Abstract
Idiopathic azoospermia (IA) is a severe form of male infertility due to unknown causes. The HSF2 gene, encoding the heat shock transcription factor 2, had been suggested to play a significant role in the spermatogenesis process since the Hsf2-knockout male mice showed spermatogenesis defects. To examine whether HSF2 is involved in the pathogenesis of IA in human, we sequenced all the exons of HSF2 in 766 patients diagnosed with IA and 521 proven fertile men. A number of coding mutations private to the patient group, which include three synonymous mutations and five missense mutations, were identified. Of the missense mutations, our functional assay demonstrated that one heterozygous mutation, R502H, caused a complete loss of HSF2 function and that the mutant suppressed the normal function of the wild-type (WT) allele through a dominant-negative effect, thus leading to the dominant penetrance of the mutant allele. These results support a role for HSF2 in the pathogenesis of IA and further implicate this transcription factor as a potential therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Among the ~15 % of couples of child-bearing age who are confronted with the problem of infertility, about half of them are due to male infertility. Idiopathic azoospermia (IA) is one of the most serious forms of male infertility and affects ~1 % of all adult men in the general population (Hu et al. 2012). Although the underlying causes of IA are still unknown, a contribution of genetic factors appears to be supported by familial case reports and by mouse models (Matzuk and Lamb 2008).
HSF2, belonging to the family of heat shock transcription factors (HSFs), had been proved to play a key role in regulating the normal spermatogenesis process in mice (He et al. 2003). At least two splice forms, HSF2a and HSF2b were identified for HSF2 and Hsf2a was predominantly expressed in mouse testis (Goodson et al. 1995). In response to various stimuli under the physiological or stress conditions, the HSFs regulate the dynamic expression of different heat shock proteins (HSPs) which are responsible for the subsequent downstream effects including stress-related cytoprotective functions, folding and assembling of nascent polypeptides and intracellular transport of proteins (Sarge and Cullen 1997). Similarly, a role has also been suggested for HSF2 in spermatogenesis by regulating the expression of different HSPs, since both Hsf2- and Hspa2-knockout mice suffer from male reproductive defects (Abane and Mezger 2010; Dix et al. 1996, 1997; Kallio et al. 2002; Wang et al. 2003, 2004). However, little is known about the exact roles of HSF2 in the etiology of male infertility in human.
Certain sequence changes of HSF2 may result in the abnormal expression of HSPs and influence the functions of HSPs. To test whether mutations in HSF2 contribute to the susceptibility to male infertility, we therefore sequenced the exons of HSF2 in patients with IA. In the present study, we identified nine synonymous mutations and five missense mutations in HSF2. Among them, we showed that the R502H mutation affected the transcriptional regulatory function of HSF2 as measured by the expression level of the target gene HSPA2 which encodes a member of HSP70 family. Thus, we proposed that mutation of HSF2 led to abnormal HSPA2 expression and potentially contributed to the onset of male infertility.
Materials and methods
Patient samples
From Jan 2007 to Oct 2011, a total of 1,880 azoospermic patients were recruited for this study in the Center of Reproductive Medicine, Tongji Medical College, Huazhong University of Science and Technology. Among of them, 776 patients fulfilled the criteria for IA diagnosis: (1) no sperm detected in the pellets of semen samples at three different occasions, (2) no obstruction, inflammation and injury of the reproductive system or pelvic cavity, (3) no endocrinological defect, and (4) no karyotypic abnormality and Y chromosome microdeletion. Testicular biopsy and histological analysis were conducted for the patients whenever possible. 524 Fertile men from the Center of Physical Examination, Peking University Shenzhen Hospital were recruited as a control, who had fathered at least one child without assisted reproductive techniques such as IVF, ICSI, IMSI. After exome sequencing and quality control steps, 766 patients aged 24–46 years (average 30.6) and 521 fertile men aged 29–51 years (average 39.6) were available for further analysis. Informed written consent was obtained from each subject and the study was approved by the local ethics committee.
Exome sequencing
Five micrograms of genomic DNA isolated from the peripheral blood samples were sent to Beijing Genomics Institute at Shenzhen for exome capture and sequencing. The capture procedure was performed in solution with a NimbleGen custom array (Roche NimbleGen, Madison, WI, USA) that is capable of enriching the exonic sequences of ~600 infertility- or subfertility-related genes. Most of these genes were reviewed by Matzuk and Lamb (2008). Besides, we also selected other genes that were shown to cause male reproductive defects in mouse models from the studies published between November 2008 and December 2010. Exome sequencing was performed on the Illumina platform with pair-end 90 bp reads. The remaining exome data are under investigation and will be published elsewhere.
FASTQ sequence files were aligned against the human reference genome (NCBI build 37.1, hg19) with the SOAPaligner software (2.21). Duplicated pair-end reads were removed from the merged data sets. Single nucleotide variants that were different from the hg19 reference genome were filtered out if they meet any of the following criteria: Phred-like quality score ≤20, overall depth ≤8×, estimated copy number ≥2 or the genomic distance between two adjacent variants <5 bp. In addition, the quality score of both the major and minor allele at heterozygous locus should be at least 20. Variants were then annotated using an in-house functional prediction tool and were compared to dbSNP132 and 1,000 Genomes databases (as of August 2010). To further refine those novel mutations that may be associated with IA, all the genetic variants detected in the fertile men were also eliminated for subsequent functional analysis.
Validation of novel missense mutations by Sanger sequencing
To validate the novel missense mutations identified by deep sequencing, PCR amplifications were carried out and the PCR products were sequenced in both directions by 3,730 DNA analyzer (Applied Biosystems). The primers for PCR and Sanger sequencing validation of HSF2 gene were listed in Supplementary Table S1.
Site-directed mutagenesis and plasmids construction
Site-directed mutagenesis was performed to generate HSF2a expression in plasmids (gifted by Jingyin Xu) bearing one of the identified missense mutations (I175T, L322V, S428L, E494K and R502H) as described previously (Zheng et al. 2004). DNA sequencing was performed to insure successful introduction of desired mutations. The PCR primers used for site-directed mutagenesis construction were shown in Supplementary Table S2.
The HSPA2 promoter was amplified from HeLa cells by PCR with the following primers: 5′-TCAGCGCTTCTCCCAAATTATGTT-3′ (forward) and 5′-AGGGGCGGCCGTTATGTAAATGAG-3′ (reverse). The PCR product was subcloned into psiCHECK™-2 vector (Promega, Madison, WI, USA) at BglII/NheI sites to construct the HSPA2-LUC plasmid. All clones were verified by DNA sequencing.
Luciferase assay
HeLa and 293FT cells (ATCC, Manassas, VA, USA) were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco BRL, Gaithersburg, MD, USA) supplemented with 10 % fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C, 95 % humidity and 5 % CO2. Cells were seeded in 24-well tissue culture plates 24 h prior to transfection. Transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Equivalent amounts (500 ng) of pPyCAGIP-HSF2a expression constructs (WT and mutant) or pPyCAGIP empty vector were cotransfected with HSPA2-LUC plasmids (200 ng). Cells were harvested 36 h after transfection and assayed for firefly and Renilla luciferase expression using the Dual Luciferase Reporter Assay System (Promega). Renilla luciferase activities were normalized to firefly luciferase activity.
Quantitative real-time RT-PCR (qRT-PCR)
Total RNA extraction and qRT-PCR reactions were performed as described previously (Mou et al. 2010). The primers for HSPA2 were 5′-CTGGGCGCGGGGAGCTGAGT-3′ (forward) and 5′-GGAAGACCCCGACGCACGAATAGG-3′ (reverse). The primers for GAPDH were 5′-AGAAGGCTGGGGCTCATTTG-3′ (forward) and 5′-AGGGGCCATCCACAGTCTTC-3′ (reverse). Data were calculated according to the Applied Biosystems Comparative CT Method (ΔΔCT Method). After normalizing to GAPDH levels, the relative expression level of HSPA2 in the HeLa or 293FT cells transfected with HSF2a R502H was compared to that in the cells transfected with HSF2a WT.
Statistical analysis
All experiments were repeated for at least three times. Data were expressed as the mean ± SD. Student’s t test was used to compare the difference in mean between two groups. A P value of less than 0.05 was considered to be statistically significant.
Results
Identification of HSF2 mutation in patients with IA
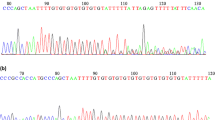
To examine whether HSF2 genetic defects were associated with IA, we screened for HSF2 exonic mutations in 766 IA patients and 521 men with proven fertility using the massively parallel sequencing technology. As shown in Table 1, nine synonymous mutations and five missense mutations were detected in HSF2. Three synonymous mutations and all the five missense mutations had not been reported in either the dbSNP135 database or the 1000 Genome Project dataset and were found to be absent in the 521 normal controls. These missense mutations were further confirmed by Sanger sequencing (Fig. 1). Alignment of the amino acid sequence of HSF2 to its orthologs in different species showed that the R502H mutation affected a highly conserved amino acid (Fig. 2).
Evolutionary conservation of amino acids affected by the missense mutations. Multiple protein alignments were performed with MegAlign (Demonstration System DNASTAR, Inc.). The identification numbers of HSF2 protein were as follows: human (NP_004497.1), chimpanzee (XP_003311472.1), rhesus (XP_001108944.2), cow (NP_001076874.1), rat (NP_113882.1), mouse (NP_032323.3), chicken (NP_001161236.1), xenopus (NP_001089021.1) and zebrafish (NP_571942.1). The mutant alleles are boxed, and the star (*) shows the conserved residue
Dominant-negative effects of mutant proteins
To evaluate whether the identified missense mutations affect the role of HSF2 in HSPA2 promoter activation, luciferase reporter constructs containing the HSF2 responsive elements (HSPA2 promoter) were tested in HeLa and 293FT cell lines. HSF2a WT, I175T, L332V, S428L and E494K mutants, but not R502H mutant, significantly increased HSPA2 promoter activity in comparison to empty vector (Fig. 3a).
Role of HSF2a in HSPA2 promoter activation and mRNA expression. a The HSPA2-LUC constructs and the WT or mutant forms of HSF2a expression vectors were cotransfected with HeLa and 293FT cells. HSPA2 promoter activity was analyzed by luciferase assay. Compared with WT and other mutants, the HSF2a R502H mutant failed to activate the HSPA2 promoter. Fold induction is shown as the ratio of WT or mutants to the average of empty plasmid. b HeLa and 293FT cells were cotransfected with HSPA2-LUC constructs (200 ng) and the WT or HSF2a R502H mutant with the indicated doses. HSPA2 promoter activities were significantly inhibited when the HSF2a R502H mutant and HSF2a WT were cotransfected. c HSF2a R502H mutant or HSF2a WT was transfected with HeLa and 293FT cells for 36 h, and the relative mRNA expression levels of HSPA2 to GAPDH were detected by qRT-PCR. The expression of HSPA2 mRNA after transfection of HSF2a R502H mutant was significantly decreased compared with the transfection of HSF2a WT (*P < 0.01)
HSF2a R502H mutants were then coexpressed with HSF2a WT in order to mimic the heterozygosity noticed in the patient. We observed that the R502H mutant was able to inhibit the activation of HSPA2 reporter induced by HSF2a WT (Fig. 3b). Meanwhile, the expression of HSPA2 mRNA after transfection of HSF2a R502H mutant was significantly decreased compared with the transfection of HSF2a WT (Fig. 3c). Collectively, these results indicated that the R502H mutant inhibited the transcriptional regulation activity of HSF2a WT through a dominant-negative effect.
Testicular biopsy analysis
Biopsy for the patient with the R502H mutation confirmed the diagnosis of non-obstructive azoospermia, and the spermatogenesis process in the patient was mainly blocked at the spermatocyte stage (Fig. 4).
Discussion
Accumulating evidence indicated that HSF2 was an essential transcriptional regulator in mouse spermatogenesis (Akerfelt et al. 2008; Goodson et al. 1995; Kallio et al. 2002; Sarge et al. 1994; Wang et al. 2003, 2004). However, no causative mutation had been identified in HSF2 in infertile men thus far. In this study, we identified five novel missense mutations of HSF2 in IA patients. One of the mutations, R502H, resulted in a heterozygous amino acid change at a conserved position. We also provided in vitro data supporting that this mutation might increase the risk of IA through abrogating the transcriptional regulatory function of HSF2. These results showed that aberration of HSF2 might be intolerable and that mutation of HSF2 might be involved in human spermatogenesis failure.
In this study, we sequenced the coding sequence of HSF2 in a large group of patients with IA. The R502H variant, localized in the transcription regulatory domain of HSF2, was found in one of the 766 patients but was absent in 521 fertile men we sequenced and other individuals reported in the public databases. Besides, local alignment analysis of the amino acid sequences of HSF2 showed that the affected arginine residue was highly conserved in multiple vertebrates, including zebrafish and xenopus. The evolutionary preservation of the entire region around this residue across multiple mammalian species indicated that mutations in this region may have great influence on the normal functions of the HSF2 protein. Unfortunately, his family is not available for genotype–phenotype correlations.
Hsf2-knockout mice (Hsf2 tm1Mmr and Hsf2 tm1Miv) developed the male hypofertile phenotype that was characterized by reduced testis size and vacuolization of the seminiferous tubules (Kallio et al. 2002; Wang et al. 2003, 2004). Besides, the synaptonemal complexes of spermatocytes in Hsf2 tm1Mmr were disorganized, and up to 90 % of the spermatocytes suffered from apoptosis, which may result in a great reduction of the sperm counts (Kallio et al. 2002). Interestingly, there is also some evidence showing that the Hsf2-knockout model (Hsf2 tm1Ijb) did not display any spermatogenesis defects, which might be associated with the different knockout strategies applied in these studies or due to the genetic background effects (McMillan et al. 2002). In our study, biopsy for the patient with the R502H mutation confirmed the diagnosis of non-obstructive azoospermia for him and further showed that the spermatogenesis process was mainly blocked at the spermatocyte stage. Therefore, the R502H mutation in the patient seems to result in a more severe phenotype than in the Hsf2-knockout mice, which may be due to the different genetic backgrounds between human and mice.
HSF2 regulates the expression of many HSP genes during development, including HSPA2, HSPH, HSPC, DNAJ, HSPB, HSP90, and HSP27 (Goodson et al. 1995; Ostling et al. 2007; Sistonen et al. 1992; Wilkerson et al. 2007). HSPs assume a molecular chaperone function to accommodate the unique set of proteins that are synthesized during spermatogenesis. These findings suggest the importance of HSF2 in spermatogenesis. When we tried to evaluate the pathogenic effect of HSF2 in infertile patients, a key question to address is whether mutation in HSF2 affects its transcriptional regulatory function. As expected, we showed that the R502H mutant affected the transcriptional regulatory function of HSF2 to HSPA2. In addition, our results also indicated that the R502H mutation not only rendered the nonfunctional transcription factor but also had a dominant-negative effect on the WT allele, which may be in consistent with the phenotypic effect of the heterozygous mutation in the IA patients.
It was reported that HSPA2, a well-defined target of HSF2, was highly expressed in human testis and appeared to have an essential role during the meiotic phase of spermatogenesis according to Son et al. (1999). Homologous HSPA2-related genes had been identified in germ cells from mammals, birds, amphibians and fish (Eddy 1999). Targeted gene disruption of Hspa2 resulted in meiosis failure, germ cell apoptosis and male infertility (Dix et al. 1996, 1997; Govin et al. 2006). Therefore, the transcriptional regulatory function of HSF2 may be essential to assure the appropriate expression of HSPA2 to protect against IA, whereas decreased transcriptional regulatory function by R502H mutation may increase the risk of IA.
In conclusion, we identified three synonymous mutations and five missense mutations private to the IA patient group by the massively parallel sequencing technology. And the functional assay confirmed that HSF2 R502H mutant suppressed the normal transcriptional regulatory function of the WT allele through a dominant-negative effect. These results suggested that HSF2 played an important role in human spermatogenesis. Our study also demonstrated that systematic analysis of the genetic mutations in large cohorts of patients complementing with subsequent functional assay may provide new insights into the cause of IA in human.
References
Abane R, Mezger V (2010) Roles of heat shock factors in gametogenesis and development. FEBS J 277:4150–4172
Akerfelt M, Henriksson E, Laiho A, Vihervaara A, Rautoma K, Kotaja N, Sistonen L (2008) Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci USA 105:11224–11229. doi:10.1073/pnas.0800620105
Dix DJ, Allen JW, Collins BW, Mori C, Nakamura N, Poorman-Allen P, Goulding EH, Eddy EM (1996) Targeted gene disruption of Hsp70-2 results in failed meiosis, germ cell apoptosis, and male infertility. Proc Natl Acad Sci USA 93:3264–3268
Dix DJ, Allen JW, Collins BW, Poorman-Allen P, Mori C, Blizard DR, Brown PR, Goulding EH, Strong BD, Eddy EM (1997) HSP70-2 is required for desynapsis of synaptonemal complexes during meiotic prophase in juvenile and adult mouse spermatocytes. Development 124:4595–4603
Eddy EM (1999) Role of heat shock protein HSP70-2 in spermatogenesis. Rev Reprod 4:23–30
Goodson ML, Park-Sarge OK, Sarge KD (1995) Tissue-dependent expression of heat shock factor 2 isoforms with distinct transcriptional activities. Mol Cell Biol 15:5288–5293
Govin J, Caron C, Escoffier E, Ferro M, Kuhn L, Rousseaux S, Eddy EM, Garin J, Khochbin S (2006) Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J Biol Chem 281:37888–37892. doi:10.1074/jbc.M608147200
He H, Soncin F, Grammatikakis N, Li Y, Siganou A, Gong J, Brown SA, Kingston RE, Calderwood SK (2003) Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J Biol Chem 278:35465–35475
Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, Jiang Y, Lu F, Wu Y, Yang X, Yao B, Lu C, Xiong C, Li Z, Gui Y, Liu J, Zhou Z, Shen H, Wang X, Sha J (2012) A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet 44:183–186. doi:10.1038/ng.1040
Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, Hiard S, Morange M, Sistonen L, Mezger V (2002) Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J 21:2591–2601. doi:10.1093/emboj/21.11.2591
Matzuk MM, Lamb DJ (2008) The biology of infertility: research advances and clinical challenges. Nat Med 14:1197–1213. doi:10.1038/nm.f.1895
McMillan DR, Christians E, Forster M, Xiao X, Connell P, Plumier JC, Zuo X, Richardson J, Morgan S, Benjamin IJ (2002) Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol 22:8005–8014
Mou L, Xu JY, Li W, Lei X, Wu Y, Xu G, Kong X, Xu GT (2010) Identification of vimentin as a novel target of HSF4 in lens development and cataract by proteomic analysis. Invest Ophthalmol Vis Sci 51:396–404. doi:10.1167/iovs.09-3772
Ostling P, Bjork JK, Roos-Mattjus P, Mezger V, Sistonen L (2007) Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem 282:7077–7086. doi:10.1074/jbc.M607556200
Sarge KD, Cullen KE (1997) Regulation of hsp expression during rodent spermatogenesis. Cell Mol Life Sci 53:191–197
Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI (1994) Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod 50:1334–1343
Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI (1992) Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol 12:4104–4111
Son WY, Hwang SH, Han CT, Lee JH, Kim S, Kim YC (1999) Specific expression of heat shock protein HspA2 in human male germ cells. Mol Hum Reprod 5:1122–1126
Wang G, Zhang J, Moskophidis D, Mivechi NF (2003) Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36:48–61. doi:10.1002/gene.10200
Wang G, Ying Z, Jin X, Tu N, Zhang Y, Phillips M, Moskophidis D, Mivechi NF (2004) Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis 38:66–80. doi:10.1002/gene.20005
Wilkerson DC, Skaggs HS, Sarge KD (2007) HSF2 binds to the Hsp90, Hsp27, and c-Fos promoters constitutively and modulates their expression. Cell Stress Chaperones 12:283–290
Zheng L, Baumann U, Reymond JL (2004) An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res 32:e115. doi:10.1093/nar/gnh110
Acknowledgments
This work was supported by grants from the National Key Scientific Program of China (No. 2011CB944303), the Promotion Program for Shenzhen Key Laboratory (CXB201005250017A), Shenzhen Foundation of Science and Technology (200901015, JC200903180681A) and the Biobank of Complex Diseases in Shenzhen (CXC201005260001A). The authors thank the patients and the family members for their cooperation during the study. The authors also thank Jing-Ying Xu at Tongji University School of Medicine for the expression plasmid of HSF2a.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mou, L., Wang, Y., Li, H. et al. A dominant-negative mutation of HSF2 associated with idiopathic azoospermia. Hum Genet 132, 159–165 (2013). https://doi.org/10.1007/s00439-012-1234-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-012-1234-7