Abstract
MicroRNAs may increase cold stress tolerance by regulating stress-related signal transduction pathways and by modulating the expression of transcription factors. However, the molecular mechanism by which microRNAs enhance cold stress tolerance is not fully understood. Here, we report that overexpression of rice microRNA156 (OsmiR156) results in increased cell viability and growth rate under cold stress in Arabidopsis, pine, and rice. OsmiR156 increases cold stress tolerance by targeting OsSPL3. OsSPL3 positively regulates the expression of OsWRKY71, a negative regulator of the transcription factors OsMYB2 and OsMYB3R-2. OsMYB2 counteracts cold stress by activating the expression of the stress-response genes OsLEA3, OsRab16A, and OsDREB2A. OsMYB3R-2 counteracts cold stress by activating the expression of OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1. In OsmiR156 transgenic rice cell lines, the transcript levels of OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 were increased by OsWRKY71 knockdown and inversely regulated by OsWRKY71 overexpression, indicating that OsmiR156 enhances cold stress tolerance by regulating the expression of transcription factor genes in plant cells. These results will increase our understanding of microRNA-related cold stress tolerance in different plant species, including monocotyledonous, dicotyledonous, and gymnosperm plant species, and will be valuable in plant molecular biotechnology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cold stress affects plant growth, development, and yield. To respond to cold stress, plants have developed capabilities to activate hormone signaling, enhance cold stress-related gene expression, and increase the activity of transcription factors to adapt to this extreme condition. Transcription factors are essential for plant cells to respond to stress signals. For example, the WRKY transcription factor gene family has been reported to be involved in biotic freezing stress through binding to the W box of the promoter of the target gene (Rushton et al. 1995). In the rice genome, 77 OsWRKY transcription factor genes have been identified (Goff et al. 2002; Yu et al. 2002). Several OsWRKY transcription factor genes, including OsWRKY71, are important in cold stress tolerance. Another type of transcription factor, the MYB transcription factor, has been reported to play central roles in plant responses to cold stress. Overexpression of OsMYB3R-2 enhances cold stress tolerance by regulating the expression of OsCycB1;1, OsCycB2;1, OsCycB2;2, and OsCDC20.1 during chilling stress (Ma et al. 2009). Both WRKY and MYB transcription factors are important for plants to respond to cold stress.
MYB transcription factors play important roles in a large number of biological processes, including cell metabolism, cell size and shape determination, cell proliferation and differentiation, and abiotic stress responses (Ma et al. 2009; Tombuloglu et al. 2013). The MYB transcription factor family, one of the largest transcription factor families in plants, regulates plant growth, development, and plant defense responses (Tombuloglu et al. 2013). In wheat (Triticum aestivum), MYB transcription factors play an important role in accumulating anthocyanin pigments in leaves by acting as a positive regulator of anthocyanin biosynthesis (Shin et al. 2016). In Chinese cabbage (Brassica rapa), genome-wide expression analysis of Myb genes demonstrated that the expression of MYB-related genes was induced under low-temperature treatment (Saha et al. 2016). In prairie cordgrass (Spartina pectinata), MYB proteins regulate freezing stress by upregulating genes involved in plasma membrane modification (Nah et al. 2016). In tomato (Solanum lycopersicum), the MYB transcription factors ANT1 and SlJAF13 are actively expressed and function in flavonoid synthesis (Lovdal et al. 2010). In Arabidopsis (Arabidopsis thaliana), MYB transcription factors function in the effect of light under low-temperature treatment and are crucial for cold acclimation (Soitamo et al. 2008). In Arabidopsis, Myb41 negatively regulates short-term transcriptional responses under osmotic stress (Lippold et al. 2009). Overexpression of MYB transcription factors results in tolerance to low temperatures and drought stress (Imtiaz et al. 2015). MYB transcription factors are also associated with plant adaptation to environmental conditions such as cold and UV stress (Hichri et al. 2011). In Arabidopsis, the MYB62L protein increases proline and chlorophyll content under salt and osmotic stress (Butt et al. 2017). Overexpression of MYB15 results in reduced expression of CBF genes and reduced freezing tolerance (Agarwal et al. 2006). MYB transcription factors play a role in freezing tolerance by modulating the expression of cold stress-related genes.
Expression of stress-related transcription factors can be regulated by microRNAs, which are 20–24 nucleotide RNA molecules that can cleave their target mRNA or cause repression of translation (Ma et al. 2009; Wang et al. 2016). MicroRNAs regulate and modulate the expression of genes that are involved in various biological functions in plants, including growth, development, and cold stress tolerance. MicroRNAs play a crucial role in plant stress tolerance by regulating the gene expression of transcription factors and signal transduction proteins. More than 2900 miRNA entries have been deposited in the miRNA database (miRbase; http://microrna.sanger.ac.uk/, release 7.0). Increasing evidence demonstrates that miRNAs play diverse roles in plant functions from development to the stress response by targeting transcription factors (Jones-Rhoades and Bartel 2004; Sunkar and Zhu 2004; Bartel 2004). In Arabidopsis thaliana, miRNAs target SCHLAFMUTZE to regulate the flowering response (Mathieu et al. 2009). In Arabis alpina, miRNAs prevent flowering before vernalization by targeting the transcription factor APETALA2 (Bergonzi et al. 2013). In Brachypodium distachyon, miRNAs target ice-binding proteins to restrict ice crystal growth and play a role in the development of a freeze-tolerant phenotype in crops (Bredow et al. 2016). In Ilex paraguariensis, miRNAs are involved in developmental processes through targeting of the SPL gene family (Debat et al. 2014). In Nicotiana tabacum, miRNAs enhance tolerance to drought, salinity, cold, and heavy metals (Frazier et al. 2014). In Hevea brasiliensis, miRNAs have been revealed to be potentially involved in the specific regulation of the control of redox status (Gebelin et al. 2012). MicroRNAs and their targets can be utilized for improving desirable plant traits (Kamthan et al. 2015). For example, amiRNA technology has been reported to be an efficient tool for the introduction of highly efficient resistance in barley plants against a DNA virus (Kis et al. 2016).
MicroRNA156 (miR156) plays an important role in improving various traits, including stress tolerance for molecular breeding in plants. The miR156 family targets promoter-binding-like (SPL) genes that encode several plant-specific transcription factors (Rhoades et al. 2002; Bonnet et al. 2004). Expression of miR156 and expression of its target genes are associated with important developmental processes and stress tolerance (Schwab et al. 2005). In the rice (Oryza sativa) genome, 11 SPL genes are putative targets of OsmiR156. In different tissues and organs, the rice SPL genes have different expression patterns. OsmiR156 genes have been reported to be expressed in the young shoots and leaves of rice. Different expression patterns of OsmiR156 and different expression patterns of its target genes in plants suggest diverse interactions between OsmiR156 and OsSPL target genes. For example, miR156 regulates IPA1 expression at the posttranscriptional level to affect plant architecture (Wang et al. 2017). In Solanum lycopersicum, miR156 promotes heterochronic development by affecting the KNOX and CLAVATA3 genes (Vendemiatti et al. 2017). In Arabidopsis thaliana, decreased expression of miR156 results in an increase in DNA methylation in adult tissues (Massoumi et al. 2017). In Panicum virgatum, overexpression of miR156 has dramatic effects on plant architecture and flowering (Baxter et al. 2017). In Medicago sativa, miR156 improves commercially important traits, including biomass, the number of branches and the time to complete growth stages under salinity stress (Arshad et al. 2017a, b) and drought stress, by silencing SPL13 (Arshad et al. 2017a, b). In Arabidopsis, miR156 modulates silique development and plant fertility by regulating ASYMMETRIC LEAVES 2 (AS2) transcript levels (Wang et al. 2016). miR156 also plays important roles in the phosphate (Pi) deficiency response (Lei et al. 2016). In the juvenile phase, miR156-mediated repression of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE (SPL) transcription factors establishes the relationship between developmental aging and photoperiodic flowering (Jung et al. 2016).
Although WRKY transcription factors and MYB transcription factors play important roles in cold stress tolerance, how microRNAs regulate the cell response to cold stress by modulating the expression of WRKY transcription factors and MYB transcription factors is not fully understood. In this investigation, we demonstrate that rice OsmiR156 increases cold stress tolerance in Arabidopsis, pine, and rice. In rice, OsmiR156 enhanced cold stress tolerance by targeting OsSPL3, a positive regulator of OsWRKY71. OsWRKY71 negatively regulates the expression of the transcription factors OsMYB2 and OsMYB3R-2. In OsmiR156 transgenic rice cell lines, the transcript levels of OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 were increased by OsWRKY71 knockdown and inversely regulated by OsWRKY71 overexpression. To the best of our knowledge, this is the first report to describe a detailed interaction between OsmiR156 and transcription factors that improves microRNA-related cold stress tolerance in different plant species, including monocotyledonous, dicotyledonous, and gymnosperm species.
Materials and methods
Plasmid constructs
Precursors of OsmiR156 were amplified from the rice genome and cloned into the expression vector pCAMBIA1301U as previously described (Xie et al. 2006; Jiao et al. 2010). The pCAMBIA1301U vector and the precursors of OsmiR156 were digested by KpnI and BamHI (Promega, Madison, WI, USA) at 37 °C. DNA was purified using a QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA, USA) and ligated to generate the expression vector pCAMBIA1301U-OsmiR156 (Tang and Newton 2005). For the overexpression of OsWRKY71 in rice, the full-length cDNA fragment of OsWRKY71 was cloned into pCAMBIA1301 (provided by CAMBIA) to generate the expression vector pCAMBIA1301-OsWRKY71. The expression vectors were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation.
Transformation of Arabidopsis, pine, and rice cells
OsmiR156 transgenic cell lines of Arabidopsis, pine (Pinus elliottii), and rice (O. sativa) were generated as described previously (Tang and Page 2013) using Agrobacterium tumefaciens strain LBA4404 carrying pCAMBIA1301U-OsmiR156 to transform the cultured cells (Tang and Newton 2005). OsWRKY71 transgenic rice cell lines were generated using Agrobacterium tumefaciens strain LBA4404 carrying pCAMBIA1301U-OsWRKY71. After transformation, cell cultures of the different cell lines were grown for 4–5 weeks, and then these cell cultures were used for further analysis.
RNA isolation and northern blot analysis
Five grams of fresh transgenic and control cell culture were used to isolate total RNA using an RNeasy Mini Plant Kit (Germantown, MD, USA) following the manufacturer’s instructions. Total RNA (6 µg) was separated by agarose gel electrophoresis. Electrophoresis of RNA and northern blotting were performed as described previously (Tang et al. 2007). The hybridization probe was digoxigenin (DIG)-labeled ZAT6 DNA (717 pb) (Roche Diagnostics). rRNA was used as the loading control for RNA samples. After northern blotting analyses, cell lines of Arabidopsis (a118, a261, and a362), pine (p169, p252, and p365), and rice (r126, r209, and r318) were used for cold stress and other stress-related gene expression experiments.
Cold treatment and determination of cell viability and growth rate
Cold treatment was performed by incubation of OsmiR156 transgenic cells and control cells of Arabidopsis, pine, and rice at − 10, 4, 12, and 24 °C in the dark for 24 h in growth chambers (Beijing ZNYT, China). Control seedlings were grown under the same conditions. After 24 h of chilling treatment, the cells were moved to a normal growth environment. The influences of cold stress on the cell growth and cell viability of Arabidopsis (a118, a261, and a362), pine (p169, p252, and p365), and rice (r126, r209, and r318) were examined as previously described (Tang and Page 2013). The average growth was expressed as mg/g FW/day. The rate of cell growth was measured 7 days after treatment. Both chilled and control samples were collected in 3–5 biological replicates.
Measurement of ion leakage
Ion leakage was measured using Arabidopsis, pine, and rice cells. Cells (1 g) were harvested from each sample of Arabidopsis, pine, and rice cell lines for measurement of ion leakage. The samples were incubated in 1000 µl of distilled water for 60 min with 60 rpm shaking. The conductivity was determined using a B-173 conductivity meter (Horiba, Kyoto, Japan). Ion leakage was calculated as the ratio of conductivity values between measurements before and after autoclaving.
Expression of the OsWRKY genes
Expression of the OsWRKY genes in the different cell lines was examined using qRT-PCR by the method of Tang et al. (Tang and Newton 2005). Expression of the stress-related WRKY transcription factors OsWRKY8 (a), OsWRKY11 (b), OsWRKY22 (c), OsWRKY30 (d), OsWRKY31 (e), OsWRKY42 (f), OsWRKY45 (g), OsWRKY71 (h), OsWRKY72 (i), OsWRKY74 (j), OsWRKY77 (k), and OsWRKY89 (l) in OsmiR156 transgenic cell lines was examined. The data were normalized to the internal control. Regular qPCR was used. The delta–delta Ct method was used to obtain the expression values. The U6 gene was used as an internal reference.
Target of OsmiR156 and the expression of OsSPL genes
By searching the rice genomic sequences using mature microRNA sequences, the target genes of OsmiR156 were obtained. Based on the characterized and deposited miRNAs in plants (Rhoades et al., 2002; Chen, 2004; Mallory et al., 2004; Guo et al., 2005; Mallory et al., 2005), rice genes that contained sequences with fewer than three mismatches to the sequence of mature miR156 were considered to be putative targets of OsmiR156. The expression of OsSPL3, OsSPL7, OsSPL11, OsSPL12, and OsSPL14 in the different cell lines was examined using qRT-PCR by the method of Tang et al. (Tang and Newton 2005; Tang and Page 2013).
Luciferase reporter assay
The promoter sequence of OsWRKY71 was cloned into a luciferase reporter vector. The GUS gene in pBI121 was removed, and the coding sequences of the OsmiR156 targets OsSPL3, OsSPL7, OsSPL11, OsSPL12, and OsSPL14 were cloned into the effector. The control effector was a plasmid expressing the GAL4 DNA-binding domain (DB). The vectors with the reporter (2 µg) and effector (2 µg) and the internal control plasmid (pPTRL, 0.04 µg) were delivered into cells by particle bombardment. Forty-eight hours after particle bombardment, the cells were lysed in lysis buffer, and the activity of luciferase was analyzed. A Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) was used to determine the luciferase activity by following the manufacturer’s instructions.
Effector construct preparation
The promoter sequences of SKP1-like protein 1B (an OsGAMYB-like 1 target), OscttP2 (an OsMYB1R target), OsLEA3 (an OsMYB2 target), KNOLLE2 (an OsMYB3R-2 target), protein phosphatase 2C (an OsMYBS3 target), OsCOS3-12-C2H2-zinc finger protein (an OsMYB4 target), flavonol synthase (an OsMYB55 target), the OsMYB transcription factor Os11g47460 (an OsMYB59 target) and the plasmid pMBL022 (Lanahan et al. 1992) were used to generate the reporter constructs pSKP1-like protein 1B-GUS, pOscttP2-GUS, pOsLEA3-GUS, pKNOLLE2-GUS, pProtein phosphatase 2C-GUS, pOsCOS3-12-C2H2-zinc finger protein-GUS, pFlavonol synthase gene-GUS, and pOsMYB transcription factor Os11g47460-GUS, respectively. The plasmid pAHC18 (Bruce et al. 1989) was used as an internal control construct (Lanahan et al. 1992; Shen et al. 1993) to normalize the GUS activity of the reporter constructs and to determine the expression of the target genes. To generate the UBI-OsWRKY71 effector construct, the coding region of OsWRKY71 was cloned into pAHC18 to generate pUBI-OsWRKY71. The coding sequences of the MYB genes OsGAMYB-like 1 (a), OsMYB1R (b), OsMYB2 (c), OsMYB3R-2 (d), OsMYBS3 (e), OsMYB4 (f), OsMYB55 (g), and OsMYB59 (h) were inserted into the cloning site of UBI-linker-NOS. The final effector constructs were designated pUBI-OsGAMYB-like1 (a), pUBI-OsMYB1R (b), pUBI-OsMYB2 (c), pUBI-OsMYB3R-2 (d), pUBI-OsMYBS3 (e), pUBI-OsMYB4 (f), pUBI-OsMYB55 (g), and pUBI-OsMYB59 (h). The insert sequences of all plasmids were confirmed by restriction enzyme digestion and sequencing.
Particle bombardment and transient expression assays
The transient expression procedure in cells was conducted using the particle bombardment technique as previously described (Shen et al. 1993). Briefly, cells were incubated for 48–72 h before particle bombardment. Reporter construct DNA and internal control construct DNA (UBI-Luciferase) were mixed in a 1:1 molar ratio, with or without an effector construct(s). Then, the DNA mixture was bombarded into cells (four replicates per test construct). For each bombardment, cells (900 mg) were arranged in a small circle with a diameter of 1.8 cm to maximize the bombarded surface area. After bombardment treatments, a GUS assay and a luciferase assay were performed as previously described (Shen et al. 1996).
Overexpression and knockdown of OsWRKY71 in rice cells
For overexpression of OsWRKY71 in rice, the maize polyubiquitin promoter (Ubi-1), the full-length cDNA fragment of OsWRKY71, and the NOS terminator were used to generate an expression vector by inserting them between the HindIII and PacI sites of the binary vector pZH1 (Shimono et al. 2007); the resultant vector was introduced into cells of rice via Agrobacterium tumefaciens strain LBA4404. Rice transformation was performed as previously described (Toki et al. 2006). To knock down OsWRKY71, the antisense sequence of OsWRKY71 was cloned into the pGEM-T Easy vector (Promega) and confirmed by sequencing. The insert was then excised with BamHI and cloned into the Ubi-NC1300 vector to generate pUbi-antisenseOsWRKy71. The resultant vector was introduced into rice via Agrobacterium tumefaciens strain LBA4404.
RNA isolation and qRT-PCR
Total RNA was isolated using the Sepasol RNA I Super reagent (Nacalai Tesque, Kyoto, Japan). First-strand cDNA was synthesized in a 50-mL reaction containing 2.5 mM oligo(dT) primers, 2.5 mM random hexamers, and 2.5 mg of total RNA with the PrimeScript™ RT reagent kit (TaKaRa Co., Ltd, Ohtsu, Japan) following the manufacturer’s instructions. RT-PCR was performed using 1X SYBR Premix Ex Taq II (TaKaRa). Each reaction mixture (5 µl) was used as a template for PCR amplification in a 25-mL mixture containing 1.5 mM MgCl2, 200 mM. The primer sequences used for OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 have been described previously (Ma et al. 2009; Yang et al. 2012). The relative levels of gene expression were determined using an MX3000P instrument following the manufacturer’s instructions (Stratagene, La Jolla, CA, USA). The data were normalized to the internal control. Regular qPCR was used. The delta–delta Ct method was used to obtain the expression values. The U6 gene was used as an internal reference.
Statistical analyses
Statistical analysis was performed using the general linear model procedure in SAS (Cary, NC, USA) with ANOVA models. Significant differences between the mean values of different cell lines derived from O. sativa, G. hirsutum, and P. elliottii were determined at the 5% level of probability. Each value for the different cell lines derived from O. sativa, G. hirsutum, and P. elliottii is presented as the mean with the standard error of the mean.
Results
Overexpression of OsmiR156 increases cold stress tolerance in Arabidopsis, pine, and rice
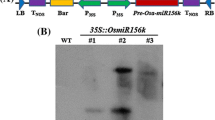
To examine the effect of OsmiR156 on cold stress tolerance in different plant species, cell suspension cultures of Arabidopsis, pine, and rice were used to produce OsmiR156 overexpression cell lines. One hundred eight OsmiR156 transgenic cell lines were produced, including 32 Arabidopsis lines, 37 pine lines, and 39 rice lines. Nine transgenic lines, including a118, a261, and a362 from Arabidopsis; p169, p252, and p365 from pine; and r126, r209, and r368 from rice, were selected for the cold stress tolerance experiment. After the integration of OsmiR156 into the genome of the transgenic cell lines was confirmed by northern blotting analysis in Arabidopsis (Fig. 1a), pine (Fig. 1b), and rice (Fig. 1c), cell viability, cell growth rates, and ion leakage rates under cold stress at − 10, 4, 12, and 24 °C were determined. The negative control cells were transgenic cells transformed with an expression vector that did not have the miR156 precursor. Overexpression of OsmiR156 increased cell viability and cell growth rates and decreased ion leakage rates in Arabidopsis (Fig. 1d, g, j), pine (Fig. 1e, h, k), and rice (Fig. 1f, i, l) cells after treatment at − 10, 4, and 12 °C, respectively. These results demonstrate that overexpression of OsmiR156 increases cold stress tolerance in the cells of different plant species, including monocotyledonous species (rice), dicotyledonous species (Arabidopsis), and gymnosperm species (pine).
Overexpression of OsmiR156 increases cell viability, cell growth rate, and decreases ion leakage rate under cold stress tolerance in Arabidopsis, pine, and rice. Overexpression of OsmiR156 was confirmed by northern blotting analysis in Arabidopsis (a), pine (b), and rice (c), respectively. Overexpression of OsmiR156 increases the cell viability, the cell growth rate, and decreases the ion leakage rate in cells of Arabidopsis (d, g, j), pine (e, h, k), and rice (f, i, l), respectively. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of three independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. *P < 0.05, **P < 0.01, ***P < 0.001, significant relative to control. N.S., no statistical significance
Expression of stress-related WRKY transcription factors in OsmiR156 transgenic rice cells
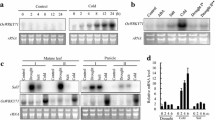
WRKY transcription factors play important roles in cold stress tolerance. To examine whether overexpression of OsmiR156 affects the expression of WRKY transcription factors, the expression of the stress-related WRKY transcription factor genes OsWRKY8 (Fig. 2a), OsWRKY11 (Fig. 2b), OsWRKY22 (Fig. 2c), OsWRKY30 (Fig. 2d), OsWRKY31 (Fig. 2e), OsWRKY42 (Fig. 2f), OsWRKY45 (Fig. 2g), OsWRKY71 (Fig. 2h), OsWRKY72 (Fig. 2i), OsWRKY74 (Fig. 2j), OsWRKY77 (Fig. 2k), and OsWRKY89 (Fig. 2l) were evaluated by qRT-PCR in OsmiR156 transgenic rice cell lines. Among the 12 WRKY transcription factor genes examined, overexpression of OsmiR156 significantly decreased the expression of OsWRKY71 (Fig. 2h) in cells treated at 4 °C and 24 °C. These results indicate that the increased cold stress tolerance caused by overexpression of OsmiR156 may be related to the expression of the transcription factor OsWRKY71.
Expression of stress-related WRKY transcription factors in OsmiR156 transgenic rice cells. Expression of stress-related WRKY transcription factors OsWRKY8 (a), OsWRKY11 (b), OsWRKY22 (c), OsWRKY30 (d), OsWRKY31 (e), OsWRKY42 (f), OsWRKY45 (g), OsWRKY71 (h), OsWRKY72 (i), OsWRKY74 (j), OsWRKY77 (k), and OsWRKY89 (l) in OsmiR156 transgenic cell lines. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. *P < 0.05, **P < 0.01, significant relative to control. NS no statistical significance
OsmiR156 regulates the expression of OsWRKY71 by targeting OsSPL3 in rice cells
Bioinformatics analysis of OsmiR156 targets showed that OsmiR156 targets the promoter-binding-like (SPL) genes and does not target OsWRKY71. Therefore, we examined the relationship between SPL genes and OsWRKY71. We examined the binding sequences for OsmiR156 in the 3′-UTR of OsSPL3, OsSPL7, OsSPL11, OsSPL12, and OsSPL14 (Fig. 3a) and examined the expression of these SPL genes in OsmiR156 transgenic rice cell lines. Our results demonstrated that overexpression of OsmiR156 decreased the expression of OsSPL3 (Fig. 3b), OsSPL7 (Fig. 3c), OsSPL11 (Fig. 3d), OsSPL12 (Fig. 3e), and OsSPL14 (Fig. 3f) in transgenic rice cell lines. To examine how these rice SPL genes affect the expression of the transcription factor OsWRKY71, we used a luciferase reporter assay. The results of the luciferase reporter assay using the OsWRKY71 promoter as a reporter and OsSPL3 as an effector (Fig. 3g) demonstrated that the luciferase activity was significantly increased (Fig. 3h). No significant difference in luciferase activity was observed when the OsWRKY71 promoter was used as a reporter and OsSPL7 was used as an effector (Fig. 3i, j), OsSPL11 was used as an effector (Fig. 3k, l), OsSPL12 was used as an effector (Fig. 3m, n), or OsSPL14 was used as an effector (Fig. 3o, p). These results demonstrate that OsSPL3 is a positive regulator of OsWRKY71 and that OsmiR156 reduces the expression of OsWRKY71 by targeting OsSPL3 in rice cells. We performed additional experiments using rice cells. Our results demonstrated that overexpression of OsSPL3 increases the expression of OsWRKY71 and that knockdown of OsSPL3 decreases the expression of OsWRKY71 (data not shown).
OsmiR156 regulates expression of OsWRKY71 by targeting OsSPL3 in rice cells. a Binding sequence of OsmiR156 in the 3′-UTR of OsSPL3, OsSPL7, OsSPL11, OsSPL12, and OsSPL14. b–f Overexpression of OsmiR156 decreased expression of OsSPL3 (b), OsSPL7 (c), OsSPL11 (d), OsSPL12 (e), and OsSPL14 (f) in transgenic rice cell lines. g, h The luciferase reporter using the OsWRKY71 promoter as reporter and the OsSPL3 as an effector (g) and the luciferase activity (h). i, j The luciferase reporter with the OsWRKY71 promoter as reporter and the OsSPL7 as an effector (i) and the luciferase activity (j). k, l The luciferase reporter with the OsWRKY71 promoter as reporter and the OsSPL11 as an effector (k) and the luciferase activity (l). m, n The luciferase reporter with the OsWRKY71 promoter as reporter and the OsSPL12 as an effector (m) and the luciferase activity (n). o, p The luciferase reporter with the OsWRKY71 promoter as reporter and the OsSPL14 as an effector (o) and the luciferase activity (p). The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. *P < 0.05, **P < 0.01, significant relative to control. NS no statistical significance
OsWRKY71 regulates the activity of MYB transcription factors in rice cells
WRKY transcription factors negatively regulate the expression of MYB transcription factors in plants. To examine how OsWRKY71 influences the expression of MYB transcription factors, the expression of eight MYB transcription factors was evaluated in OsWRKY transgenic rice cell lines. Our results demonstrated that OsWRKY71 does not block the activity of the MYB transcription factors OsGAMYB-like 1 (Fig. 4a), OsMYB1R (Fig. 4b), OsMYBS3 (Fig. 4e), OsMYB4 (Fig. 4f), OsMYB55 (Fig. 4g), and OsMYB59 (Fig. 4h) in transgenic rice cell lines. However, OsWRKY71 suppresses the OsMYB2-activated expression of OsLEA3 (Fig. 4c) and the OsMYB3R-2-activated expression of OsKNOLLE2 (Fig. 4d). These results show that OsWRKY71 is a negative regulator of the transcription factors OsMYB2 and OsMYB3R-2. We performed additional experiments using rice cells. Our results demonstrated that overexpression of OsWRKY71 decreases the expression of MYB3R2 and MYB2 and that knockdown of OsWRKY71 increases the expression of MYB3R2 and MYB2 (data not shown).
OsWRKY71 regulates the activity of MYB transcription factors in rice cells. a, b, e, f, g, h OsWRKY71 does not block the activity of MYB transcription factor OsGAMYB-like1 (a), OsMYB1R (b), OsMYB2 (c), OsMYB3R-2 (d), OsMYBS3 (e), OsMYB4 (f), OsMYB55 (g), and OsMYB59 (h) in transgenic rice cell lines (upper: Schematic diagrams of the reporter and effector constructs used in the co-bombardment experiment; lower: normalized GUS activity). c, d OsWRKY71 suppress OsMYB2-activated expression of OsLEA3 (c) and OsMYB3R-2-activated expression of OsKNOLLE2 (d), The reporter construct (such as OsLEA3-GUS), effector constructs (such as UBI-OsMYB2 and UBI-OsWRKY71), and the internal construct (UBI-Luciferase), at an equal molar ratio, were co-bombarded into rice cells. Transformed rice cells were incubated for 24 h. GUS activity was normalized in every independent transformation relative to the luciferase activity. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. ***P < 0.001, significant relative to control. NS no statistical significance
The expression of OsLEA3, OsRab16A, and OsDREB2A was regulated by the transcription factors OsWRKY71 and OsMYB2 in rice cells
The transcription factor OsMYB2 regulates the expression of the cold stress-related genes OsLEA3, OsRab16A, and OsDREB2. Therefore, we examined the expression of these cold stress-related genes in OsmiR156 transgenic rice cell lines. Our results showed that overexpression of OsmiR156 increased the expression of OsLEA3 (Fig. 5a), OsRab16A (Fig. 5b), and OsDREB2A (Fig. 5c). To determine whether the OsmiR156-mediated increase in OsLEA3 (Fig. 5a), OsRab16A (Fig. 5b), and OsDREB2A (Fig. 5c) expression is related to OsWRKY71, we performed overexpression and knockdown of OsWRKY71 in OsmiR156 transgenic rice cell lines. Overexpression of OsWRKY71 decreased the expression of OsLEA3 (Fig. 5d), OsRab16A (Fig. 5e), and OsDREB2A (Fig. 5f) in OsmiR156 transgenic cell lines. Knockdown of OsWRKY71 resulted in increased expression of OsLEA3 (Fig. 5g), OsRab16A (Fig. 5h), and OsDREB2A (Fig. 5i) in OsmiR156 transgenic cell lines.
Expression of OsLEA3, OsRab16A, and OsDREB2A regulated by transcription factor OsWRKY71 in rice cells. Overexpression of OsmiR156 increases expression of OsLEA3 (a), OsRab16A (b), and OsDREB2A (c). Overexpression of OsWRKY71 decreases expression of OsLEA3 (d), OsRab16A (e), and OsDREB2A (f) in OsmiR156 transgenic cell lines. Knockdown of OsWRKY71 results in increased expression of OsLEA3 (g), OsRab16A (h), and OsDREB2A (i) in OsmiR156 transgenic cell lines. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. *P < 0.05, **P < 0.01, ***P < 0.001, significant relative to control. NS no statistical significance
The expression of OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 was regulated by the transcription factors OsWRKY71 and OsMYB3R-2 in rice cells
The transcription factor OsMYB3R-2 regulates the expression of the cold stress-related genes OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1. Therefore, we examined the expression of these cold stress-related genes in OsmiR156 transgenic rice cell lines. Our results showed that overexpression of OsmiR156 increased the expression of OsKNOLLE2 (Fig. 6a), OsCTP1 (Fig. 6d), OsCycB1.1 (Fig. 6g), OsCycB2.1 (Fig. 6j), and OsCDC20.1 (Fig. 6m). To determine whether the OsmiR156-mediated increase in OsKNOLLE2 (Fig. 6a), OsCTP1 (Fig. 6d), OsCycB1.1 (Fig. 6g), OsCycB2.1 (Fig. 6j), and OsCDC20.1 (Fig. 6m) expression is related to OsWRKY71, we performed overexpression and knockdown of OsWRKY71. Overexpression of OsWRKY71 decreased the expression of OsKNOLLE2 (Fig. 6b), OsCTP1 (Fig. 6e), OsCycB1.1 (Fig. 6h), OsCycB2.1 (Fig. 6k), and OsCDC20.1 (Fig. 6n) in OsmiR156 transgenic cell lines. Knockdown of OsWRKY71 resulted in increased expression of OsKNOLLE2 (Fig. 6c), OsCTP1 (Fig. 6f), OsCycB1.1 (Fig. 6i), OsCycB2.1 (Fig. 6l), and OsCDC20.1 (Fig. 6o) in OsmiR156 transgenic cell lines.
Expression of OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 regulated by transcription factor OsWRKY71 in rice cells. Overexpression of OsmiR156 increases expression of OsKNOLLE2 (a), OsCTP1 (d), OsCycB1.1 (g), OsCycB2.1 (j), and OsCDC20.1 (m). Overexpression of OsWRKY71 decreases expression of OsKNOLLE2 (b), OsCTP1 (e), OsCycB1.1 (h), OsCycB2.1 (k), and OsCDC20.1 (n) in OsmiR156 transgenic cell lines. Knockdown of OsWRKY71 results in increased expression of OsKNOLLE2 (c), OsCTP1 (f), OsCycB1.1 (i), OsCycB2.1 (l), and OsCDC20.1 (o) in OsmiR156 transgenic cell lines. The statistically significant difference between groups was determined by one-way ANOVA. Data are presented as means of five independent experiments. Error bars represent standard error. The asterisk indicates significant differences compared to the control, as assessed by a t test. *P < 0.05, **P < 0.01, ***P < 0.001, significant relative to control. NS no statistical significance
Discussion
Cold stress is one of the factors that affects plant growth, development, and productivity (Agarwal et al. 2006; Bredow et al. 2016). Plants counteract cold stress through signal transduction and molecular genetic regulation (Bredow et al. 2016). It has been reported that miRNAs improve cold stress tolerance in many plant species, including rice, Triticum aestivum, Zea mays, Panicum virgatum, and Arabidopsis. In rice, miR156, miR168, miR169, miR171, miR319, miR396, and miR397 have been found to participate in the drought response. In addition, miR156, miR164, miR166, miR167, miR169, miR171, miR444, miR397, miR528, miR1425, miR827, miR319a.2, and miR408 have been identified to participate in responses to drought stress and Cd treatment (Xie et al. 2006; Jiao et al. 2010). It has been reported that OsmiR528 is upregulated, especially in roots, and that OsmiR827 is strongly induced by phosphate (Pi) starvation (Jian et al. 2016). In our study, genetic and biochemical assays were performed to explore the effect of an miRNA in response to cold stress in different plant species. Overexpression of OsmiR156 increased cell viability and cell growth rates and decreased ion leakage rates in Arabidopsis cells (Fig. 1d, g, j), pine cells (Fig. 1e, h, k), and rice cells (Fig. 1f, i, l) after treatment at − 10, 4, and 12 °C, respectively. These results demonstrate that overexpression of OsmiR156 enhances cold stress tolerance in the cells of different plant species, including monocotyledonous species (rice), dicotyledonous species (Arabidopsis), and gymnosperm species (pine).
MicroRNAs decrease the expression of target messenger RNAs to regulate various biological processes (Agarwal et al. 2006; Bredow et al. 2016). MicroRNAs play pivotal roles in plant adaptation. Overexpression and underexpression of miRNAs are related to various types of stress tolerance (Agarwal et al. 2006; Bredow et al. 2016; Jian et al. 2016). Cold stress has been the subject of intense investigation to reveal the complex mechanisms responsible for cold tolerance. In Arabidopsis thaliana, miR394 responds to drought and ABA stress, and miR399f functions in maintaining phosphate homeostasis (Baek et al. 2016). In rice, miR171 is enriched in the roots during drought stress. MicroRNA silencing of the transcription factor ASR5 regulates gene expression in response to Al (Arenhart et al. 2016). In Solanum tuberosum, miR171 participates in the response to drought stress. In Medicago truncatula, miR171h reduces mycorrhizal colonization and reduces nodule numbers. RNA sequencing of Saccharum sp. cultivars has shown that the majority of the predicted miRNA target genes are transcription factors and genes involved in hormone signaling (Carnavale Bottino et al. 2013). It has been reported that miRNA activity could be important for rapid changes in miRNA expression profiles (Chatterjee and Grosshans 2009). In tobacco, miR395 is most sensitive to stress and is upregulated by NaCl (Frazier et al. 2011). In cotton (Gossypium hirsutum), miRNVL5 is important in the regulation of the plant response to salt stress (Gao et al. 2016). In this study, we found that OsmiR156 increases cold stress tolerance by targeting OsSPL3. OsSPL3 positively regulates the expression of OsWRKY71, a negative regulator of the transcription factors OsMYB2 and OsMYB3R-2.
MicroRNA-regulated auxin signaling might be involved in plant acclimation to abiotic stress. In Arabidopsis, expression of miR393 can be induced by salt stress during acclimation to salinity (Iglesias et al. 2014). In tomato, microRNAs 165 and 166 target the transcription factor HD-ZIP III to regulate cell responses to abiotic stress (Jia et al. 2015). In Brassica napus, miR156, miR169, miR860, miR399, miR171, and miR395 modulate plant hormone signaling to counteract salt and drought stress (Jian et al. 2016). It has been reported that miR402 could be a positive regulator of seed germination under abiotic stress (Kim et al. 2010). MicroRNA knockdown of nitrate transporter 1 (NPF2.4) results in a change in the long-distance transport of Cl− in Arabidopsis under abiotic stress (Li et al. 2016a, b). In Oryza sativa indica, expression of miR820 is induced in the salt-susceptible Pusa Basmati 1 and salt-tolerant Pokkali varieties (Sharma et al. 2015). Expression profiling of microRNAs indicates that a large number of microRNAs regulate abiotic stress responses (Shen et al. 2010). In addition, ROS production might modulate the expression of miRNAs to affect transcription (Srivastava et al. 2017). In sugarcane plants under drought and salt stress, miRNAs play critical roles in regulatory pathways (Thiebaut et al. 2012). In cotton (Gossypium hirsutum L.), miR156-SPL2, miR162-DCL1, miR159-TCP3, miR395-APS1 and miR396-GRF1 play significant roles in response to salinity stress (Wang et al. 2013). In rice, overexpression of OsmiR393 results in hyposensitivity to auxin treatments (Xia et al. 2012). Here, we identified that in OsmiR156 transgenic rice cell lines, the transcript levels of OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 are increased by OsWRKY71 knockdown and inversely regulated by OsWRKY71 overexpression.
RNA sequencing and bioinformatics analysis have demonstrated that a large number of miRNAs involved in abiotic stress tolerance affect plant growth and productivity. In cotton, a total of 155 miRNAs were identified to be differentially expressed under drought and salinity stress (Xie et al. 2015). In date palm, 153 miRNAs were identified to regulate the expression of genes that are important for adaptation to salinity (Yaish et al. 2015). In Arabidopsis, more than 90 miRNAs were found to participate in abiotic stress responses (Zhan et al. 2012). Between Populus cathayana L. (a salt-sensitive plant) and Salix matsudana Koidz (a highly salt-tolerant plant), 193 miRNAs were identified to be differentially expressed (Zhou et al. 2012). In common wheat, a total of 201 miRNAs were identified to be associated with floral development under cold stress (Song et al. 2017). MicroRNA-mediated gene regulation may be important to improve plant stress tolerance. It has been reported that microRNA398 regulates plant responses to oxidative stress, ultraviolet stress, and phosphate deficiency (Zhu et al. 2011). In creeping bentgrass, overexpression of miR319a enhances drought and salt tolerance by increasing leaf wax (Zhou et al. 2013). In watermelon, miR159-5p, miR858, and miR8029-3p are involved in melatonin-related cold tolerance (Li et al. 2016a, b). In this investigation, our results demonstrated that OsmiR156 increases cold stress tolerance by targeting OsSPL3, which regulates the expression of WRKY directly and MYB indirectly.
In this investigation, we identified a multiple-step mechanism of miRNA-enhanced cold stress tolerance. We found that overexpression of rice OsmiR156 resulted in increased cell viability and growth rates under cold stress in Arabidopsis, pine, and rice. OsmiR156 increases cold stress tolerance by targeting OsSPL3. OsSPL3 positively regulates the expression of OsWRKY71, a negative regulator of the transcription factors OsMYB2 and OsMYB3R-2. OsMYB2 counteracts cold stress by activating the expression of the stress-response genes OsLEA3, OsRab16A, and OsDREB2A. OsMYB3R-2 counteracts cold stress by activating the expression of OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1. In OsmiR156 transgenic rice cell lines, the transcript levels of OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 were increased by OsWRKY71 knockdown and inversely regulated by OsWRKY71 overexpression, indicating that OsmiR156 enhances cold stress tolerance by regulating the expression of transcription factor genes in plant cells. These results will increase our understanding of microRNA-related cold stress tolerance in different plant species, including monocotyledonous, dicotyledonous, and gymnosperm plant species, and will be valuable in plant molecular biotechnology.
Conclusion
We report that OsmiR156 increases cold stress tolerance by targeting OsSPL3. OsSPL3 positively regulates the expression of OsWRKY71. OsWRKY71 negatively regulates the expression of the transcription factors OsMYB2 and OsMYB3R-2. OsMYB2 activates the expression of OsLEA3, OsRab16A, and OsDREB2A. OsMYB3R-2 activates the expression of OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1. Overexpression of OsmiR156 results in increased cell viability and growth rate under cold stress in Arabidopsis, pine, and rice. In OsmiR156 transgenic rice cell lines, the transcript levels of OsLEA3, OsRab16A, OsDREB2A, OsKNOLLE2, OsCTP1, OsCycB1.1, OsCycB2.1, and OsCDC20.1 were increased by OsWRKY71 knockdown and inversely regulated by OsWRKY71 overexpression, indicating that OsmiR156 enhances cold stress tolerance by regulating the expression of transcription factor genes in plant cells. These findings may provide new information for our understanding of microRNA-related cold stress tolerance in different plant species, including monocotyledonous, dicotyledonous, and gymnosperm plant species. Under normal conditions, miR156 does not greatly reduce the expression of the transcription factor WRKY71. The expression of MYB2 and MYB3R-2 is inhibited by the transcription factor WRKY71. Decreased expression of both MYB2 and MYB3R-2 results in cold stress sensitivity. Overexpression of miR156 reduces the expression of OsWRKY71, leading to increased function of MYB2 and MYB3R-2 and increased expression of their target genes, which enhances cold stress tolerance.
Data availability
This article does not contain any large-scale data.
References
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Arenhart RA, Schunemann M, Bucker Neto L, Margis R, Wang ZY, Margis-Pinheiro M (2016) Rice ASR1 and ASR5 are complementary transcription factors regulating aluminium responsive genes. Plant Cell Environ 39:645–651
Arshad M, Feyissa BA, Amyot L, Aung B, Hannoufa A (2017a) MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci 258:122–136
Arshad M, Gruber MY, Wall K, Hannoufa A (2017b) An insight into microRNA156 role in salinity stress responses of Alfalfa. Front Plant Sci 8:356
Baek D, Chun HJ, Kang S, Shin G, Park SJ, Hong H, Kim C, Kim DH, Lee SY, Kim MC, Yun DJ (2016) A role for Arabidopsis miR399f in salt, drought, and ABA signaling. Mol Cells 39:111–118
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baxter HL, Mazarei M, Dumitrache A, Natzke J, Rodriguez M, Gou J, Fu C, Sykes RW, Turner GB, Davis MF, Brown S, Davison B, Wang ZY, Stewart CN Jr (2017) Transgenic miR156 switchgrass in the field: growth, recalcitrance, and rust susceptibility. Plant Biotechnol J 16:39–49
Bergonzi S, Albani MC, Ver Loren van Themaat E, Nordstrom KJ, Wang R, Schneeberger K, Moerland PD, Coupland G (2013) Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340:1094–1097
Bredow M, Vanderbeld B, Walker VK (2016) Knockdown of ice-binding proteins in Brachypodium distachyon demonstrates their role in freeze protection. PLoS One 11:e0167941
Butt HI, Yang Z, Chen E, Zhao G, Gong Q, Yang Z, Zhang X, Li F (2017) Functional characterization of cotton GaMYB62L, a novel R2R3 TF in transgenic Arabidopsis. PLoS One 12:e0170578
Carnavale Bottino M, Rosario S, Grativol C, Thiebaut F, Rojas CA, Farrineli L, Hemerly AS, Ferreira PC (2013) High-throughput sequencing of small RNA transcriptome reveals salt stress regulated microRNAs in sugarcane. PLoS One 8:e59423
Chatterjee S, Grosshans H (2009) Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461:546–549
Debat HJ, Grabiele M, Aguilera PM, Bubillo RE, Otegui MB, Ducasse DA, Zapata PD, Marti DA (2014) Exploring the genes of yerba mate (Ilex paraguariensis A. St.-Hil.) by NGS and de novo transcriptome assembly. PLoS One 9:e109835
Frazier TP, Sun G, Burklew CE, Zhang B (2011) Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Mol Biotechnol 49:159–165
Frazier TP, Burklew CE, Zhang B (2014) Titanium dioxide nanoparticles affect the growth and microRNA expression of tobacco (Nicotiana tabacum). Funct Integr Genomics 14:75–83
Gao S, Yang L, Zeng HQ, Zhou ZS, Yang ZM, Li H, Sun D, Xie F, Zhang B (2016) A cotton miRNA is involved in regulation of plant response to salt stress. Sci Rep 6:19736
Gebelin V, Argout X, Engchuan W, Pitollat B, Duan C, Montoro P, Leclercq J (2012) Identification of novel microRNAs in Hevea brasiliensis and computational prediction of their targets. BMC Plant Biol 12:18
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Iglesias MJ, Terrile MC, Windels D, Lombardo MC, Bartoli CG, Vazquez F, Estelle M, Casalongue CA (2014) MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS One 9:e107678
Imtiaz M, Yang Y, Liu R, Xu Y, Khan MA, Wei Q, Gao J, Hong B (2015) Identification and functional characterization of the BBX24 promoter and gene from chrysanthemum in Arabidopsis. Plant Mol Biol 89:1–19
Jia X, Ding N, Fan W, Yan J, Gu Y, Tang X, Li R, Tang G (2015) Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci 233:11–21
Jian H, Wang J, Wang T, Wei L, Li J, Liu L (2016) Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front Plant Sci 7:658
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jung JH, Lee HJ, Ryu JY, Park CM (2016) SPL3/4/5 Integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol Plant 9:1647–1659
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:208
Kim JY, Kwak KJ, Jung HJ, Lee HJ, Kang H (2010) MicroRNA402 affects seed germination of Arabidopsis thaliana under stress conditions via targeting DEMETER-LIKE Protein3 mRNA. Plant Cell Physiol 51:1079–1083
Kis A, Tholt G, Ivanics M, Varallyay E, Jenes B, Havelda Z (2016) Polycistronic artificial miRNA-mediated resistance to wheat dwarf virus in barley is highly efficient at low temperature. Mol Plant Pathol 17:427–437
Lei KJ, Lin YM, Ren J, Bai L, Miao YC, An GY, Song CP (2016) Modulation of the phosphate-deficient responses by microRNA156 and its targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol 57:192–203
Li B, Byrt C, Qiu J, Baumann U, Hrmova M, Evrard A, Johnson AA, Birnbaum KD, Mayo GM, Jha D, Henderson SW, Tester M, Gilliham M, Roy SJ (2016a) Identification of a stelar-localized transport protein that facilitates root-to-shoot transfer of chloride in Arabidopsis. Plant Physiol 170:1014–1029
Li H, Dong Y, Chang J, He J, Chen H, Liu Q, Wei C, Ma J, Zhang Y, Yang J, Zhang X (2016b) High-throughput microRNA and mRNA sequencing reveals that micrornas may be involved in melatonin-mediated cold tolerance in Citrullus lanatus L. Front Plant Sci 7:1231
Lippold F, Sanchez DH, Musialak M, Schlereth A, Scheible WR, Hincha DK, Udvardi MK (2009) AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol 149:1761–1772
Lovdal T, Olsen KM, Slimestad R, Verheul M, Lillo C (2010) Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 71:605–613
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256
Massoumi M, Krens FA, Visser RG, De Klerk GM (2017) Azacytidine and miR156 promote rooting in adult but not in juvenile Arabidopsis tissues. J Plant Physiol 208:52–60
Mathieu J, Yant LJ, Murdter F, Kuttner F, Schmid M (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol 7:e1000148
Nah G, Lee M, Kim DS, Rayburn AL, Voigt T, Lee DK (2016) Transcriptome analysis of Spartina pectinata in response to freezing stress. PLoS One 11:e0152294
Rushton PJ, Macdonald H, Huttly AK, Lazarus CM, Hooley R (1995) Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol Biol 29:691–702
Saha G, Park JI, Ahmed NU, Kayum MA, Kang KK, Nou IS (2016) Characterization and expression profiling of MYB transcription factors against stresses and during male organ development in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Physiol Biochem 104:200–215
Sharma N, Tripathi A, Sanan-Mishra N (2015) Profiling the expression domains of a rice-specific microRNA under stress. Front Plant Sci 6:333
Shen J, Xie K, Xiong L (2010) Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol Genet Genomics 284:477–488
Shin DH, Choi MG, Kang CS, Park CS, Choi SB, Park YI (2016) A wheat R2R3-MYB protein PURPLE PLANT1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis. Biochem Biophys Res Commun 469:686–691
Soitamo AJ, Piippo M, Allahverdiyeva Y, Battchikova N, Aro EM (2008) Light has a specific role in modulating Arabidopsis gene expression at low temperature. BMC Plant Biol 8:13
Song G, Zhang R, Zhang S, Li Y, Gao J, Han X, Chen M, Wang J, Li W, Li G (2017) Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genom 18:212
Srivastava AK, Sablok G, Hackenberg M, Deshpande U, Suprasanna P (2017) Thiourea priming enhances salt tolerance through co-ordinated regulation of microRNAs and hormones in Brassica juncea. Sci Rep 7:45490
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Tang W, Newton RJ (2005) Transgenic Christmas trees regenerated from Agrobacterium tumefaciens-mediated transformation of zygotic embryos using the green fluorescence protein as a reporter. Mol Breeding 16:235–246
Tang W, Page M (2013) Transcription factor AtbZIP60 regulates expression of Ca2+ -dependent protein kinase genes in transgenic cells. Mol Biol Rep 40:2723–2732
Tang W, Newton RJ, Weidner DA (2007) Genetic transformation and gene silencing mediated by multiple copies of a transgene in eastern white pine. J Exp Bot 58:545–554
Thiebaut F, Grativol C, Carnavale-Bottino M, Rojas CA, Tanurdzic M, Farinelli L, Martienssen RA, Hemerly AS, Ferreira PC (2012) Computational identification and analysis of novel sugarcane microRNAs. BMC Genom 13:290
Tombuloglu H, Kekec G, Sakcali MS, Unver T (2013) Transcriptome-wide identification of R2R3-MYB transcription factors in barley with their boron responsive expression analysis. Mol Genet Genomics 288:141–155
Vendemiatti E, Zsogon A, Silva G, de Jesus FA, Cutri L, Figueiredo CRF, Tanaka FAO, Nogueira FTS, Peres LEP (2017) Loss of type-IV glandular trichomes is a heterochronic trait in tomato and can be reverted by promoting juvenility. Plant Sci 259:35–47
Wang M, Wang Q, Zhang B (2013) Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530:26–32
Wang Z, Wang Y, Kohalmi SE, Amyot L, Hannoufa A (2016) SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Mol Biol 92:661–674
Wang J, Yu H, Xiong G, Lu Z, Jiao Y, Meng X, Liu G, Chen X, Wang Y, Li J (2017) Tissue-specific ubiquitination by IPA1 interacting Protein1 modulates IPA1 protein levels to regulate plant architecture in rice. Plant Cell 29:697–707
Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M (2012) OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7:e30039
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factor and microRNA156 in rice. Plant Physiol 142:280–293
Xie F, Wang Q, Sun R, Zhang B (2015) Deep sequencing reveals important roles of microRNAs in response to drought and salinity stress in cotton. J Exp Bot 66:789–804
Yaish MW, Sunkar R, Zheng Y, Ji B, Al-Yahyai R, Farooq SA (2015) A genome-wide identification of the miRNAome in response to salinity stress in date palm (Phoenix dactylifera L.). Front Plant Sci 6:946
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296:79–92
Zhan X, Wang B, Li H, Liu R, Kalia RK, Zhu JK, Chinnusamy V (2012) Arabidopsis proline-rich protein important for development and abiotic stress tolerance is involved in microRNA biogenesis. Proc Natl Acad Sci USA 109:18198–18203
Zhou J, Liu M, Jiang J, Qiao G, Lin S, Li H, Xie L, Zhuo R (2012) Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol Biol Rep 39:8645–8654
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161:1375–1391
Zhu C, Ding Y, Liu H (2011) MiR398 and plant stress responses. Physiol Plant 143:1–9
Acknowledgements
We acknowledge internal grants. We are also thankful to the central instrumentation facility of NIPGR for providing the necessary experimental setup. We thank the Council of Scientific Research and the University Commission. We further thank Dr. Prasad for support. The authors are grateful to Dr. Neale, Dr. Page, Dr. Bradshaw, Dr. Lischewski, Dr. Thompson, and Dr. Andersen-Ranberg for their critical reading and suggestions during the preparation of this manuscript.
Funding
This work was supported by a grant from the Education Committee of Hubei Providence of China and by the National Natural Science Foundation of China (31270740).
Author information
Authors and Affiliations
Contributions
WT and MZ conceived and designed the experiments. WT wrote the paper. WT and MZ performed the experiment and analyzed the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Zhou, M., Tang, W. MicroRNA156 amplifies transcription factor-associated cold stress tolerance in plant cells. Mol Genet Genomics 294, 379–393 (2019). https://doi.org/10.1007/s00438-018-1516-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1516-4