients with azoospermia or severe oligozoospermia. Despite a throughout diagnostic work-up, in up to the 72% of the male partners of infertile couples, no etiological factor can be found; hence, the cause of infertility remains unclear. Recently, several novel genetic causes of spermatogenic failure (SPGF) have been described. The aim of this review was to collect all the available evidence of SPGF genetics, matching data from in-vitro and animal models with those in human beings to provide a comprehensive and updated overview of the genes capable of affecting spermatogenesis. By reviewing the literature, we provided a list of 60 candidate genes for SPGF. Their investigation by Next Generation Sequencing in large cohorts of patients with apparently idiopathic infertility would provide new interesting data about their racial- and ethnic-related prevalence in infertile patients, likely raising the diagnostic yields. We propose a phenotype-based approach to identify the genes to look for.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Genetic anomalies are known to affect about 15% of infertile male patients and in particular those with azoospermia or severe oligozoospermia (Asero et al. 2014). At moment, the genetic diagnostic work-up of male infertility includes the screening of some candidate genes (whose list is progressively increasing) when a central hypogonadism is diagnosed, karyotype analysis, and/or Yq microdeletion in the presence of a primary testicular failure, CFTR gene evaluation for the diagnosis of congenital bilateral agenesia of the vas deferens when an obstructive form is suspected (Asero et al. 2014). This work flow has remained unchanged in the last 20 years. Worryingly, despite a throughout diagnostic work-up (including genetic testing), in up to 72% of the male partners of infertile couples no etiological factor can be found and the cause of infertility is elusive (Tüttelmann et al. 2018). Accordingly, a prospective single Centre study on 1737 patients with low sperm count has esteemed the rate of idiopathic oligozoospermia up to 75% (Punab et al. 2017).
Recently, much effort has been made in the attempt to identify novel candidate genes responsible for spermatogenic failure (SPGF) through genome-wide association studies (GWAS). As such, the number of reports of monogenic forms of SPGF is progressively increasing. Whether this genetic selection may impact on survival, as it appears for cancer (Voskarides 2018), has to be demonstrated.
Therefore, the aim of this review was to gather all the available data on the recently identified genetic causes of SPGF, including the clinical feature of the affected patients and the evidence coming up from in-vitro and animal studies, in the attempt to suggest a comprehensive and updated panel of genes to look for. To better understand the possible mechanisms through which such genes may act, the current knowledge on the genetic regulation of spermatogenesis is discussed below.
Physiology of spermatogenesis
Spermatogenesis is a 74 day-long process occurring in seminiferous tubules through which diploid cells develop into haploid mature spermatozoa. It includes three main phases: proliferation of spermatogonia, meiotic division of spermatocytes and changes in shape and nuclear content of spermatids (spermiogenesis). Such processes require intimate interaction between germ and somatic cells, such as Sertoli and peritubular myoid cells (Neto et al. 2016; Potter and Falco 2017).
Spermatogonial stem cells (SSCs) are undifferentiated spermatogonia defined by the ability to maintain the potential to self-renew. They differentiate into type Apaired and type Aaligned spermatogonia which have loss the self-renewal. They latter divide into differentiating spermatogonia that include type A1–4 and type B spermatogonia and this takes 16 days. Type B spermatogonia subsequently divide into primary spermatocyte in other 16 days (Chen and Liu 2015).
Several markers are known to be expressed in these cells and act in the regulation of their proliferation or differentiation. In particular, PAX7 and ID4 are expressed in mice SSCs, and among these, PAX7 is detectable also in human SSCs (Chen and Liu 2015). Furthermore, PLZF, a transcription factor required for SSCs self-renewal (Chen and Liu 2015) and SALL4, which removes PLZF from its targets (Liao et al. 2014) have been detected in human spermatogonia (Fischer et al. 2008; Vigueras-Villaseñor et al. 2015). Evidence in mice suggests that the competition PLZF/SALL4 may to be involved in the balance of quiescent and cycling SSCs/progenitor cells (Liao et al. 2014). In addition, POU5F1, c-Kit, PLAP, and AP2γ are required for gonocytes pluripotency in humans (Vigueras-Villaseñor et al. 2015).
Mice undifferentiated spermatogonia (SSCs, type Apaired and type Aaligned) express GFRA1, LIN28, NANOS2, NGN3, NANOS3, PLZF, SALL4, and CHD1proteins (Chen and Liu 2015). Among these, studies on human testicular tissue provide data for NANOS2 being expressed in prenatal germ cells and in spermatogonia, spermatocytes, and round spermatids of the adult testis (Kusz et al. 2009), NANOS3, detected in type A spermatogonia, primary spermatocytes, round and elongated spermatids (Julaton and Pera 2011), PLZF (Fischer et al. 2008), and SALL4 (Kohlhase et al. 2002).
Finally, SOHLH1 and SOHLH2 are selectively expressed in differentiating spermatogonia (Chen and Liu 2015) and coordinate their differentiation in the human (Suzuki et al. 2012). The transcriptional regulator DMRT1, localized in human spermatogonia and spermatocytes (Ying et al. 2007), has been found to induce Sohlh1 expression in spermatogonia and to repress their commitment to meiosis (Matson et al. 2010).
Primary spermatocytes undergo to the first meiotic division (a 16 day-long process) and differentiate into secondary spermatocytes. The first meiotic division occurs through a complex series of key molecular events happening in prophase, such as programmed double-strand breaks (DSBs) (leptotene), pairing of homologue chromosomes (zygotene), and crossing over (CO) (pachytene) (Pawlowski and Cande 2005).
At first, centriole duplication, which is needed to ensure proper chromosome segregation, happens. It is regulated by Polo-like kinase 4 (PLK-4) (Harris et al. 2011). In mice, Plk-4 expression is detected starting from pachytene spermatocyte stage and up to the end of spermatogenesis (Harris et al. 2011). Centrosome and spindle integrity, required for chromosome alignment, are ensured by the human augmin complex (Lawo et al. 2009). DSBs are induced by a specific DNA topoisomerase. In detail, SPO11β is the catalytic subunit of the DNA topoisomerase VI-like protein complex, that is indispensable for meiotic recombination (Robert et al. 2016). The human and the mouse SPO11β proteins share the 82% of amino acid identity. In mice, SPO11β is selectively expressed in pachitene spermatocytes when CO occurs (Shannon et al. 1999). After DSBs’ formation, the single-strand DNA (ssDNA) is coated by the trimeric replication protein A (RPA) complex, which includes the RPA1, RPA2, and RPA3 subunits. It prevents ssDNA from degradation (Bochkareva et al. 2002).
Pairing of homologous chromosomes requires the synaptonemal complex, a tripartite protein structure of bivalent chromosomes, whose assembly is indispensable for successful progression of the first meiotic division (Fraune et al. 2012). It is made up of two lateral and one central element and requires MEIOB, RAD51, and SPATA22 proteins (Xu et al. 2017). Early during leptotene stage, it is formed along each chromosome and is required for CO.
CO originates from the recombination of double-strand breaks (DSBs) of homologues chromosomes (Souquet et al. 2013), which needs DSBs repair. The complex RAD51/DMC1 is specific for meiotic DSBs repair (Neale and Keeney 2006). MEIOB and SPATA22 ensure RAD51/DMC1 complex stability (Souquet et al. 2013; Xu et al. 2017). Many other proteins seem to play a role in chromosome recombination, such as MSH4, MSH5, TEX11, TEX 15, MLH1/MLH3, SYCE, HSF2, HEI10, RNF212, and CNTD1 (Xu et al. 2017; Xing et al. 2005; Yang et al. 2008).
Once chromosomal recombination occurs, homologous chromosomes split up. This process is called segregation and needs that kinetochores attach to microtubules stemming from the same pole. Ensuing sister chromosomes segregation, daughter cells divide, but they remain connected through intercellular bridges (Greenbaum et al. 2006). The latter have been proposed to play a role in germ-cell communication, synchronization, and chromosome dosage compensation in haploid cells (Greenbaum et al. 2006). TEX14, a testis-specific protein sharing 64% of identity with the mouse homologue, localizes into the intercellular bridges of differentiating spermatogonia until their maturation into spermatozoa and seems to be required for their formation (Greenbaum et al. 2006).
Secondary spermatocytes then undergo to the second meiotic division (taking few hours) during which segregation of sister chromatids occurs (Muller et al. 2013). In addition, after chromatids segregation, cell division results in intercellular bridges (Greenbaum et al. 2006).
Later, spermiogenesis takes place. It is a 26-day-long process during which DNA packaging, acrosome biogenesis, midpiece formation, flagella organization, and cytoplasm expulsion occur. These events enable differentiation of round spermatids in mature spermatozoa (Tournaye et al. 2017). Molecular mechanisms occurring in spermiogenesis are largely unknown. Evidence suggests a role for DPY19L2 (Harbuz et al. 2011) and SIRT1 (Chao et al. 2017) in acrosome synthesis. Genes involved in human flagella and axonema organization have been previously reviewed (Ji et al. 2017).
The disruption of the fine and complex mechanisms regulating spermatogenesis may result in SPGF (Fig. 1).
Candidate genes
We classified candidate genes according to their location in autosomes or heterochromosomes. A complete list of genes thought to cause human SPGF is reported in Table 1. We discussed only the newly identified ones.
X-linked genes
TEX11(Xq13.1) is a conserved gene among vertebrates encoding for a 104 kDa protein that mediates protein–protein interactions. TEX11 is considered a meiosis-specific factor playing a role in DNA DSBs repair (Adelman et al. 2008) and showing the highest level of expression in zygotene spermatocytes and the lowest one in late leptotene spermatocytes (Blatch et al. 1999). In a cohort of azoospermic patients undergone to testis histology, mutations have been detected in the 15% (5/33) of patients with meiotic arrest and in none (0/63) with Sertoli cell only syndrome (SCOS). All patients with TEX11 mutations had FSH and total testosterone (TT) within the normal range, aside of one patient with partial meiotic arrest [p.A698T (c.2092G>A)] showing few post meiotic cells and high FSH levels (28 IU/l) (Yatsenko et al. 2015). LH and testicular volumes (TV) were not reported (Yatsenko et al. 2015). TEX11 mutations, most of which causing azoospermia, have already been reviewed (Sha et al. 2018a, b, c). Recently, a novel TEX11 missense mutation [p.W856C (c.2653G>T)] has been described in two brothers with azoospermia due to meiotic arrest, having hormone values and a mean TV of about 15 ml (Sha et al. 2018a, b, c). These results confirm that TEX11 is required for human meiosis. Its disruption in expression or function causes the SPGF X-linked, 2 (SPGFX2) (OMIM 309120).
HAUS7(Xq28) encodes for a component of the human augmin complex, a 8-subunit compound ensuring centrosome and spindle integrity, critical structures for chromosome alignment and separation during mitosis and meiosis (Lawo et al. 2009). The hemizygous variant c.G386T, p.G129V (a highly pathogenic mutation by in silico analysis) has recently been described in two brothers with severe oligozoospermia. They had normal hormone values. TV were not reported (Li et al. 2018a, b).
USP26(Xq26.2) encodes for the ubiquitin-specific protease 26, a testis-specific expressed protein belonging to the family of deubiquitinating enzymes (Wosnitzer et al. 2014). In mice, this protein localizes in blood–testis barrier and near the Sertoli-germ-cell interface (Lin et al. 2011a, b). Stouff and colleagues described three USP26 variants (c494T-C, p.L165S, c.370_371insACA, c.1423C-T, p.H475Y) in 8/111 (7.2%) patients with SCOS (Stouff et al. 2005a, b). However, among these, two (494T-C and 370_371insACA) were present with significant frequencies in sub-Saharian African and South and East Asian population, including fertile men (Ravel et al. 2006), and therefore, they do not seem to affect spermatogenesis. The c.1082G-A, p.R344W USP26 transition has been firstly detected in two Han Chinese azoospermic men with arrest at the spermatocyte stage (Ma et al. 2016). The study included a cohort of 776 NOA patients and 709 fertile men. The mutation was not detected in any of controls. Co-immunoprecipitation analysis demonstrated a decreased deubiquitinating activity of the R344W variant and its incapacity to bind the androgen receptor (AR). The authors concluded that this variant may play a role in human SPGF through a disruption in AR function (Ma et al. 2016).
Y-linked genes
DBY(or DDX3Y)(Yq11.221) encodes for a conserved RNA DDX3 helicase specifically expressed in pre-meiotic spermatogonial cells, which acts in RNA unwinding, annealing and remodeling of ribonucleoprotein complexes (for review, see Kotov et al. 2017). It maps within the AZFa region, whose deletion causes SCOS. Studies indicate that DBYexpression is required for human fetal germ-cell proliferation (Kotov et al. 2017). To investigate the role of DBY in the AZFa microdeletion phenotype, Ramathal and coworker developed a line of pluripotent stem cells lacking the AZFa and complemented with DBY. These cells were transplanted in mice seminiferous tubules, resulting in the rescuing of spermatogenesis, in contrast with the findings in the AZFa-deleted cell line used as control (Ramathal et al. 2015). These findings strongly support the role of the lack of DBY in the determination of the phenotype of patients with AZFa deletion. Accordingly, DBY deletions have been identified in patients with SCOS or severe hypospermatogenesis (Foresta et al. 2000).
USP9Y(Yq11.221) encodes for the ubiquitin-specific protease 9 and maps inside the AZFa region. Despite mutations and deletions in this gene have been reported in azoospermic patients (Sun et al. 1999; Foresta et al. 2000), a 513,594-bp deletion in the AZFa region encompassing the USP9Y gene has been described in a normozoospermic men, his brother, and father (Luddi et al. 2009). Therefore, it does not seem to be required for normal sperm production.
Genes on autosomes
MEIOB(16p13.3) is an evolutionary conserved protein among vertebrates containing single-strand DNA (ssDNA)-binding sites (Souquet et al. 2013). In mice, its D383 single residue seems to be indispensable for MEIOB, SPATA 22, and PRA interaction (Xu et al. 2017). Meiob-deficientmice do not form CO in germ cells and are infertile due to a meiotic arrest (Souquet et al. 2013; Luo et al. 2013). In line with these findings, the homozygosity for a missense mutation of the evolutionary conserved asparagine 64 (N64I) of MEIOB protein (belonging to the RPA1-binding domain) has been reported in patients with NOA. The analysis has been carried out in four brothers having azoospermia. Hormonal data and TV were available only in three of them. Two brothers had FSH and TV with the normal range. The last one had the right TV measuring 5 ml, the left 15 ml and his FSH levels were 13.5 IU/l. Histologic analysis (available only in one brother) showed complete spermatocyte maturation arrest (Gershoni et al. 2017). These results suggest that MEIOB is required for meiosis I and that its mutation is associated with complete spermatocyte maturation arrest in humans. MEIOB gene mutations are thought to be responsible for SPGF 22 (OMIM 617706).
TEX 14(17q22) encodes for a testis-specific protein of 1497 aminoacidic residues, which is highly conserved among mammals. It includes an ankyrin repeated domain and a protein kinase-C domain. It localizes in germ-cell intercellular bridges (Greenbaum et al. 2006) and acts in the inactivation of germ-cell division (Kim et al. 2015). In Tex 14−/− mice, spermatogenesis halts before the achievement of the first meiotic division (Greenbaum et al. 2006). In addition, a 51-bp (bp) exonic insertion in TEX 14 (resulting in a truncated protein) has been reported to cause maturation arrest at early meiotic phase of spermatogenesis in pigs (Sironen et al. 2011). A 10 bp deletion in TEX 14 (exon 16: 2668-2678del), resulting in a truncated protein, has recently been reported in two azoospermic brothers. In accordance with the findings in animals, histologic section showed spermatogonia in all the tubules and few spermatocyte cells, thus suggesting that also in humans, TEX 14 may be required for the early phase of the first meiotic division. Hormonal findings and TV were reported only in one brother, showing normal TV (29 ml, bilaterally) and FSH levels of 12.2 U/l (Gershoni et al. 2017). LH and TT values were within the normal range. TEX 14 mutations cause the SPGF 23 (OMIM 617707).
DNAH6(2p11.2) is a poorly characterized gene encoding for a testis-specific expressed protein (belonging to the dynein family proteins) that has very recently been identified in the neck region of spermatozoa (Li et al. 2018a, b). Although its role in spermatogenesis is unknown, the homozygosity for the H3471Q missense mutation has been identified in two azoospermic brothers showing complete spermatocyte maturation arrest. They displayed normal FSH serum levels and borderline TV (right TV measured 8 and 10 ml, respectively, left TV measured 15 ml in both patients). One patient had an history positive for right cryptorchidism. LH and TT values were in the normal range. The other members of the family were carries of the mutation but did not show NOA. At the light of such findings, this gene has been hypothesized to play a role in rapid prophase movements (Gershoni et al. 2017). Furthermore, compound heterozygous variants in DNAH6 have been described in a patient with globozoospermia and acephalic spermatozoa (Li et al. 2018a, b).
NR5A1(9q33.3), also called steroidogenic factor, encodes for a transcription factor involved in the expression and regulation of genes known to play a role in steroidogenesis, sexual differentiation, and reproduction. Its mutations have been associated with a wide spectrum of phenotypes, such as 46,XY partial and complete gonadal dysgenesis with or without adrenal failure, hypospadias, micropenis and anorchia, and 46,XX primary ovarian insufficiency (Lin et al. 2008; Lourenco et al. 2009).
Among a cohort of infertile patients, NR5A1 mutations have been identified in the 3.9% (4/103) azoospermic or cryptozoospermic patients, in the 4.3% (2/48) of the patients with severe oligozoospermia (sperm concentration < 1 million/ml), and in the 2% (1/50) of the patients with moderate oligozoospermia (sperm concentration from 1 to 10 million/ml). No mutations have been reported in fertile and normozoospermic men or in patients with mild oligozoospermia (sperm count > 10 million/ml) (Bashamboo et al. 2010). Similar findings have been reported also elsewhere (Ferlin et al. 2015).
In contrast with these results, no NR5A1 mutation has been found in a cohort of 414 Indian patients with NOA (Sudhakar et al. 2018). From the histologic point of view, NR5A1 missense mutations result in SCOS, severe hypospermatogenesis or partial spermatocyte arrest in NOA patients (Bashamboo et al. 2010; Ferlin et al. 2015). Considering all cases reported, azoospermic patients with NR5A1 mutations more frequently display high FSH levels (> 10.7 UI/ml) and may exhibit also high LH levels but normal TT values. The mean TV has been reported ranging from 7 to 12.2 ml (Bashamboo et al. 2010; Ferlin et al. 2015). The point mutations p.Arg191Cys (c.571C>T), p.Gly212Ser (c.634G>A), and p.Asp238Asn (c.712G>A) have been described in patients with severe oligozoospermia (< 1 million/ml) and high FSH serum levels. p.Gly123Ala (c.368G>C)/p.Pro129Leu (c.386C>T) have been detected in patients with moderate oligozoospermia (1–10 million/ml) and normal FSH levels (Bashamboo et al. 2010). Currently, NR5A1 mutations cause the SPGF 8 (OMIM 613957).
SOHLH1(9q34.3) encodes for the spermatogenesis- and oogenesis-specific basic helix–loop–helix protein 1, which is involved in spermatogonia differentiation with the SOHLH2 (13q13.3) as well, likely by the regulation of genes involved in SSC pluripotency maintenance (Suzuki et al. 2012). The heterozygosis for the c.346-1G-A splice site mutation has been reported in two of 96 Korean (Choi et al. 2010) and in two of 40 Japanese (Nakamura et al. 2017) NOA patients. No SOHLH2 mutation in human SPGF has been reported so far.
Nanos gene encodes for a translational regulator of specific mRNA involved in Drosophila germ-cell development. Nanos 2 and Nanos 3 play a role in mice spermatogenesis. The human orthologous NANOS 1(10q26.11) seems to be involved in germ-cell development. Indeed, the heterozygosis for the deletion of two single aminoacidic residues (p.Ser78del or p.Ala173del) has been identified in Polish azoospermic patients with low TV (6–10 ml) and high FSH (15.4–18.2 IU/l) levels. The histologic feature, available only in one patient, showed SCOS (Kusz-Zamelczyket al. 2013). Furthermore, the NANOS1 missense mutations pArg246His and Arg276Tyr have been reported in patients with severe oligozoospermia (Kusz-Zamelczyk et al. 2013). Mutations of this gene cause the SPGF 12 (OMIM 615413).
TAF4B(17p13.2), also called TAFII105, encodes for a protein of 862 amino acidic residues, belonging to the family of TATA box-binding proteins (TAFs). Gathering with the TATA-binding protein (TBP), TAFs form the TFIID complex, that is involved in the initiation of gene transcription by RNA polymerase II. TAF4B is predominantly expressed in testis (Freiman et al. 2001). Although its exact role in spermatogenesis is unclear, Taf4b-null male mice became infertile by 3 months, due to an impaired gonocytes proliferation (Falender et al. 2005).
In humans, the nonsense mutation TAF4B p.R611X has been observed in four brothers, three with azoospermia, and one with oligozoospermia (6 million/ml) (Ayhan et al. 2014). This mutation results in a truncated protein of 252 residues, lacking the histone fold domain (residues 653–702) (which increases the DNA-binding activity of TAFs) and the TAF12 interaction domain (residues 830–862) (that promotes the interaction with TAF12). FSH levels of affected men were increased (its values have not been reported). TV and testis histology are not available (Ayhan et al. 2014). TAF4B is involved in SPGF 13 (OMIM 601689).
ZMYND15(17p13.2) encodes for a testis-specific histone deacetylase-dependent transcriptional repressor which has been suggested to control normal temporal expression of haploid cell genes (e.g., Prm1, Tnp1, Spem1, and Catsper3) during spermatogenesis.
In mice, its inactivation results in late spermatids depletion (Yan et al. 2010). Testis histology from azoospermic patients with a nonsense ZMYND15 mutation (p.K507Sfs*3) showed maturation arrest at the spermatid stage. Many primary and secondary spermatocytes and very few spermatids were observed (Ayhan et al. 2014). This mutation resulted in a truncated protein lacking the Pro-rich domain which is essential for the binding of signal transduction and cytoskeleton proteins (Kay et al. 2000).
The mutation has been identified in three azoospermic brothers. Only in one, TESA resulted in twins. FSH levels of affected men were increased. TVs were not available. These findings suggest that, in humans, ZMYND15 may play a role in meiosis II, which is required for spermatid differentiation. Finally, its expression in testis of azoospermic patients has recently been shown to predict a successful sperm retrieval among patients with NOA (Hashemi et al. 2018). ZMYND15 mutations are responsible for SPGF 14 (OMIM 615842).
SYCP3(12q23.2) encodes for the synaptonemal complex protein 3, which has a key role in the first meiotic division (Fraune et al. 2012). In mice, a null mutation of Sycp3 causes azoospermia with meiotic arrest (Yuan et al. 2000).
A deletion (643delA) of SYCP3 resulting in a truncated protein has been described in 11% (2/19) azoospermic patients with maturation arrest and in no fertile controls (0/75). FSH and TV values have not been reported (Miyamoto et al. 2003). Such prevalence may reasonably be overestimated, since in a subsequent study, no SYCP3 mutations have been reported among a cohort of 58 patients with azoospermia and maturation arrest (Stouffs et al. 2005a, b). This gene is currently believed to be responsible for SPGF 4 (OMIM 270960).
HSF2(6q22.31) encodes for a protein belonging to the HSFs family, which activates target genes under stress conditions. In mice, Hsf2 has been shown to interact with the promoter region of the hsp70i gene. Its blocking reduced cell survival (Xing et al. 2005). Hsf2-null mice showed structural defects in the synaptonemal complex of pachytene spermatocytes and developing spermatocytes were eliminated via apoptosis (Kallio et al. 2002). In humans, three synonymous mutations and five missense mutations of HSF2 have been identified in azoospermic patients with maturation arrest at the spermatocyte stage. FSH and TV levels of azoospermic patients with an HSF2 mutation have not been reported (Mou et al. 2013).
SYCE1(10q26.3) encodes for the synaptonemal complex central element protein 1, a testis-specific 329-aminoacidic protein, which is essential during meiotic prophase I (Fraune et al. 2012). In Syce-null mice, meiosis is arrested at prophase I (Bolcun-Filas et al. 2009).
The homozygosis for a splice site mutation resulting in a premature stop codon has been identified in two azoospermic brothers of Iranian–Jewish origin with spermatocytes maturation arrest. No abnormalities in FSH serum levels and TV were found (Maor-Sagie et al. 2015). Furthermore, the homozygosis for a 134-kb deletion on chromosome 10 encompassing the SYCE gene has been described in three Chinese patients with spermatocyte maturation arrest. The heterozygosis for this deletion was observed both in patients with NOA and in fertile controls (Huang et al. 2015). These findings support a role for SYCE disrupted function in the pathogenesis of NOA in humans. Accordingly, its mutations cause SPGF 15 (OMIM 616950).
TEX15(8p12) encodes for a testis-specific protein of 2.785-aminoacidic residues playing a role in DNA DSBs repair (Yang et al. 2008). Male (but not female) Tex15-null mice have early meiotic arrest (Yang et al. 2008).
A nonsense mutation (c.2130T>G, p.Y710*) leading to a premature stop codon has been described in patients with azoospermia (Okutman et al. 2015). Their testicular histology showed maturation arrest at the primary spermatocyte stage. The mean TV was 10–12 ml. FSH serum levels are not available. The compound heterozygosis for a nonsense mutation (c.2419A>T, p.Lys807*) and a single-nucleotide deletion (c.3040delT, p.Ser1014Leufs*5) leading to premature stop codons has been identified in two brothers with NOA and high FSH (16.7 and 14.1 IU/l; respectively), high LH (12.5 and 11.3 IU/l), and borderline-low TT (3.1 and 3.5 ng/ml) levels. The other family members carrying the single mutation were fertile (Colombo et al. 2017). TEX 15 gene is currently believed to be responsible for SPGF 25 (OMIM 605795).
PLK-4(4p28.1) seem to play a role in germ-cell maintenance. In mice, its point mutation causes germ-cell loss (Harris et al. 2011). In humans, among 81 patients with histologically documented SCOS and different serum FSH levels, a heterozygous 13 bp deletion in the Ser/Thr domain of PLK4 was identified in one patient (Miyamoto et al. 2016).
DMRT1(9p24.3) encodes for a testis-specific transcription factor playing a role in testis differentiation. Deletions of the 9p chromosome including DMRT1 are well known associated with the 9p deletion syndrome and XY gonadal dysgenesis (Vinci et al. 2007). Smaller deletions (Lopes et al. 2013) and mutations (Tüttelmann et al. 2018) of this gene have been identified in NOA patients (Tüttelmann et al. 2018) without XY gonadal dysgenesis.
SEPT12(16p13.3) encodes for a testis-specific GTB-binding protein, which concentrates in the acrosome in the first step of spermiogenesis (Lin et al. 2011a, b). The heterozygosity for a transition mutation (c.584G>A/p.Asp197Asn) interfering with GTP binding has been described in a patient with a severe oligoasthenozoospermia. FSH and TV values have not been reported (Kuo et al. 2012). SEPT12 gene is the hallmark of SPGF10 (OMIM 611562).
KLHL10(17q21.2) encodes for an evolutionary conserved protein exclusively expressed in spermatids. The c.647A>C (Q216P) (p.Gln216Pro) and the c.937G>A (A313T) p.Ala313Thr missense mutations, both affecting KLHL10 homodimerization, have been identified in patients with mild and moderate oligozoospermia. FSH and TV values have not been reported (Yatsenko et al. 2006). KLHL10 gene is the hallmark of SPGF 10 (OMIM 608778).
CATSPER1(11q13.1) encodes for a protein which mediates calcium entry into spermatozoa. The c.539-540insT (p.Lys180LysfsX8) and c.948-949insATGGC (p.Asp317MetfsX18) mutations, which are predicted to lead to frameshifts and premature stop codons, have been described in patients with oligozoospermia. Their FSH and TV values have not been reported (Avenarius et al. 2009). CATSPER1is responsible for SPGF 7 (OMIM 612997).
A complete and updated list of genes is available in Table 1.
Racial disparities
Genetic basis of racial disparity has emerged as a significant unexpected aspect of medical diagnosis. Several studies have addressed the role of racial and ethnic differences in female reproductive potential (Butts et al. 2010). In men, due to genetic inheritance, specific polymorphisms and/or genetic variations of genes influencing spermatogenesis may gather together in some ethnic groups more than in others. On this account, the SPO11 gene (OMIM 605114) C631T polymorphism predisposes to male infertility in Chinese, but not in Iranian people (Ren et al. 2017). Similarly, MEI1 gene (OMIM 608797) C1791A and C2397T polymorphisms associate with azoospermia in European American but not in Israeli and Japanese (Sato et al. 2006). Limited to the evidence coming from cohort studies, NR5A1 mutations might occur with a higher frequency in Caucasian compared to Indian men (Bashamboo et al. 2010; Ferlin et al. 2015; Sudhakar et al. 2018). In addition, SOHLH1 variants have been described in Asiatic countries (Choi et al. 2010; Nakamura et al. 2017), but no other ethnic groups have been analyzed as far. SYCE mutations have also been reported in Iranian–Jewish men and Chinese patients (Maor-Sagie et al. 2015; Huang et al. 2015). Finally, studies indicate two USP26 variants as frequently occurring among sub-Saharian African and South and East Asian population, including fertile men (Ravel et al. 2006). Therefore, it is recommended to extend the evaluation of the key genes hypothesized to be responsible of SPGF (Table 1) to large cohorts of different race and ethnicity, including fertile men. This may shed light into the understanding of the impact of racial disparities in male infertility, prevalence of genetic SPGF, and the real pathogenic role of the investigate genes.
Population origin of gene responsible for SPGF as well as gene variants is reported in the Supplementary Table 1.
Cryptorchidism and mini-puberty: genetic background and impact on spermatogenesis
Cryptorchidism often associates with SPGF (Kalfa et al. 2019). The reason of such association might be the insufficient testosterone secretion during mini-puberty in cryptorchidic children (Živković and Fratrić 2014). Studies indicate that the GnRH-induced raise in LH and testosterone levels induce the maturation of gonocytes into type A spermatogonia during mini-puberty. The lack of secretion of these hormones disrupts both type A spermatogonia differentiation and Sertoli cell proliferation. Mini-puberty modulates Sertoli cell gene expression. Indeed, GnRH administration in testis negative for type A spermatogonia at the histologic levels increases FASLG and GDNF expression, thus possibly promoting Sertoli cell proliferation and germ-cell self-renewal (Gegenschatz-Schmid et al. 2018).
Genetic factors, as well as intra-uterine environment, play a role in the pathogenesis of cryptorchidism and in the modulation of its prevalence due to genetic–environment interaction. Accordingly, the prevalence of cryptorchidism is higher in patients with a familiar history positive for cryptorchidism compared to those with a negative one (Urh et al. 2018). Nevertheless, the specific genetic abnormality causing cryptorchidism is rarely identified. Table 2 provides a list of the currently known genes responsible for isolated cryptorchidism, associated with germ-cell tumors or syndromic forms. It further shows those genes identified in mice and rat models, which may represent candidate ones for the human being (Urh et al. 2018; Wang et al. 2018; Kalfa et al. 2019).
Despite the association between cryptorchidism and SPGF (Kalfa et al. 2019), there is a lack of knowledge on the possible implication of these genes (Table 2) in human SPGF. To overcome this, such genes might be encompassed in the list of those to search for in patients with SPGF.
Conclusion and future perspectives
In conclusion, male infertility is a widespread disease and it has been esteemed up to 75% the rate of idiopathic oligozoospermia (Punab et al. 2017). Genetic diagnostic work-up is usually suggested in case of azoospermia and severe oligozoospermia, having these forms a greater probability to hide a genetic abnormality compared to the other less severe sperm parameter abnormalities (Asero et al. 2014).
In recent years, mutations of several genes have been firstly identified in men with SPGF. Preliminary data seem to support that looking for mutations of such candidate genes in patients with unexplained azoospermia or severe oligozoospermia would improve patient care (Tüttelmann et al. 2018). Indeed, the analysis of TEX11, NR5A1, and DMRT1 genes in 80 patients with non-obstructive azoospermia (NOA), normal karyotype, and no Yq microdeletions resulted in the discovery of likely etiopathogenic mutations in 4 patients (accounting for the 5% of the whole cohort), thus increasing the diagnostic rate up to 25% (Tüttelmann et al. 2018). On this account, the detection of a broaden diagnostic panel of genes in patients with apparently idiopathic NOA or severe oligozoospermia may increase the probability to achieve a diagnosis.
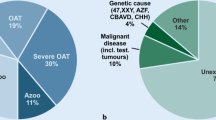
By reviewing the literature, we provided a list of 60 candidate genes for SPGF (Table 1). Their investigation by NGS in large cohorts of patients with apparently idiopathic infertility would provide new interesting data about their prevalence in infertile patients (also according with race and ethnicity), likely raising the diagnostic yields (Tüttelmann et al. 2018). The assessment of mutations in candidate genes might be performed on the basis of the clinical phenotype (Fig. 2). Furthermore, hopefully multicenter, studies are needed.
References
Adelman CA, Petrini JH (2008) ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 4(3):e1000042
Amiri-Yekta A, Coutton C, Kherraf ZE, Karaouzène T, Le Tanno P, Sanati MH, Sabbaghian M, Almadani N, Sadighi Gilani MA, Hosseini SH, Bahrami S, Daneshipour A, Bini M, Arnoult C, Colombo R, Gourabi H, Ray PF (2016) Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum Reprod 31(12):2872–2880
Asero P, Calogero AE, Condorelli RA, Mongioì L, Vicari E, Lanzafame F, Crisci R, La Vignera S (2014) Relevance of genetic investigation in male infertility. J Endocrinol Invest 37(5):415–427
Avenarius MR, Hildebrand MS, Zhang Y, Meyer NC, Smith LLH, Kahrizi K, Najmabadi H, Smith RJH (2009) Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet 84:505–510
Ayhan O, Balkan M, Guven A, Hazan R, Atar M, Tok A, Tolun A (2014) Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet 51:239–244
Bashamboo A, Ferraz-de-Souza B, Lourenco D, Lin L, Sebire NJ, Montjean D, Bignon-Topalovic J, Mandelbaum J, Siffroi J-P, Christin-Maitre S, Radhakrishna U, Rouba H, Ravel C, Seeler J, Achermann JC, McElreavey K (2010) Human male infertility associated with mutations in NR5A1 encoding steroidogenic factor 1. Am J Hum Genet 87:505–512
Ben Khelifa M, Zouari R, Harbuz R, Halouani L, Arnoult C, Lunardi J, Ray PF (2011) A new AURKC mutation causing macrozoospermia: implications for human spermatogenesis and clinical diagnosis. Molec Hum Reprod 17:762–768
Ben Khelifa M, Coutton C, Zouari R, Karaouzene T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J, Hennebicq S, Grunwald D, Escalier D, Pernet-Gallay K, Jouk P-S, Thierry-Mieg N, Toure A, Arnoult C, Ray PF (2014) Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 94:95–104
Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays 21(11):932–939
Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A (2002) Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J 21:1855–1863
Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ (2009) Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet 5:e1000393
Butts SF, Seifer DB (2010) Racial and ethnic differences in reproductive potential across the life cycle. Fertil Steril 93(3):681–690
Chao Liu Z, Song L, Wang H, Yu W, Liu Y, Shang Z, Xu H, Zhao F, Gao J, Wen L, Zhao Y, Gui J, Gao JF, Li W (2017) Sirt1 regulates acrosome biogenesis by modulating autophagic flux during spermiogenesis in mice. Development 144:441–451
Chen SR, Liu YX (2015) Regulation of spermatogonial stem cell self-renewal and spermatocyte meiosis by Sertoli cell signaling. Reproduction 149(4):R159–R167
Choi Y, Jeon S, Choi M, Lee M, Park M, Lee DR, Jun KY, Kwon Y, Lee O-H, Song SH, Kim JY, Lee K-A, Yoon TK, Rajkovic A, Shim SH (2010) Mutations in SOHLH1 gene associate with nonobstructive azoospermia. Hum Mutat 31:788–793
Colombo R, Pontoglio A, Bini M (2017) Two novel TEX15 mutations in a family with nonobstructive azoospermia. Gynecol Obstet Invest 2(3):283–286
Coutton C, Vargas AS, Amiri-Yekta A, Kherraf Z-E, Ben Mustapha SF, Le Tanno P, Wambergue-Legrand C, Karaouzene T, Martinez G, Crouzy S, Daneshipour A, Hosseini SH, Mitchell V et al (2018) Mutations in CFAP43 and CFAP44 cause male infertility and flagellum defects in trypanosoma and human. Nat Commun 9:686
Dam AHDM, Koscinski I, Kremer JAM, Moutou C, Jaeger AS, Oudakker AR, Tournaye H, Charlet N, Lagier-Tourenne C, van Bokhoven H, Viville S (2007) Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet 81:813–820
Dieterich K, Rifo RS, Faure AK, Hennebicq S, Amar BB, Zahi M, Perrin J, Martinez D, Sele B, Jouk PS, Ohlmann T, Rousseaux S, Lunardi J, Ray PF (2007) Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet 39:661–665
Dieterich K, Zouari R, Harbuz R, Vialard F, Martinez D, Bellayou H, Prisant N, Zoghmar A, Guichaoua MR, Koscinski I, Kharouf M, Noruzinia M et al (2009) The aurora kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet 18:1301–1309
Dirami T, Rode B, Jollivet M, Da Silva N, Escalier D, Gaitch N, Norez C, Tuffery P, Wolf J-P, Becq F, Ray PF, Dulioust E, Gacon G, Bienvenu T, Toure A (2013) Missense mutations in SLC26A8, encoding a sperm-specific activator of CFTR, are associated with human asthenozoospermia. Am J Hum Genet 92:760–766
Dong FN, Amiri-Yekta A, Martinez G, Saut A, Tek J, Stouvenel L, Lores P, Karaouzene T, Thierry-Mieg N, Satre V, Brouillet S, Daneshipour A et al (2018) Absence of CFAP69 cause male infertility due to multiple morphological abnormalities of the flagella in human and mouse. Am J Hum Genet 102:636–648
El Khouri E, Thomas L, Jeanson L, Bequignon E, Vallette B, Duquesnoy P, Montantin G, Copin B, Dastot-Le Moal F, Blanchon S, Papon JF, Lorès P, Yuan L, Collot N, Tissier S, Faucon C, Gacon G et al (2016) Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. Am J Hum Genet 99(2):489–500
Ellnati E, Kuentz P, Redin C, Jaber S, Vanden Meerschaut F, Makarian J, Koscinski I, Nasr-Esfahani MH, Demirol A, Gurgan T, Louanjli N, Iqbal N et al (2012) Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum Mol Genet 21:3695–3702
Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS (2005) Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19(7):794–803
Ferlin A, Rocca MS, Vinanzi C, Ghezzi M, Di Nisio A, Foresta C (2015) Mutational screening of NR5A1 gene encoding steroidogenic factor 1 in cryptorchidism and male factor infertility and functional analysis of seven undescribed mutations. Fertil Steril 104(1):163.e1–119.e1
Fischer S, Kohlhase J, Bohm D, Schweiger B, Hoffmann D, Heitmann M, Horsthemke B, Wieczorek D (2008) Biallelic loss of function of the promyelocytic leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet 45:731–737
Foresta C, Moro E, Garolla A, Onisto M, Ferlin A (1999) Y chromosome microdeletions in cryptorchidism and idiopathic infertility. J Clin Endocr Metab 84:3660–3665
Foresta C, Ferlin A, Moro E (2000) Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum Mol Genet 9:1161–1169
Fraune J, Schramm S, Alsheimer M, Benavente R (2012) The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 318(12):1340–1346
Freiman RN, Albright SR, Zheng S, Sha WC, Hammer RE, Tjian R (2001) Requirement of tissue-selective TBP-associated factor TAFII105 in ovarian development. Science 293(5537):2084–2087
Gegenschatz-Schmid K, Verkauskas G, Demougin P, Bilius V, Dasevicius D, Stadler MB, Hadziselimovic F (2018) Curative GnRHa treatment has an unexpected repressive effect on Sertoli cell specific genes. Basic Clin Androl 28:2
Gershoni M, Hauser R, Yogev L, Lehavi O, Azem F, Yavetz H, Pietrokovski S, Kleiman SE (2017) A familial study of azoospermic men identifies three novel causative mutations in three new human azoospermia genes. Genet Med 19(9):998–1006
Greenbaum MP, Yan W, Wu MH, Lin YN, Agno JE, Sharma M, Braun RE, Rajkovic A, Matzuk MM (2006) TEX14 is essential for intercellular bridges and fertility in male mice. Proc Natl Acad Sci USA 103(13):4982–4987
Harbuz R, Zouari R, Pierre V, Ben Khelifa M, Kharouf M, Coutton C, Merdassi G, Abada F, Escoffier J, Nikas Y, Vialard F, Koscinski I, Triki C, Sermondade N, Schweitzer T, Zhioua A et al (2011) A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am J Hum Genet 88:351–361
Harris RM, Weiss J, Jameson JL (2011) Male hypogonadism and germ cell loss caused by a mutation in polo-like kinase 4. Endocrinology 152:3975–3985
Hashemi MS, Mozdarani H, Ghaedi K, Nasr-Esfahani MH (2018) Expression of ZMYND15 in testes of azoospermic men and association with sperm retrieval. Urology 114:99–104
He WB, Tu CF, Liu Q, Meng LL, Yuan SM, Luo AX, He FS, Shen J, Li W, Du J, Zhong CG, Lu GX, Lin G, Fan LQ, Tan YQ (2018) DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J Med Genet 55(3):198–204
Huang N, Wen Y, Guo X, Li Z, Dai J, Ni B, Yu J, Lin Y, Zhou W, Yao B, Jiang Y, Sha J, Conrad DF, Hu Z (2015) A screen for genomic disorders of infertility identifies MAST2 duplications associated with nonobstructive azoospermia in humans. Biol Reprod 93:61
Ji ZY, Sha YW, Ding L, Li P (2017) Genetic factors contributing to human primary ciliary dyskinesia and male infertility. Asian J Androl 19(5):515–520
Julaton VT, Reijo Pera RA (2011) NANOS3 function in human germ cell development. Hum Mol Genet 20(11):2238–2250
Kalfa N, Gaspari L, Ollivier M, Philibert P, Bergougnoux A, Paris F, Sultan C (2019) Molecular genetics of hypospadias and cryptorchidism recent developments. Clin Genet 95(1):122–131
Kallio M, Chang Y, Manuel M, Alastalo T-P, Rallu M, Gitton Y, Pirkkala L, Loones M-T, Paslaru L, Larney S, Hiard S, Morange M, Sistonen L, Mezger V (2002) Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J 21:2591–2601
Kasak L, Punab M, Nagirmaja L, Grigorova M, Minajeva A, Lopes AM, Punab AM, Aston KI, Carvalho F, Laasik E, Smith LB, GEMINI Consortium, Conrad DF, Laan M (2018) Bi-allelic recessive loss-of-function variants in FANCM cause non-obstructive azoospermia. Am J Hum Genet 103:200–212
Kay BK, Williamson MP, Sudol M (2000) The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J 14(2):231–241
Kherraf Z-E, Christou-Kent M, Karaouzene T, Amiri-Yekta A, Martinez G, Vargas AS, Lambert E, Borel C, Dorphin B, Aknin-Seifer I, Mitchell MJ, Metzler-Guillemain C et al (2017) SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med 9:1132–1149
Kherraf ZE, Amiri-Yekta A, Dacheux D, Karaouzene T, Coutton C, Christou-Kent M, Martinez G, Landrein N, Le Tanno P, Mustapha SFB, Halouani L, Marrakchi O et al (2018) A homozygous ancestral SVA-insertion-mediated deletion in WDR66 induces multiple morphological abnormalities of the sperm flagellum and male infertility. Am J Hum Genet 103:400–412
Kim HJ, Yoon J, Matsuura A, Na JH, Lee WK, Kim H, Choi JW, Park JE, Park SJ, Kim KT, Chang R, Lee BI, Yu YG, Shin YK, Jeong C, Rhee K, Lee HH (2015) Structural and biochemical insights into the role of testis-expressed gene 14 (TEX14) in forming the stable intercellular bridges of germ cells. Proc Natl Acad Sci USA 112(40):12372–12377
Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, Winter RM, Reardon W (2002) Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 11:2979–2987
Koscinski I, ElInati E, Fossard C, Redin C, Muller J, Velez de la Calle JV, Schmitt F, Ben Khelifa M, Ray PF, Kilani Z, Barratt CLR, Viville S (2011) DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet 88:344–350
Kotov AA, Olenkina OM, Godneeva BK, Adashev VE, Olenina LV (2017) Progress in understanding the molecular functions of DDX3Y (DBY) in male germ cell development and maintenance. Biosci Trends 11(1):46–53
Kumar TR (2017) The SO(H)L(H) ‘O’ drivers of oocyte growth and survival but not meiosis I. J Clin Invest 127:2044–2047
Kuo YC, Lin YH, Chen HI, Wang YY, Chiou YW, Lin HH, Pan HA, Wu CM, Su SM, Hsu CC, Kuo PL (2012) SEPT12 mutations cause male infertility with defective sperm annulus. Hum Mutat 33:710–719
Kusz KM, Tomczyk L, Sajek M, Spik A, Latos-Bielenska A, Jedrzejczak P, Pawelczyk L, Jaruzelska J (2009) The highly conserved NANOS2 protein: testis-specific expression and significance for the human male reproduction. Mol Hum Reprod 15:165–171
Kusz-Zamelczyk K, Sajek M, Spik A, Glazar R, Jedrzejczak P, Latos-Bielenska A, Kotecki M, Pawelczyk L, Jaruzelska J (2013) Mutations of NANOS1, a human homologue of the Drosophila morphogen, are associated with a lack of germ cells in testes or severe oligo-astheno-teratozoospermia. J Med Genet 50:187–193
Lawo S, Bashkurov M, Mullin M, Gomez Ferreria M, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JRA, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L (2009) HAUS, the 8-subunit human augmin complex, regulates centrosome and spindle integrity. Curr Biol 19:816–826
Li L, Sha Y, Wang X, Li P, Wang J, Kee K, Wang B (2017) Whole-exome sequencing identified a homozygous BRDT mutation in a patient with acephalic spermatozoa. Oncotarget 8:19914–19922
Li L, Sha YW, Su ZY, Mei LB, Ji ZY, Zhang Q, Lin SB, Wang X, Qiu PP, Li P, Yin C (2018a) A novel mutation in HAUS7 results in severe oligozoospermia in two brothers. Gene 639:106–110
Li L, Sha YW, Xu X, Mei LB, Qiu PP, Ji ZY, Lin SB, Su ZY, Wang C, Yin C, Li P (2018b) DNAH6 is a novel candidate gene associated with sperm head anomaly. Andrologia. https://doi.org/10.1111/and.12953 (Epub ahead of print)
Liao HF, Chen WS, Chen YH, Kao TH, Tseng YT, Lee CY, Chiu YC, Lee PL, Lin QJ, Ching YH (2014) DNMT3L promotes quiescence in postnatal spermatogonial progenitor cells. Development 141:2402–2413
Lin L, Achermann JC (2008) Steroidogenic factor-1 (SF-1, Ad4BP, NR5A1) and disorders of testis development. Sex Dev 2:200–209
Lin YW, Hsu TH, Yen PH (2011a) Localization of ubiquitin specific protease 26 at blood-testis barrier and near Sertoli cell-germ cell interface in mouse testes. Int J Androl 34(5 Pt 2):e368–e377
Lin YH, Chou CK, Hung YC, Yu IS, Pan HA, Lin SW, Kuo PL (2011b) SEPT12 deficiency causes sperm nucleus damage and developmental arrest of preimplantation embryos. Fertil Steril 95:363–365
Lopes AM, Aston KI, Thompson E et al (2013) Human spermatogenic failure purges deleterious mutation load from the autosomes and both sex chromosomes, including the gene DMRT1. PLoS Genet 9:e1003349
Lores P, Coutton C, El Khouri E, Stouvenel L, Givelet M, Thomas L, Rode B, Schmitt A, Louis B, Sakheli Z, Chaudhry M, Fernandez-Gonzales A et al (2018) Homozygous missense mutation L673P in adenylate kinase 7 (AK7) leads to primary male infertility and multiple morphological anomalies of the flagella but not to primary ciliary dyskinesia. Hum Mol Genet 27:1196–1211
Lourenco D, Brauner R, Lin L, De Perdigo A, Weryha G, Muresan M, Boudjenah R, Guerra-Junior G, Maciel-Guerra AT, Achermann JC, McElreavey K, Bashamboo A (2009) Mutations in NR5A1 associated with ovarian insufficiency. N Engl J Med 360:1200–1210
Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, Balestri P, Piomboni P (2009) Spermatogenesis in a man with complete deletion of USP9Y. New Eng J Med 360:881–885
Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ (2013) MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun 4:2788
Ma Q, Li Y, Guo H, Li C, Chen J, Luo M, Jiang Z, Li H, Gui Y (2016) A novel missense mutation in USP26 gene is associated with nonobstructive azoospermia. Reprod Sci 23:1434–1441
Maor-Sagie E, Cinnamon Y, Yaacov B, Shaag A, Goldsmidt H, Zenvirt S, Laufer N, Richler C, Frumkin A (2015) Deleterious mutation in SYCE1 is associated with non-obstructive azoospermia. J Assist Reprod Genet 32:887–891
Martinez G, Kherraf Z-E, Zouari R, Fourati Ben Mustapha S, Saut A, Pernet-Gallay K, Bertrand A, Bidart M, Hograindleur JP, Amiri-Yekta A, Kharouf M, Karaouzene T, Thierry-Mieg N, Dacheux-Deschamps D, Satre V, Bonhivers M, Toure A, Arnoult C, Ray PF, Coutton C (2018) Whole-exome sequencing identifies mutations in FSIP2 as a recurrent cause of multiple morphological abnormalities of the sperm flagella. Hum Reprod 33:1973–1984
Mata M, Lluch-Estelles J, Armengot M, Sarrion I, Carda C, Cortijo J (2012) New adenylate kinase 7 (AK7) mutation in primary ciliary dyskinesia. Am J Rhinol Allergy 26:260–264
Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D (2010) The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19:612–624
Miller MP, Amon A, Ünal E (2013) Meiosis I: when chromosomes undergo extreme makeover. Curr Opin Cell Biol 25(6):687–696
Miyamoto T, Hasuike S, Yogev L, Maduro MR, Ishikawa M, Westphal H, Lamb DJ (2003) Azoospermia in patients heterozygous for a mutation in SYCP3. Lancet 362(9397):1714–1719
Miyamoto T, Bando Y, Koh E, Tsujimura A, Miyagawa Y, Iijima M, Namiki M, Shiina M, Ogata K, Matsumoto N, Sengoku K (2016) A PLK4 mutation causing azoospermia in a man with Sertoli cell-only syndrome. Andrology 4(1):75–81
Mou L, Wang Y, Li H, Huang Y, Jiang T, Huang W, Li Z, Chen J, Xie J, Liu Y, Jiang Z, Li X, Ye J, Cai Z, Gui Y (2013) A dominant-negative mutation of HSF2 associated with idiopathic azoospermia. Hum Genet 132(2):159–165
Mozdarani H, Ghoraeian P, Mozdarani S, Fallahi P, Mohseni-Meybodi A (2018) High frequency of de novo DAZ microdeletion in sperm nuclei of subfertile men: possible involvement of genome instability in idiopathic male infertility. Hum Fertil (Camb) 21(2):137–145
Nakamura S, Miyado M, Saito K, Katsumi M, Nakamura A, Kobori Y, Tanaka Y, Ishikawa H, Yoshida A, Okada H, Hata K, Nakabayashi K, Okamura K, Ogata H, Matsubara Y, Ogata T, Nakai H, Fukami M (2017) Next-generation sequencing for patients with non-obstructive azoospermia: implications for significant roles of monogenic/oligogenic mutations. Andrology 5(4):824–831
Neale MJ, Keeney S (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442:153–158
Neto FT, Bach PV, Najari BB, Li PS, Goldstein M (2016) Genetics of male infertility. Curr Urol Rep 17(10):70
Okutman O, Muller J, Baert Y, Serdarogullari M, Gultomruk M, Piton A, Rombaut C, Benkhalifa M, Teletin M, Skory V, Bakircioglu E, Goossens E, Bahceci M, Viville S (2015) Exome sequencing reveals a nonsense mutation in TEX15 causing spermatogenic failure in a Turkish family. Hum Mol Genet 24(19):5581–5588
Paff T, Loges NT, Aprea I, Wu K, Bakey Z, Haarman EG, Daniels JMA, Sistermans EA, Bogunovic N, Dougherty GW, Höben IM, Große-Onnebrink J, Matter A, Olbrich H, Werner C, Pals G, Schmidts M, Omran H, Micha D (2017) Mutations in PIH1D3 cause X-linked primary ciliary dyskinesia with outer and inner dynein arm defects. Am J Hum Genet 100(1):160–168
Pawlowski WP, Cande WZ (2005) Coordinating the events of the meiotic prophase. Trends Cell Biol 15:674–681
Potter SJ, De Falco T (2017) Role of the testis interstitial compartment in spermatogonial stem cell function. Reproduction 153:151–162
Punab M, Poolamets O, Paju P, Vihljajev V, Pomm K, Ladva R, Korrovits P, Laan M (2017) Causes of male infertility: a 9-year prospective monocentre study on 1737 patients with reduced total sperm counts. Hum Reprod 32(1):18–31
Ramathal C, Angulo B, Sukhwani M, Cui J, Durruthy Durruthy J, Fang F, Schanes P, Turek PJ, Orwig KE, Reijo Pera R (2015) DDX3Y gene rescue of a Y chromosome AZFa deletion restores germ cell formation and transcriptional programs. Sci Rep 5:15041
Ravel C, El Houate B, Chantot S, Lourenco D, Dumaine A, Rouba H, Bandyopadahyay A, Radhakrishna U, Das B, Sengupta S, Mandelbaum J, Siffroi JP, McElreavey K (2006) Haplotypes, mutations and male fertility: the story of the testis-specific ubiquitin protease USP26. Mol Hum Reprod 12:643–646
Ren ZJ, Ren PW, Yang B, Liao J, Liu SZ, Fang K, Ren SQ, Liu LR, Dong Q (2017) The SPO11-C631T gene polymorphism and male infertility risk: a meta-analysis. Ren Fail 39(1):299–305
Robert T, Nore A, Brun C, Maffre C, Crimi B, Guichard V, Bourbon HM, de Massy B (2016) The TopoVIB-like protein family is required for meiotic DNA double-strand break formation. Science 351:943–949
Sato H, Miyamoto T, Yogev L, Namiki M, Koh E, Hayashi H, Sasaki Y, Ishikawa M, Lamb DJ, Matsumoto N, Birk OS, Niikawa N, Sengoku K (2006) Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J Hum Genet 51:533–540
Sha YW, Xu X, Mei LB, Li P, Su ZY, He XQ, Li L (2017a) A homozygous CEP135 mutation is associated with multiple morphological abnormalities of the sperm flagella (MMAF). Gene 633:48–53
Sha YW, Wang X, Xu X, Su ZY, Cui Y, Mei LB, Huang XJ, Chen J, He XM, Ji ZY, Bao H, Yang X, Li P, Li L (2017b) Novel mutations in CFAP44 and CFAP43 cause multiple morphological abnormalities of the sperm flagella (MMAF). Reprod Sci 2017:1933719117749756
Sha YW, Wang X, Su ZY, Wang C, Ji ZY, Mei LB, Zhang L, Deng BB, Huang XJ, Yan W, Chen J, Li P, Cui YQ, Qu QL, Yin C, He XM (2018a) TDRD6 is associated with oligoasthenoteratozoospermia by sequencing the patient from a consanguineous family. Gene 659:84–88
Sha YW, Sha YK, Ji ZY, Mei LB, Ding L, Zhang Q, Qiu PP, Lin SB, Wang X, Li P, Xu X, Li L (2018b) TSGA10 is a novel candidate gene associated with acephalic spermatozoa. Clin Genet 93:776–783
Sha Y, Zheng L, Ji Z, Mei L, Ding L, Lin S, Wang X, Yang X, Li P (2018c) A novel TEX11 mutation induces azoospermia: a case report of infertile brothers and literature review. BMC Med Genet 19(1):63
Shannon M, Richardson L, Christian A, Handel MA, Thelen MP (1999) Differential gene expression of mammalian SPO11/TOP6A homologs during meiosis. FEBS Lett 462:329–334
Sironen A, Uimari P, Venhoranta H, Andersson M, Vilkki J (2011) An exonic insertion within Tex14 gene causes spermatogenic arrest in pigs. BMC Genom 12:591
Souquet B, Abby E, Hervé R, Finsterbusch F, Tourpin S, Le Bouffant R, Duquenne C, Messiaen S, Martini E, Bernardino-Sgherri J, Toth A, Habert R, Livera G (2013) MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet 9(9):e1003784
Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I (2005a) SYCP3 mutations are uncommon in patients with azoospermia. Fertil Steril 84(4):1019–1020
Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I (2005b) Possible role of USP26 in patients with severely impaired spermatogenesis. Eur J Hum Genet 13:336–340
Sudhakar DVS, Nizamuddin S, Manisha G, Devi JR, Gupta NJ, Chakravarthy BN, Deenadayal M, Singh L, Thangaraj K (2018) NR5A1 mutations are not associated with male infertility in Indian men. Andrologia 50:3
Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, Oates R, Page DC (1999) An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet 23:429–432
Suzuki H, Ahn HW, Chu T, Bowden W, Gassei K, Orwig K, Rajkovic A (2012) SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol 361:301–312
Tang S, Wang X, Li W, Yang X, Li Z, Liu W, Li C, Zhu Z, Wang L, Wang J, Zhang L, Sun X, Zhi E, Wang H, Li H, Jin L, Luo Y, Wang J, Yang S, Zhang F (2017) Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet 100:854–864
Tewes AC, Ledig S, Tuttelmann F, Kliesch S (2014) DMRT1 mutations are rarely associated with male infertility. Fertil Steril 102:816–820
Tournaye H, Krausz C, Oates RD (2017) Novel concepts in the aetiology of male reproductive impairment. Lancet Diabetes Endocrinol 5(7):544–553
Tüttelmann F, Ruckert C, Röpke A (2018) Disorders of spermatogenesis: perspectives for novel genetic diagnostics after 20 years of unchanged routine. Med Genet 30(1):2–20
Urh K, Kolenc Ž, Hrovat M, Svet L, Dovč P, Kunej T (2018) Molecular mechanisms of syndromic cryptorchidism: data synthesis of 50 studies and visualization of gene-disease network. Front Endocrinol (Lausanne) 9:425
Vigueras-Villaseñor RM, Cortés-Trujillo L, Chávez-Saldaña M, Vázquez FG, Carrasco-Daza D, Cuevas-Alpuche O, Rojas-Castañeda JC (2015) Analysis of POU5F1, c-Kit, PLAP, AP2γ and SALL4 in gonocytes of patients with cryptorchidism. Acta Histochem 117(8):752–761
Vinci G, Chantot-Bastaraud S, El Houate B et al (2007) Association of deletion 9p, 46,XY gonadal dysgenesis and autistic spectrum disorder. Mol Hum Reprod 13:685–689
Voskarides K (2018) Combination of 247 genome-wide association studies reveals high cancer risk as a result of evolutionary adaptation. Mol Biol Evol 35(2):473–485
Wang X, Jin H, Han F, Cui Y, Chen J, Yang C, Zhu P, Wang W, Jiao G, Wang W, Hao C, Gao Z (2017) Homozygous DNAH1 frameshift mutation causes multiple morphological anomalies of the sperm flagella in. Chin Clin Genet 91:313–321
Wang Y, Gray DR, Robbins AK, Crowgey EL, Chanock SJ, Greene MH, McGlynn KA, Nathanson K, Turnbull C, Wang Z, Devoto M, Barthold JS, Testicular Cancer Consortium (2018) Subphenotype meta-analysis of testicular cancer genome-wide association study data suggests a role for RBFOX family genes in cryptorchidism susceptibility. Hum Reprod 33(5):967–977
Wosnitzer MS, Mielnik A, Dabaja A, Robinson B, Schlegel PN, Paduch DA (2014) Ubiquitin specific protease 26 (USP26) expression analysis in human testicular and extragonadal tissues indicates diverse action of USP26 in cell differentiation and tumorigenesis. PLoS One 9:e98638
Xing H, Wilkerson DC, Mayhew CN, Lubert EJ, Skaggs HS, Goodson ML, Hong Y, Park-Sarge O-K, Sarge KD (2005) Mechanism of hsp70i gene bookmarking. Science 307:421–423
Xu Y, Greenberg RA, Schonbrunn E, Wang PJ (2017) Meiosis-specific proteins MEIOB and SPATA22 cooperatively associate with the single-stranded DNA-binding replication protein A complex and DNA double-strand breaks. Biol Reprod 6(5):1096–1104
Yan W, Si Y, Slaymaker S, Li J, Zheng H, Young DL, Aslanian A, Saunders L, Verdin E, Charo IF (2010) Zmynd15 encodes a histone deacetylase-dependent transcriptional repressor essential for spermiogenesis and male fertility. J Biol Chem 85:31418–31426
Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ (2008) Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180(4):673–679
Yatsenko AN, Roy A, Chen R, Ma L, Murthy LJ, Yan W, Lamb DJ, Matzuk MM (2006) Non-invasive genetic diagnosis of male infertility using spermatozoal RNA: KLHL10 mutations in oligozoospermic patients impair homodimerization. Hum Mol Genet 15:3411–3419
Yatsenko AN, Georgiadis AP, Röpke A, Berman AJ, Jaffe T, Olszewska M, Westernströer B, Sanfilippo J, Kurpisz M, Rajkovic A, Yatsenko SA, Kliesch S, Schlatt S, Tüttelmann F (2015) X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372(22):2097–2107
Yin H, Ma H, Hussain S, Zhang H, Xie X, Jiang L, Jiang X, Iqbal F, Bukhari I, Jiang H, Ali A, Zhong L et al (2018) A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med (Epub ahead of print)
Ying M, Chen B, Tian Y, Hou Y, Li Q, Shang X, Sun J, Cheng H, Zhou R (2007) Nuclear import of human sexual regulator DMRT1 is mediated by importin-beta. Biochim Biophys Acta 1773:804–813
Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C (2000) The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol Cell 5:73–83
Zhu F, Wang F, Yang X, Zhang J, Wu H, Zhang Z, Zhang Z, He X, Zhou P, Wei Z, Gecz J, Cao Y (2016) Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am J Hum Genet 99:942–949
Živković D, Fratrić I (2014) Disturbances of sperm maturation and minipuberty: is there a connection? Biomed Res Int 2014:912746
Funding
This research did not receive any specific Grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
RC: conception and design of the study, data acquisition, analysis and interpretation of data, drafting and writing of the article, and final approval of the manuscript. RAC: data acquisition, analysis and interpretation of data, and final approval of the manuscript. YD: data acquisition, analysis and interpretation of data, and final approval of the manuscript. SLV: data acquisition, analysis and interpretation of data, and final approval of the manuscript. AEC: conception and design of the study, analysis and interpretation of data, critical final revision of the manuscript, and final approval of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interests in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cannarella, R., Condorelli, R.A., Duca, Y. et al. New insights into the genetics of spermatogenic failure: a review of the literature. Hum Genet 138, 125–140 (2019). https://doi.org/10.1007/s00439-019-01974-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-019-01974-1