Abstract
Publicly available microarray data and RNA gel blot analysis identified a rice WRKY transcription factor, OsWRKY71, that is highly upregulated in response to cold stress. Experiments with OsWRKY71 promoter:GFP transgenic rice confirmed its cold-inducible expression. Transient expression of OsWRKY71-GFP in maize mesophyll protoplasts indicated that it is localized predominantly in the nucleus and to a lesser extent in the cytosol. Transcriptional activation assays revealed that OsWRKY71 suppresses luciferase reporter activity in maize protoplasts, suggesting that it functions as a transcriptional repressor in rice. To characterize the function of OsWRKY71 in rice, we generated transgenic rice plants carrying CaMV35S promoter:OsWRKY71. Upon cold (4 °C) treatment, two selected OsWRKY71 transgenic lines, OX12 and OX21, recovered much better with respect to survival rate, photosynthetic ability, fresh weight, and dry weight than the control lines. RT-PCR analysis of known cold-responsive genes found that expression of OsTGFR and WSI76 was increased in OsWRKY71 transgenic lines in response to cold stress. Our results suggest that OsWRKY71 has a positive function in cold tolerance by regulating downstream target genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Unlike animals, plants cannot move away from disadvantageous environmental conditions such as cold, drought, and high salinity. Cold exposure (low temperature or chilling) represents one of the major environmental stresses that adversely affect rice productivity through poor germination, yellowing leaves, growth retardation, and improper microspore development (Andaya and Mackill 2003; Baruah et al. 2009; Shimono et al. 2011a; de Los Reyes et al. 2013). Therefore, screening for cold tolerance genes is particularly important in light of the current increase in occasional extreme weather events.
To cope with diverse environmental stresses plants regulate the expression of a large number of stress-associated genes. The gene products include water channel proteins, the enzymes required for the biosynthesis of osmoprotectants, lipid desaturases for membrane modification, protective proteins, such as antifreeze proteins and chaperones, detoxification proteins, and transcription factors (Shinozaki and Yamaguchi-Shinozaki 1996; Singh et al. 2002). In particular, transcription factors play an essential role in the regulation of the plant response to abiotic stresses through the binding of transcription factors to cognate cis-acting elements present in the promoter region of their target genes (Casaretto and Ho 2003; Fujita et al. 2005; Yamaguchi-Shinozaki and Shinozaki 2006; Hu et al. 2008; Zou et al. 2010; Yang et al. 2012; Peng et al. 2013). One class of proteins that is unique to plants and plays a vital role in abiotic stress response is the WRKY family of proteins that bind to the consensus cis-element W box (TTGACT/C) (Eulgem et al. 2000; Cai et al. 2008; Ciolkowski et al. 2008; Rushton et al. 2010). Previous studies have identified a large number of WRKYs, including 74 WRKY-encoding genes in Arabidopsis (Ulker and Somssich 2004) and 125 in rice (Rice WRKY working group 2012).
Until now, the functions of WRKY proteins have been mainly analyzed with a focus on defense responses upon pathogen infection. For instance, the Arabidopsis WRKYs AtWRKY18, AtWRKY40, and AtWRKY60 are negative regulators of resistance to the hemibiotrophic bacterial pathogen Pseudomonas syringae (Xu et al. 2006) and the barley WRKYs HvWRKY1 and HvWRKY2 are negative regulators of the basal defense response against the fungal pathogen Blumeria graminis (Shen et al. 2007). The rice WRKY OsWRKY45 enhances resistance to Magnaporthe oryzae and Xanthomonas oryzae pathovar oryzae (Xoo) (Shimono et al. 2007; Jiang et al. 2010; Shimono et al. 2011b; Matsushita et al. 2012). Overexpression of OsWRKY62 and OsWRKY76 compromises XA21-mediated resistance to Xoo (Peng et al. 2008; Seo et al. 2011) and overexpression of OsWRKY28 enhances rice susceptibility to M. oryzae (Chujo et al. 2013). Consistent with these findings, reduced expression of these rice WRKYs enhances resistance to Xoo and M. oryzae (Peng et al. 2010; Delteil et al. 2012).
Notably, a number of WRKY genes are involved in the coordination of multiple biological processes. For instance, AtWRKY33 regulates disease resistance, NaCl tolerance and thermotolerance (Birkenbihl et al. 2012; Jiang and Deyholos 2009; Li et al. 2011), and a pepper WRKY, CaWRKY40, modulates resistance to Ralstonia solanacearum and tolerance to heat stress (Dang et al. 2012). These findings suggest that WRKYs can serve as nodes in the crosstalk between different physiological processes; however, the functions of the majority of WRKY family members and their possible roles in signaling crosstalk remain poorly understood.
Rice OsWRKY71 is upregulated by several defense-signaling molecules, such as methyl jasmonate, salicylic acid and 1-aminocyclo-propane-1-carboxylic acid, as well as by biotic elicitors, wounding and pathogen infection. Accordingly, OsWRKY71 overexpression enhances resistance to Xoo (Liu et al. 2007; Chujo et al. 2008). On the other hand, OsWRKY71 acts as a transcriptional repressor of gibberellin-responsive genes (Zhang et al. 2004). In the present study, we show that OsWRKY71 expression is induced by cold stress. In an attempt to investigate the in vivo function of OsWRKY71, we analyzed the responses of OsWRKY71 overexpression lines upon cold treatment. Our data indicate that OsWRKY71 may have an important role in cold tolerance in rice.
Materials and methods
Plant materials and exposure of rice seedlings to chilling stress
Rice (Oryza sativa japonica cultivar [cv.] Dongjin) plants were grown in a greenhouse under a 14/10-h light/dark period, at a temperature of 24–28 °C and 70–80 % humidity. To investigate the expression of OsWRKY71 in rice, seedlings and mature leaves and panicles of flowering plants were subjected to cold (4 °C), abscisic acid (ABA, 100 µM), salt (300 mM NaCl), and drought (20 % polyethylene glycol and air-drying) treatments. To analyze the response to cold stress, homozygous transgenic plants of the T2 generation were used. Five-week-old seedlings were treated at 4 °C in a low-temperature environment room, and thereafter moved to the greenhouse for 6 days. Surviving seedlings were photographed and characterized to investigate the response of OsWRKY71 overexpressing transgenic plants (OsWRKY71-OX) to chilling stress. To analyze the expression of cold-responsive genes in seedlings in response to cold stress, 5-week-old seedlings of transgenic plants of T2 generation were exposed to 4 °C in the low-temperature environment room. At 0, 12, and 24 h of chilling treatment, leaf blades of cold-stressed seedlings were sampled for analysis.
RNA gel blot and RT-PCR analysis
Total RNA was prepared from various rice tissues after different abiotic stress treatments using Trizol reagent (Invitrogen, Carlsbad, CA, USA). For RNA gel blot analysis, 20 μg of isolated total RNA was fractionated on a 1.3 % agarose gel and transferred onto Hybond-N+ nylon membranes (Amersham Biosciences, Pittsburgh, PA, USA). Hybridization was then carried out with [α-32P] dCTP-labeled gene-specific probe according to standard procedures under high-stringency hybridization conditions (Cho et al. 2006). Gene-specific probes for OsWRKY71 (LOC_Os02g08440) and SalT (LOC_Os01g24710) were amplified using the primers listed in Supplementary Table S1. PCR amplification was performed in a final volume of 40 µL (100 pmol of each primer, 20 µM each of dNTPs, 10 mM Tris–HCl pH 9.0, 2 mM MgCl2, 50 mM KCl, 0.1 % Triton X-100, and 0.5 U of Taq polymerase) using 50 ng of genomic DNA as template.
For RT-PCR analysis, total RNA isolated from wild type (WT) and transgenic rice plants was reverse transcribed with oligo-dT primer and a first-strand cDNA synthesis kit (Roche, Mannheim, Germany). The synthesized first-strand cDNA was used in subsequent PCR reactions as described above with gene-specific primers and control primers for OsUBQ5 (Jain et al. 2006; Supplementary Table S1). The cold-responsive genes analyzed included OsDREB1A (LOC_Os09g35030), Glutamate decarboxylase (LOC_Os03g13300), TPP1 (Trehalose-6-phosphate phosphatase1; LOC_Os02g44230), OsTGFR (LOC_Os01g68510), WSI76 (LOC_Os07g48830), OsCORTM1 (Cold-regulated 413 thylakoid membrane1; LOC_Os05g49170) and OsMAT1 (Methionine adenosyltransferase1; LOC_Os05g04510).
Analysis of OsWRKY71 promoter:GFP expression
The 1943-bp promoter region was isolated from rice genomic DNA by PCR using pOsWRKY71-F/R primers (Supplementary Table S1). After adapter-conjugating PCR with attB1 and attB2 adaptors, the isolated promoter fragment was linked to the Green fluorescent protein (GFP) gene by the Gateway LR clonase (Invitrogen). The transformation vector contained the bar gene between the CaMV35S promoter and the terminator of nopaline synthase gene (Park et al. 2010). Through Agrobacterium-mediated rice transformation and propagation of the transgenic lines, several independent lines of the T2 generation were obtained. Among them, one representative transgenic line was selected for further study.
Total RNA was extracted from 1-month-old leaves that were subjected to air-drying (drought), low temperature (4 °C) or high salinity (300 mM NaCl) for 0 to 6 h, and reverse transcribed with an oligo-dT primer and a first-strand cDNA synthesis kit (Roche). Real-time qRT-PCR was carried out with 50 ng of cDNA template using the GFP-specific primers GFP-F/R. Fluorescence from the amplicon was detected by a Stratagene Mx3000p Real-Time PCR machine and Mx3005P software v2.02 (Stratagene, USA). All results were measured from triplicate reactions for each sample. To normalize the real-time qRT-PCR data, the OsUBI1 (LOC_Os06g46770) gene primers OsUbi1-F/R were used as a reference.
Subcellular localization of OsWRKY71-GFP
Full-length cDNA of OsWRKY71 was amplified by PCR using full-length F/R primers (Supplementary Table S1) and the PCR product was cloned into the pENTR™/D-TOPO vector (Invitrogen). The insert was confirmed by sequencing and then subcloned into the destination vector p2FGW7 for N-terminal GFP fusion (Karimi et al. 2002) using the Gateway LR clonase (Invitrogen). The resulting construct was introduced into maize mesophyll protoplasts using PEG-calcium mediated transformation method (Cho et al. 2009). After incubation for 24 h, expression of the GFP fusion protein was monitored using a confocal microscope (LSM510 META, Carl Zeiss, Germany). GFP fluorescence was excited at 488 nm and detected in the emission range of 500–525 nm. OsABF1-RFP was used as a nuclear marker (Amir Hossain et al. 2010).
Assay for transcriptional activity of OsWRKY71
For the transient expression assay, an effector vector was constructed by fusing the full-length cDNA of OsWRKY71, amplified by PCR using primers containing NdeI and EcoRI sites (Supplementary Table S1), to the GAL4 binding domain (BD) sequence under the control of the CaMV35S promoter and the Ω sequence. The previously described GAL4-responsive reporter construct (Ohta et al. 2000) was used as a reporter vector, and contains 5X GAL4, a minimal TATA, the Ω sequence and the Luciferase (LUC) gene. The maize ubiquitin promoter:β-Glucuronidase (GUS) construct (ZmUBQ:GUS) was used as an internal control (Han et al. 2013). Maize mesophyll protoplasts isolated from the second leaves of dark grown plants were transfected with the effector, reporter and internal control vectors as described previously (Han et al. 2013). Transfected protoplasts were incubated in W5 solution for 12 h (Zhang et al. 2011). LUC and GUS activity assays were performed as previously described (Han et al. 2013). All experiments were repeated three times with similar results.
Production of OsWRKY71 overexpressing transgenic plants
Full-length cDNA of OsWRKY71 was amplified by PCR using full-length F/R primers (Supplementary Table S1), cloned into pENTR™/D-TOPO vectors (Invitrogen), and then subcloned into the binary plant vector pH2GW7 (Karimi et al. 2002) using the gateway LR clonase (Invitrogen). To produce transgenic rice plants expressing CaMV35S:OsWRKY71, the Agrobacterium tumefaciens LBA4404 strain harboring this vector was grown on AB media supplemented with 10 mg L−1 streptomycin and 50 mg L−1 hygromycin for 3 days at 28 °C, and transgenic calli were obtained via the Agrobacterium-mediated co-cultivation method as described previously (Jeon et al. 2000). OsWRKY71-OX transgenic plants were selected on medium containing 50 mg L−1 hygromycin and 250 mg L−1 cefotaxime. T2 homozygous lines were selected by testing for hygromycin resistance of progeny plants and analyzed for further characterization.
Phenotype analysis of OsWRKY71-OX lines in response to cold stress
To examine the cold tolerance of OsWRKY71-OX lines, we analyzed both the chlorophyll fluorescence and survival ratio of transgenic plants (Lee et al. 2004). In brief, the youngest leaves of 10-day-old seedlings were cut and floated on distilled water at 4 °C in a cold chamber under continuous light of 110 μmol m−2 s−1 for up to 40 h and the chlorophyll fluorescence was measured using the Plant Efficiency Analyzer (Hansatech, Kings Lynn, UK). The ratio Fv/Fm was calculated to examine damage in the photosystem (Genty et al. 1989). For survival analysis, 10-day-old seedlings were treated in the cold chamber for up to 50 h. After recovery for 6 days under normal growth conditions, the third-leaf survived plants were counted and their fresh weights were measured. Dry weights were measured after completely drying at 80 °C for 2 days.
Results
OsWRKY71 expression in response to cold treatment
Analysis of publicly available microarray data revealed that expression of OsWRKY71 is greatly increased when rice plants are challenged by cold stress (Supplementary Fig. S1). This prompted us to further investigate the expression of OsWRKY71 in rice in response to cold and other environmental stresses by RNA blot analysis (Fig. 1). OsWRKY71 expression was examined by subjecting rice seedlings and panicles of flowering plants to cold (4 °C), ABA (100 µM), salt (300 mM NaCl), and drought (20 % PEG and air-drying) treatments. OsWRKY71 expression was rapidly increased in cold-treated seedlings as early as 2 h, but was barely detectable in untreated samples (Fig. 1a). Notably, none of the ABA, salt, and drought treatments increased OsWRKY71 expression relative to that of untreated leaves, even 8 h after each treatment (Fig. 1b), indicating its unique responsiveness to cold treatment. The cold-responsive expression of OsWRKY71 was consistently observed in the mature leaf and panicle after 3 days (I) and 7 days (II) exposure to cold stress (Fig. 1c). In contrast, the gene expression of SalT was upregulated in response to salt and drought, as previously reported (Claes et al. 1990).
Analysis of cold-responsive expression of OsWRKY71. a RNA blot analysis of the time course of OsWRKY71 expression under control or cold-treated conditions. Two-week-old seedlings were transferred to the 4 °C growth chamber for 0, 2, 4, 8, 12, and 24 h of cold treatment and then sampled for RNA blot analysis. Untreated seedlings were sampled at the same time as controls. rRNAs are shown as a loading control. b RNA blot analysis of OsWRKY71 under control, ABA (100 µM), salt (300 mM NaCl), cold (4 °C), and drought (air dry* and 20 % PEG**) conditions. All stresses were applied to 2-week-old seedlings for 8 h. c RNA blot analysis of OsWRKY71 in mature leaves and panicles under control, drought, salt, and cold conditions. Mature leaves and panicles a week before anthesis were sampled after 3 (I) or 7 days (II) of various stress treatments. SalT expression was included as a drought- and salt-responsive marker gene. d Promoter activity analysis of pOsWRKY71:GFP in transgenic rice. A strong GFP signal was detected only after cold (4 °C) treatment, in contrast to drought (air dry) and salt (300 mM NaCl) treatments. The fold values were represented as a graph relative to the level of GFP mRNA in the lowest-expressing transgenic plants, as indicated by an asterisk
To further confirm that OsWRKY71 is regulated by cold at the transcriptional level, we constructed a fusion vector containing the approximately 2-kb promoter sequence of OsWRKY71 and the reporter gene GFP (pOsWRKY71:GFP) and introduced the vector into rice. pOsWRKY71:GFP transgenic rice plants were treated with cold, drought and salt. GFP expression analysis indicated that OsWRKY71 promoter activity was dramatically increased after only 2 h of cold treatment (Fig. 1d).
Determination of subcellular localization of OsWRKY71
To determine the subcellular localization of OsWRKY71, an OsWRKY71-GFP fusion construct under the control of the CaMV35S promoter was generated. In a transient expression assay with maize mesophyll protoplasts, OsWRKY71-GFP was detected mostly in the nucleus and to a lesser extent in the cytosol, as confirmed by the nuclear marker construct, OsABF1-RFP (Fig. 2a). Therefore, we concluded that the OsWRKY71 protein might be targeted to the nucleus.
Subcellular localization of OsWRKY71 and transcriptional repression assay. a Expression of OsWRKY71-GFP fusion protein in maize protoplast cells. The OsWRKY71-GFP signal is green; OsABF1-RFP is used as a nuclear marker. Bright light field and merged images are also shown. b Structures of the reporter and effector vectors used for the transcriptional activity assay of OsWRKY71. ZmUBQ:GUS was used as an internal control. Nos, nopaline synthase terminator. c Luciferase activity assay. The effector vector, either empty vector (EV) or BD-OsWRKY71 (OsWRKY71), was co-infected with the reporter LUC vector and the internal control vector ZmUBQ:GUS. The fold change was calculated as the LUC/GUS activity ratio. The values are the averages with standard deviation of the results from three independent experiments. Normalized LUC activity recorded after transfection with the empty vector control alone was arbitrarily set at 1. *P < 0.05
Transcriptional repression ability of OsWRKY71
Next, we examined the transcriptional activation ability of OsWRKY71 using the maize protoplast transient expression system (Han et al. 2013). The effector constructs for this system contain the CaMV35S promoter, the TMV translation enhancer Ω sequence, and either the GAL4 DNA BD or the OsWRKY71 cDNA insert fused to the BD (Fig. 2b). As a reporter, we used a GAL4-responsive reporter construct (Ohta et al. 2000) that contains five copies of the GAL4 binding site in tandem and a minimal TATA region of the CaMV35S promoter, the Ω sequence, and the firefly LUC gene (Fig. 2b). In the maize protoplasts, the reporter LUC activity was considerably reduced when co-infected with the effector BD-OsWRKY71 fusion vector compared with the empty vector control (Fig. 2c). This result suggests that OsWRKY71 may function as a transcriptional repressor to regulate downstream target genes.
Characterization of transgenic rice plants overexpressing OsWRKY71
To characterize the function of OsWRKY71 in rice, transgenic rice plants constitutively overexpressing OsWRKY71 under the control of the CaMV35S promoter were generated using Agrobacterium-mediated transformation. The transcript level of OsWRKY71 was much higher in two independent transgenic T2 homozygous lines, OsWRKY71-OX12 (OX12) and OsWRKY71-OX21 (OX21), than in the transformation background WT (cv. Dongjin) and the segregated non-transgenic line (NT) (Fig. 3a). There was no apparent difference in agricultural phenotypic characteristics such as plant height and yield potential among these transgenic lines (data not shown).
Production and characterization of OsWRKY71-OX transgenic rice plants. a RT-PCR analysis of OsWRKY71 expression in OsWRKY71-OX12 (OX12) and OsWRKY71-OX21 (OX21) transgenic lines. OsUBQ5 was amplified as an internal RT-PCR control. b OsWRKY71 overexpression confers cold tolerance in rice. Five-week-old rice seedlings of wild type (WT), segregant non-transgenic (NT), and OsWRKY71-OX transgenic lines (OX12 and OX21) were exposed to cold stress (4 °C) for 50 h and then allowed to recover for 6 days under normal conditions
To investigate whether OsWRKY71 overexpression correlated with cold tolerance in rice, two 5-week-old control lines, WT and NT, and two OsWRKY71-OX lines, OX12 and OX21, were exposed to cold stress (4 °C) for 50 h and then allowed to recover their growth for 6 days under normal growth conditions. We found that OX12 and OX21 transgenic plants recovered much better, maintaining a greater number of green leaves, than WT and NT control plants, which displayed severely dried leaves (Fig. 3b).
Cold stress tolerance was examined by measuring the third-leaf survival ratio of 10-day-old seedlings. Most plants of the OsWRKY71-OX lines OX12 and OX21, 94 and 97 %, respectively, survived after cold stress treatment followed by recovery. In contrast, only 54 and 75 % of the control WT and NT plants, respectively, survived (Table 1). Our results clearly indicate that OsWRKY71 overexpression in rice plants increases tolerance to low-temperature stress.
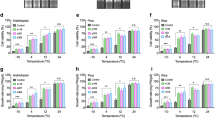
The elevated tolerance of OsWRKY71-OX transgenic rice plants to cold stress was further evaluated by measuring chlorophyll fluorescence in two control and two transgenic lines that were subjected to low temperature (4 °C) for 20 or 40 h. The ratio F v/F m between the variable fluorescence (F v) and the maximum fluorescence (F m) is used to estimate the quantum yield of PSII photochemistry (Krause and Weis 1991). Under normal growth conditions (0 h), all tested plants showed an F v/F m of approximately 0.85, which is close to the value of 0.8 observed in all plant species studied (Fig. 4a). When exposed to cold stress, the control plants showed a significant reduction in the F v/F m values: 0.57 and 0.58 after 20 h and 0.2 and 0.18 after 40 h in WT and NT, respectively. In contrast, OsWRKY71-OX lines maintained relatively high F v/F m values of 0.7 and 0.71 after 20 h and 0.3 and 0.37 after 40 h for OX12 and OX21, respectively (Fig. 4a). Concordant with these findings, OX12 and OX21 plants retained higher levels of both fresh weight and dry weight than control plants (Fig. 4b, c). These results further suggest that overexpression of OsWRKY71 enhances cold tolerance in transgenic rice.
Phenotype analysis of OsWRKY71-OX transgenic rice seedlings in response to cold treatment. a Changes in chlorophyll fluorescence of wild type (WT), segregant non-transgenic (NT), and OsWRKY71-OX transgenic lines (OX12 and OX21) at 20 h and 40 h of cold stress treatment. Fresh weight (b) and dry weight (c) after 6 days of recovery. *P < 0.05
Expression of cold-responsive genes in OsWRKY71-OX lines
To obtain clues regarding the molecular mechanism of OsWRKY71-mediated cold tolerance, we monitored the expression of previously known cold-responsive genes in OsWRKY71-OX lines by RT-PCR analysis. Transcription levels of the selected cold marker genes were examined in both control and transgenic lines under cold (4 °C) treatment for 12 or 24 h. We found remarkable upregulation of the rice DREB gene OsDREB1A (Dubouzet et al. 2003; Ito et al. 2006) in the control, WT, NT, and transgenic OX12 and OX21 plants. These plants showed no difference in the pattern of OsDREB1A expression (Fig. 5), which peaked 12 h after cold treatment. Likewise, expression of two marker genes, Glutamate decarboxylase (Su et al. 2010) and TPP1 (Pramanik and Imai 2005), reached a similar peak 12 h after cold treatment in control and transgenic lines and thereafter decreased. OsCORTM1 (OsCOR413-TM1; Ma et al. 2009) and OsMAT1 (Ma et al. 2009), as well as the PCR control gene OsUBQ5, did not respond to cold treatment in control and transgenic lines.
Expression analysis of cold-responsive marker genes in OsWRKY71-OX lines in response to cold stress. Leaves of 5-week-old seedlings exposed to 4 °C were harvested at 0, 12, and 24 h. OsUBQ5 was used as an internal control. WT wild type, NT segregant non-transgenic line, OX12 and OX21 OsWRKY71-OX transgenic rice lines
Interestingly, two cold-responsive genes, OsTGFR and WSI76, which are the rice homologs of target genes of Arabidopsis DREBs (Dubouzet et al. 2003; Chen et al. 2008; Saito and Yoshida 2011), showed differential induction in OsWRKY71-OX plants under normal and cold stress conditions (Fig. 5). OsTGFR expression was detected in OsWRKY71-OX plants before and after cold treatment. WSI76 expression was more highly induced after cold treatment in OX12 and OX21 than in control lines. This result suggests that OsWRKY71 may have a regulatory function in the expression of OsTGFR and WSI76 in response to cold treatment.
Discussion
Among several families of transcription factors that are involved in the transcriptional regulation of plant stress-responsive genes, increasing attention has recently been paid to the role of WRKY transcription factors in regulating the response to abiotic and biotic stresses. However, relatively little progress has been made in understanding the function of WRKY proteins under abiotic stresses (Chen et al. 2012).
Here, we provide evidence that OsWRKY71 positively regulates cold tolerance in rice plants. First, we showed that OsWRKY71 was induced specifically by cold (Fig. 1). Second, we found that it functioned as a nuclear transcriptional repressor (Fig. 2). Third, OsWRKY71 overexpression enhanced cold tolerance in transgenic rice, and this elevated tolerance was further confirmed by measurement of chlorophyll fluorescence, F v/F m (Figs. 3, 4; Table 1). Finally, we identified two potential downstream targets of OsWRKY71, OsTGFR and WSI76, in response to cold treatment (Fig. 5). Together, these data suggest that OsWRKY71 may have an important transcriptional regulatory role in response to cold stress, and that the OsWRKY71-mediated pathway is likely essential for cold tolerance in rice. It is noteworthy that OsWRKY71 was not upregulated by ABA and other abiotic stresses such as salt and drought in rice seedlings (Fig. 1), implying that OsWRKY71 may function in an ABA-independent cold response pathway.
Numerous transcriptional expression data have revealed cold-responsive genes in common with those under observation in the present study that are associated with cold stress in different plant species (Fowler and Thomashow 2002; Maruyama et al. 2004; Seki et al. 2002; Dubouzet et al. 2003; Wang et al. 2004; Fowler et al. 2005; Skinner et al. 2005; Ito et al. 2006; Badawi et al. 2007; Cheng et al. 2007; Zhou et al. 2008; Mittal et al. 2012; Zhang et al. 2012). For instance, DREB1s act as upstream regulators during cold stress and are rapidly induced several fold after cold treatment. Our experimental observation of expression patterns of cold-responsive genes other than OsCORTM1 and OsMAT1, such as OsDREB1A, Glc decarboxylase, and TPP1, indicated that many of the rice homologs of cold-responsive genes are also rapidly regulated in cold-exposed rice (Fig. 5), supporting conserved regulatory mechanism(s) in a range of plant species.
In Arabidopsis there are seven members belonging to the GolS gene family, of which GolS3 is controlled by DREB1A and induced by cold stress (Taji et al. 2002). Expression of the rice homolog of GolS3, WSI76, was found to be increased under chilling stress (Saito and Yoshida 2011). In this context, it is noteworthy that we found a remarkable increase in the transcripts of WSI76 in OsWRKY71-OX transgenic rice compared with the controls WT and NT after cold treatment (Fig. 5). In Boea hygrometrica, a WRKY is involved in the regulation of GolS expression (Wang et al. 2009). OsTGFR is a homolog of target genes of Arabidopsis DREBs (Dubouzet et al. 2003; Chen et al. 2008; Ma et al. 2009; Saito and Yoshida 2011). We found an alteration in OsTGFR expression in OsWRKY71-OX transgenic rice in response to cold compared with expression in the controls. The enhancement of stress tolerance in OsWRKY71 transgenic plants likely results from changes in expression of these cold-responsive genes. In previous studies, OsWRKY28 (Chujo et al. 2013) and OsWRKY76 (Yokotani et al. 2013), closely related paralogues of OsWRKY71, exhibited transcriptional repressor activity. Consistent with these findings, we found that OsWRKY71 functioned as a transcriptional repressor (Fig. 2c). This result suggests that OsWRKY71 exerts its function through transcriptional suppression of target genes that may negatively regulate the downstream genes OsTGFR and WSI76. How the transcriptional repressor OsWRKY71 regulates downstream target genes in response to cold stress remains to be determined.
Strong constitutive overexpression of transcription factors often results in negative growth phenotypes in transgenic plants. For example, growth retardation has been observed in transgenic rice plants overexpressing stress-responsive transcription factors such as OsDREB1A, OsDREB1B, OsWRKY89 and OsNAC6 (Ito et al. 2006; Nakashima et al. 2007; Wang et al. 2007). Although constitutive expression of these transgenes produced the best result in respect to cold tolerance, the overall advantage was reduced by significant differences in agronomical traits such as flowering time and yield potentials of transgenic lines. In the present study, OsWRKY71 overexpression resulted in no obvious phenotypic alteration, and plant architecture was indistinguishable from that of WT under normal growth conditions. Given that OsWRKY71 overexpression improved cold tolerance with an unaltered growth phenotype under normal conditions, our data imply the potential application of OsWRKY71 for genetic improvement of cold tolerance in rice. To further understand the molecular mechanism of enhanced cold tolerance responses mediated by OsWRKY71, it would be useful to analyze global gene expression changes in OsWRKY71-OX in response to cold stress and to identify and characterize downstream targets in future studies.
References
Amir Hossain MA, Cho JI, Han M, Ahn CH, Jeon JS, An G, Park PB (2010) The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J Plant Physiol 167:1512–1520
Andaya VC, Mackill DJ (2003) Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J Exp Bot 54:2579–2585
Badawi M, Danyluk J, Boucho B, Houde M, Sarhan F (2007) The CBF gene family in hexaploid wheat and its relationship to the phylogenetic complexity of cereal CBFs. Mol Genet Genom 277:533–554
Baruah A, Ishigo-Oka N, Adachi M, Oguma Y, Tokizono Y, Onishi K, Sano Y (2009) Cold tolerance at the early growth stage in wild and cultivated rice. Euphytica 165:459–470
Birkenbihl RP, Diezel C, Somssich IE (2012) Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol 159:266–285
Cai M, Qiu D, Yuan T, Ding X, Li H, Duan L, Xu C, Li X, Wang S (2008) Identification of novel pathogen-responsive cis-elements and their binding proteins in the promoter of OsWRKY13, a gene regulating rice disease resistance. Plant Cell Environ 31:86–96
Casaretto J, Ho TH (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15:271–284
Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30:2191–2198
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. BBA-Gene Regul 1819:120–128
Cheng C, Yun KY, Ressom HW, Mohanty B, Bajic VB, Jia Y, Yun SJ, de los Reyes BG (2007) An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom 8:175
Cho JI, Ryoo N, Ko S, Lee SK, Lee J, Jung KH, Lee YH, Bhoo SH, Winderickx J, An G et al (2006) Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.). Planta 224:598–611
Cho JI, Ryoo N, Eom JS, Lee DW, Kim HB, Jeong SW, Lee YH, Kwon YK, Cho MH, Bhoo SH et al (2009) Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol 149:745–759
Chujo T, Kato T, Yamada K, Takai R, Akimoto-Tomiyama C, Minami E, Nagamura Y, Shibuya N, Yasuda M, Nakashita H et al (2008) Characterization of an elicitor-induced rice WRKY gene, OsWRKY71. Biosci Biotech Biochem 72:240–245
Chujo T, Miyamoto K, Shimogawa T, Shimizu T, Otake Y, Yokotani N, Nishizawa Y, Shibuya N, Nojiri H, Yamane H et al (2013) OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol Biol 82:23–37
Ciolkowski I, Wanke D, Birkenbihl R, Somssich I (2008) Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY domain function. Plant Mol Biol 68:81–92
Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, Caplan A (1990) Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell 2:19–27
Dang FF, Wang YN, Yu L, Eulgem T, Lai Y, Liu ZQ, Wang X, Qiu AL, Zhang TX, Lin J et al (2012) CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ 36:757–774
de Los Reyes BG, Yun SJ, Herath V, Xu F, Park MR, Lee JI, Kim KY (2013) Phenotypic, physiological, and molecular evaluation of rice chilling stress response at the vegetative stage. Methods Mol Biol 956:227–241
Delteil A, Blein M, Faivre-Rampant O, Guellim A, Estevan J, Hirsch J, Bevitori R, Michel C, Morel JB (2012) Building a mutant resource for the study of disease resistance in rice reveals the pivotal role of several genes involved in defence. Mol Plant Pathol 13:72–82
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Fowler SG, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Fowler SG, Cook D, Thomashow MF (2005) Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol 137:961–968
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA-signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17:3470–3488
Genty B, Briantais JM, Baker NR (1989) The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta 990:87–92
Han M, Ryu HS, Kim CY, Park DS, Ahn YK, Jeon JS (2013) OsWRKY30 is a transcription activator that enhances rice resistance to the Xanthomonas oryzae pathovar oryzae. J Plant Biol 56:258–265
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181
Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47:141–153
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J et al (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69:91–105
Jiang CJ, Shimono M, Sugano S, Kojima M, Yazawa K, Yoshida R, Inoue H, Hayashi N, Sakakibara H, Takatsuji H (2010) Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice–Magnaporthe grisea interaction. Mol Plant Microbe Interact 23:791–798
Karimi M, Inze D, Depicker A (2002) Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7:193–195
Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: the basic. Ann Rev Plant Physiol Plant Mol Biol 42:313–349
Lee SC, Huh KW, An K, An G, Kim SR (2004) Ectopic expression of a cold-inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.). Mol Cells 18:107–114
Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233:1237–1252
Liu XQ, Bai XQ, Wang XJ, Chu CC (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164:969–979
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X et al (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 150:244–256
Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K (2004) Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J 38:982–993
Matsushita A, Inoue H, Goto S, Nakayama A, Sugano S, Hayashi N, Takatsuji H (2012) The nuclear ubiquitin proteasome degradation affects WRKY45 function in the rice defense program. Plant J 73:302–313
Mittal D, Madhyastha DA, Grover A (2012) Genome-wide transcriptional profiles during temperature and oxidative stress reveal coordinated expression patterns and overlapping regulons in rice. PLoS One 7:e40899
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Ohta M, Ohme-Takagi M, Shinshi H (2000) Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J 22:29–38
Park SH, Yi N, Kim YS, Jeong MH, Bang SW, Choi YD, Kim JK (2010) Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J Exp Bot 61:2459–2467
Peng Y, Bartley LE, Chen X, Dardick C, Chern M, Ruan R, Canlas PE, Ronald PC (2008) OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant 1:446–458
Peng Y, Bartley LE, Canlas PE, Ronald PC (2010) OsWRKY IIa transcription factors modulate rice innate immunity. Rice 3:36–42
Peng X, Zhang L, Zhang L, Liu Z, Cheng L, Yang Y, Shen S, Chen S (2013) The transcriptional factor LcDREB2 cooperates with LcSAMDC2 to contribute to salt tolerance in Leymus chinensis. Plant Cell Tiss Org Cult 113:245–256
Pramanik MH, Imai R (2005) Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol 58:751–762
Rice WRKY Working Group (2012) Nomenclature report on rice WRKY’s—conflict regarding gene names and its solution. Rice 5:1–3
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Saito M, Yoshida M (2011) Expression analysis of the gene family associated with raffinose accumulation in rice seedlings under cold stress. J Plant Physiol 168:2268–2271
Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J 31:279–292
Seo YS, Chern M, Bartley LE, Han M, Jung KH, Le I, Walia H, Richter T, Xu X, Cao P et al (2011) Towards establishment of a rice stress response interactome. PLoS Genet 7:e1002020
Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315:1098–1103
Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H (2007) Rice WRKY45 plays a crucial role in benzothiazole-inducible blast resistance. Plant Cell 19:2064–2076
Shimono H, Ishii A, Kanda E, Suto M, Nagano K (2011a) Genotypic variation in rice cold tolerance responses during reproductive growth as a function of water temperature during vegetative growth. Crop Sci 51:290–297
Shimono M, Koga H, Akagi A, Hayashi N, Goto S, Sawada M, Kurihara T, Matsushita A, Sugano S, Jiang CJ et al (2011b) Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Mol Plant Pathol 13:83–94
Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. Curr Opin Biotech 7:161–167
Singh KB, Foley RC, Oñate-Sánchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Skinner JS, von Zitzewitz J, Szucs P, Marquez-Cedillo L, Filichkin T, Amundsen K, Stockinger EJ, Thomashow MF, Chen TH, Hayes PM (2005) Structural, functional, and phylogenetic characterization of a large CBF gene family in barley. Plant Mol Biol 59:533–551
Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153:145–158
Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29:417–426
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Wang X, Liu S, Liu X, Chen Z, Liu X, Pang Y, Sun X, Tang K (2004) Molecular cloning and characterization of a CBF gene from Capsella bursapastoris. DNA Seq 15:180–187
Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z (2007) Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol 65:799–815
Wang Z, Zhu Y, Wang L, Liu X, Liu Y, Phillips J, Deng X (2009) A WRKY transcription factor participates in dehydration tolerance in Boea hygrometrica by binding to the W-box elements of the galactinol synthase (BhGolS1) promoter. Planta 230:1155–1166
Xu XP, Chen CH, Fan BF, Chen ZX (2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18:1310–1326
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63:2541–2556
Yokotani N, Sato Y, Tanabe S, Chujo T, Shimizu T, Okada K, Yamane H, Shimono M, Sugano S, Takatsuji H et al (2013) WRKY76 is a rice transcriptional repressor playing opposite roles in blast disease resistance and cold stress tolerance. J Exp Bot 64:5085–5097
Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol 134:1500–1513
Zhang CQ, Xu Y, Lu Y, Yu HX, Gu MH, Liu QQ (2011) The WRKY transcription factor OsWRKY78 regulates stem elongation and seed development in rice. Planta 234:541–554
Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274
Zhou QY, Tian AG, Zou HF, Xie ZM, Lei G, Huang J, Wang CM, Wang HW, Zhang JS, Chen SY (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503
Zou CS, Jiang WB, Yu DQ (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61:3901–3914
Acknowledgments
This work was supported by grants from the Mid-Career Researcher Program (NRF-2013R1A2A2A01068887 to J.-S. Jeon) and Basic Science Research Program (NRF-2014R1A1A1003903 to S.-K. Lee) of the National Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
C.-Y. Kim, K. T. X. Vo, and C. D. Nguyen have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, CY., Vo, K.T.X., Nguyen, C.D. et al. Functional analysis of a cold-responsive rice WRKY gene, OsWRKY71 . Plant Biotechnol Rep 10, 13–23 (2016). https://doi.org/10.1007/s11816-015-0383-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-015-0383-2