Abstract
Expansins are pH-dependent cell wall loosening proteins which form a large family in plants. They have been shown to be involved in various developmental processes and been implicated in enabling plants’ ability to absorb nutrients from the soil as well as conferring biotic and abiotic stress resistances. It is therefore clear that they can be potential targets in genetic engineering for crop improvement. Tobacco (Nicotiana tabacum) is a major crop species as well as a model organism. Considering that only a few tobacco expansins have been studied, a genome-wide analysis of the tobacco expansin gene family is necessary. In this study, we identified 52 expansins in tobacco, which were classified into four subfamilies: 36 NtEXPAs, 6 NtEXPBs, 3 NtEXLAs and 7 NtEXLBs. Compared to other species, the NtEXLB subfamily size was relatively larger. Phylogenetic analysis showed that the 52 tobacco expansins were divided into 13 subgroups. Gene structure analysis revealed that genes within subfamilies/subgroups exhibited similar characteristics such as gene structure and protein motif arrangement. Whole-genome duplication and tandem duplication events may have played important roles in the expanding of tobacco expansins. Cis-Acting element analysis revealed that each expansin gene was regulated or several expansin genes were co-regulated by both internal and environmental factors. 35 of these genes were identified as being expressed according to a microarray analysis. In contrast to most NtEXPAs which had higher expression levels in young organs, NtEXLAs and NtEXLBs were preferentially expressed in mature or senescent tissues, suggesting that they might play different roles in different organs or at different developmental stages. As the first step towards genome-wide analysis of the tobacco expansin gene family, our work provides solid background information related to structure, evolution and expression as well as regulatory cis-acting elements of the tobacco expansins. This information will provide a strong foundation for cloning and functional exploration of expansin genes in tobacco.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Expansins comprise a group of proteins that can induce a rapid pH-dependent cell wall extension (acid growth) and stress relaxation (McQueen-Mason et al. 1992; Cosgrove 2000a, b). They are considered to bind to glucan-coated cellulose in the cell wall causing a reversible disruption of hydrogen bonding between cellulose microfibrils and glucan matrix and thus loosening the cell wall in a pH-dependent manner (Sampedro and Cosgrove 2005; Cosgrove 2015). Expansins are commonly found in plants and are well known to play important roles in diverse biological processes related to cell wall modification, including seed germination (Yan et al. 2014), root growth and development (Guo et al. 2011; Yu et al. 2011), stem growth and internode elongation (Cho and Kende 1997; Kuluev et al. 2014), leaf initiation and expansion (Fleming et al. 1997; Kuluev et al. 2013), flowering and flower size (Kuluev et al. 2012), pollen germination and fertilization (Cosgrove 2000a), fruit growth and/or ripening (Rose et al. 1997; Brummell et al. 1999; Civello et al. 1999; Ishimaru et al. 2007). They have also been associated with nutrient uptake and efficiency (Zhou et al. 2014), abiotic and biotic stress tolerance (Li et al. 2011; Zhao et al. 2011; Lü et al. 2013). After looking closely at the various roles played by expansins in plants, Marowa et al. (2016) have concluded that inclusion of expansins in plant breeding programs presents an opportunity to improve crops in various aspects. These include but are not limited to improving germination, leaf size, fruit growth and ripening and tolerance to abiotic and biotic stresses (Marowa et al. 2016).
Expansins which are encoded by a multi-gene family have been reported in many plant species and organs, including Arabidopsis and rice (Lee et al. 2001), maize (Zhang et al. 2014a, b), soybean (Zhu et al. 2014), tomato (Lu et al. 2016), apple (Zhang et al. 2014a, b) to mention but a few. They have also been reported in bacteria and fungi, most of which colonize plant surfaces (Georgelis et al. 2015). Genome-wide analysis has revealed a large expansin superfamily (EXP) in plants comprising of four subfamilies defined as α-expansin (EXPA), β-expansin (EXPB), expansin-like A (EXLA) and expansin-like B (EXLB) according to phylogenetic sequence analysis and a standardized nomenclature (Kende et al. 2004; Sampedro and Cosgrove 2005). EXPA was the first group of expansins to be identified as cell wall loosening proteins with their enigmatic effects on cell wall rheology (Cosgrove 2000a), which can rapidly induce creep and stress relaxation of primary cell walls without any lytic activity but are acid-dependent. EXPB contain two subgroups, group-1 grass pollen allergens which are known to facilitate pollen tube invasion of the stigma by dissolving the middle lamella (Sampedro et al. 2015) and a group we know very little (Cosgrove 2015). Both EXPA and EXPB are the two major subfamilies of the plant expansin gene family which are involved in cell expansion and other developmental processes without detectable enzymatic activities (Li and Cosgrove 2001; Yennawar et al. 2006; Cosgrove 2015). Even though there is no direct evidence to show that the other two smaller subfamilies, EXLA and EXLB, could have cell wall loosening function as well, the ectopic expression of AtEXLA2 increased the length of root and hypocotyls of Arabidopsis plants (Boron et al. 2015).
Canonical plant expansins have been defined as small proteins, about 250–275 amino acids in size, which are considered to typically contain two conserved domains, DPBB_1 and Pollen_allerg_1, that are preceded by a signal peptide of 20–30 amino acids in length (Sampedro and Cosgrove 2005; Yennawar et al. 2006). The first domain is a six-stranded double-psi beta-barrel, which has a distant homology to the catalytic domain of a glycoside hydrolase GH45, but lack its β-1, 4-glucanase activity (Yennawar et al. 2006). It also contains several conserved cysteine (Cys) residues. The second domain, also classified as a family-63 carbohydrate binding module (CBM63), contains a β-sandwich fold and is homologous to a group-1 grass pollen allergen, which might be a polysaccharide binding domain because it has the conserved aromatic and polar residues-tryptophan (Trp) on its surface (Georgelis et al. 2012; Cosgrove 2015).
Although genome-wide studies on expansins have been reported in many plant species, a systematic analysis of this gene family in tobacco (Nicotiana tabacum) has not yet been published to date, except expression studies for a few tobacco expansin genes (Kuluev et al. 2013, 2014). The completion of high-quality draft genomes of tobacco (Sierro et al. 2014) provides us the opportunity to uncover the expansin gene family and its characteristics in this allotetraploid crop. In this study, we identified the expansin genes in the tobacco genome, analyzed their structure and evolution relationships, and detected the cis-acting elements involved in their expression regulation under internal and environmental conditions. We also detected the expression patterns of the tobacco expansin genes with both the Tobacco Expression Atlas (TobEA) and quantitative real time PCR (qRT-PCR). Our results are expected to pave the way for further functional researches on this gene family in tobacco.
Materials and methods
Identification and sequence analysis of the tobacco expansin genes
The genome sequences of tobacco, tomato (Solanum lycopersicum; SL 2.40 ITAG2.3) and potato (Solanum tuberosum; PGSC_DM_v3.4) were all downloaded from sol genomics network (http://solgenomics.net/). The local genomes, coding sequences and protein sequences were constructed from these databases with the blast-2.2.9 programs, which were downloaded from the national center for biotechnology information (NCBI) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/release/2.2.9/blast-2.2.9-ia32-win32.exe). To identify the expansin gene family in these three species, BLASTN was performed against the local databases for each species using the 36 expansin gene sequences of Arabidopsis (AtEXPs) as queries.
Protein domain analysis for all the protein sequences was performed with Pfam 28.0 (http://pfam.xfam.org/) based on hidden Markov model. The online software SMART (http://smart.embl-heidelberg.de/) was used to confirm the presence of the two classical domains, DPBB_1 (PF03330) and Pollen_allerg_1 (PF01357) with default parameters. Only genes with integrated open reading frames (ORFs) and containing both DPBB_1 and Pollen_allerg_1 domains were reserved for further analysis. Redundant sequences were manually removed. The gene names were nominated according to Kende et al. (2004).
The DNAMAN v8.0 software (http://www.lynnon.com/) was used to calculate the number of amino acids, molecular weight (MW) and isoelectric point (pI) for each expansin gene. The signal peptide cleavage sites were predicted with SignalP v4.1 server (http://www.cbs.dtu.dk/services/SignalP/). To classify which subfamily the expansin genes belonged to, a phylogenetic tree was constructed together with the AtEXPs for each species with MEGA v5.0 using the neighbor-joining (NJ) method with 500 bootstrap replicates (Tamura et al. 2011). Before the phylogenetic tree construction, a multi-sequence alignment was performed with the clustalW program (Thompson et al. 1994).
Gene structures of the tobacco expansins were obtained by comparing the predicted CDS with their corresponding genomic DNA sequences using online program GSDS (http://gsds.cbi.pku.edu.cn/). The online program MEME (http://meme-suite.org/) was used to detect other conserved motifs contained for each gene besides the two well-known conserved domains with optimum motif width ≥6 and ≤200 and maximum of 20 motifs (Lin et al. 2011). To analyze gene locations, the start and end positions of each ORF were obtained from the tobacco genome database using the blast-2.2.9 programs. Based on the physical locations, contig regions containing two or more homologous genes within 200 kb were defined as gene clusters (Cheng et al. 2012).
We searched each tobacco and Arabidopsis expansin gene in InterproScan 5 for GO category annotations (http://www.ebi.ac.uk/interpro/). With the InterproScan output file, BGI WEGO was used to draw the GO figures (http://wego.genomics.org.cn/cgi-bin/wego/index.pl). According to the physical location on contigs of the tobacco expansin genes, the 1.5 kb upstream regions preceding the start codon were cut out with the blast-2.2.9 programs. Cis-Acting element analysis in the 1.5 kb upstream regions for each expansin gene was performed with PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Expression analysis of the tobacco expansin genes
TobEA, a custom designed Affymetrix tobacco expression microarray was generated from a set of over 40,000 unigenes and used to measure gene expression in 19 different samples throughout the tobacco lifecycle from seed to senescence (Edwards et al. 2010), providing researchers the opportunity to acquaint the expression profiles for genes of interest. The 19 samples include different organs as well as the same organ but at different developmental stages. They are seed, young root and mature root, young shoot, cotyledons, vegetable shoot apex, lower stem and upper stem, floral shoot apex, closed bud and open bud, flower, young leaf, cauline leaf, mature leaf, early senescent leaf, mid/early senescent leaf, mid/late senescent leaf, and late senescent leaf. The microarray data of gene expression in tobacco were downloaded from the EMBL-EBI ArrayExpress with accession number E-MTAB-176 (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-176/). The identified tobacco expansin gene sequences were used as queries against the tobacco SGN unigene databases to find out their corresponding Unigene IDs used in the microarray data using BLASTN. With a Perl script, the microarray data were compiled into a database, which were then clustered with Cluster 3.0 (http://rana.lbl.gov/eisenSoftware.htm) using the Euclidean distance and the hierarchical clustering method of complete linkage cluster. Finally, the clustering tree was constructed and viewed with Java Treeview (http://jtreeview.sourceforge.net/).
Plant materials
The tobacco variety K326 was used as the plant materials to investigate the expression profiles of expansin genes. The tobacco K326 seeds were grown in a culturing room at 23 ± 1 °C under a 16-h light/8-h dark cycle. The seedlings were then planted in pots in greenhouse under natural conditions until flowering period. In total, six types of tissues were sampled at the following stages: germinating seeds, root (fast growing period), stem (fast growing period), leaf (fast growing period), blooming flowers and senescent leaf. The samples were frozen immediately in liquid nitrogen for at least 10 min and stored at −80 °C for total RNA extraction.
RNA isolation and qRT-PCR analysis
Total RNA was isolated from each sample using the Trizol reagent (Roche), and treated with the DNase I (Fermentas). After quality and quantity analysis with gel electrophoresis and NanoDrop 2000, the first cDNA strand was synthesized from 3 μg total RNA using the Transcriptor First Strand cDNA Synthesis Kit version 6.0 (Roche) according to the user protocol. The cDNA was diluted five times before being used in downstream experiments. The ABI 7500 Real-Time System (Applied biosystems) was used for qRT-PCR experiments. The tobacco actin housekeeping gene was used as an internal control. The gene-specific primers for qRT-PCR were designed with Primer premier 6.0. Reactions contained 2 μL of cDNA as template, 400 nM forward and reverse primers, 10 μL Roche FastStart Universal SYBR Green Master (ROX), and sterile water to a total volume of 20 μL. Thermal cycling for qRT-PCR was as follows: 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 15 s, and 60 °C for 34 s, with melting curves constructed by increasing the final temperature from 60 to 95 °C for each gene to analyze their specificity. Relative expression levels were analyzed using ABI 7500 software v2.0.1.
Results
Identification of expansin gene family in tobacco
With the AtEXPs as quarries, through tobacco genome blast and protein motif identification, a total of 52 putative expansin gene sequences were identified in the tobacco genome (Table 1). All the 52 putative expansin genes (NtEXPs) had the two conserved domains, DPBB_1 and Pollen_allerg_1, in their protein sequences. Protein sequences that contained only one of these domains or did not have integrated ORFs were excluded. The coding sequences (CDS, Additional file 1), genomic sequences (Additional file 2), protein sequences (Additional file 3) and 1.5 kb nucleotide sequences upstream of the initiation codon for transcription (Additional file 4) for the 52 NtEXPs were all downloaded from ftp://ftp.solgenomics.net/genomes/Nicotiana_tabacum/.
The expansin gene family is a large family with four subfamilies namely EXPA, EXPB, EXLA and EXLB. To group these 52 tobacco expansins into their respective subfamilies, we performed a phylogenetic analysis based on their full length protein sequences. To achieve the desired results, we included expansins from Arabidopsis (36 AtEXPs) and rice (Oryza sativa; 58 OsEXPs) which have already been classified. Basing on the phylogenetic tree, we could classify the NtEXPs according to the clusters exhibited on the tree (Additional file 5). The tobacco expansin gene family was accordingly classified into the four subfamilies: NtEXPA, NtEXPB, NtEXLA and NtEXLB, each containing 36, 6, 3 and 7 members, respectively (Table 2). Since the chromosome loci of the 52 NtEXPs are still unclear, we nominated the genes sequentially according to the subfamily they belonged to on the basis of the nomenclature rules proposed by Kende et al. (2004).
The isolated NtEXPs encode proteins ranging from 240 to 332 amino acids in size with an average length of 260 amino acids. The molecular weight of the 52 expansins proteins ranges from 26.2 to 35.6 kDa. Forty two of the 52 expansins contain signal peptides ranging from 19 to 29 amino acids in length. The pI value ranges from 4.47 to 10.19 for the NtEXPs. Interestingly, all of the members in the NtEXPA (except NtEXPA25), NtEXPB, and NtEXLA subfamilies have pI values above 7.0, while the pI values of most members in NtEXLB are below 7.0. All the tobacco expansin genes have orthologous in Arabidopsis (Table 1).
To gain insights into the size characteristics of the four subfamilies, expansin genes from seven species, Arabidopsis, soybean (Glycine max), rice, maize (Zea mays), tobacco and two other solanum plants, tomato and potato, were compared. Expansin data for Arabidopsis, soybean, rice and maize were obtained from published results (Lee et al. 2001; Zhang et al. 2014a, b; Zhu et al. 2014). Genome-wide identification of expansin gene family was executed in tomato and potato following the same way we used to identify the tobacco expansin genes. A total of 38 and 39 putative expansin gene sequences were identified in the tomato and potato genomes, respectively, which were also grouped into the four subfamilies (Table 2). The number of genes contained in each subfamily of the seven species is rather uneven. In most cases, EXLA has the smallest subfamily size, whereas EXPA has the largest subfamily size except in maize. In monocotyledons, rice and maize, EXLB has the smallest subfamily size and the size of their EXPB subfamily is much larger than in dicotyledons, whereas in Arabidopsis EXLB also has a smallest subfamily size. In solanum plants, the sizes of EXPB and EXLB subfamilies are almost equal. In addition, soybean has a much larger EXLB subfamily size than other species.
Phylogenetic and structural analysis of NtEXPs
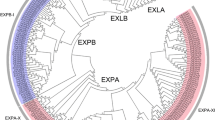
Phylogenetic analysis showed that members of the NtEXPA, NtEXPB, NtEXLA and NtEXLB subfamilies were clustered together according to their evolutionary relationships (Fig. 1). In the unrooted tree, all the bootstrap values were above 50 %. Eighteen sister pairs of genes were supported by bootstrap values of 100 %. Basing on the grouping methods used for Arabidopsis and rice expansin gene family (Sampedro et al. 2005), we found that the tobacco expansin gene family contained 8 subgroups for NtEXPA, 2 subgroups for NtEXPB, 1 subgroup for NtEXLA and 2 subgroups for NtEXLB (Additional file 5). Among these subgroups, EXPA-IV constituted the largest clade with 9 NtEXPAs. In contrast, EXPA-V, EXPA-VIII, EXPA-XI and EXPA-XII subgroups which were identified in rice and/or Arabidopsis did not contain any tobacco expansin genes.
a Phylogenetic relationships of the tobacco expansins; b exon–intron organizations of tobacco expansion genes; c motif location in the protein sequences of tobacco expansins. In the phylogenetic tree constructed with MEGA v5.0, NtEXPA, NtEXPB, NtEXLA and NtEXLB subfamilies were marked in red, blue, pink and green lines, respectively. In the expansin gene structures, exons were represented by yellow boxes, whereas introns were shown in black lines. The motif compositions were detected with MEME, in which 11 different colored boxes were found in the 52 NtEXPs, with each gene containing six to eight motifs. The scale underneath was to measure nucleotides or amino acids length (color figure online)
Gene structures of the NtEXPs were obtained by comparing the predicted CDS with their corresponding genomic DNA sequences using GSDS. Gene structure analysis showed that members within each subfamily/subgroup had similar exon–intron organizations (Fig. 1). In most cases, NtEXPAs contained two introns, except NtEXPA22 and NtEXPA24, which contained three introns and only one intron, respectively. All the NtEXPB and NtEXLA subfamily members contained three and four introns, respectively (Table 1). Like NtEXLA, one subgroup of NtEXLB had four introns, whereas the other two subgroups had three which were consistent with NtEXPB. These results showed that NtEXPB and NtEXLA are more conserved in exon–intron structure than NtEXPA and NtEXLB, and reflected the gene structure divergence of different subfamilies during evolution. NtEXLB subfamily has seemingly a transitional type of gene structure between NtEXPB and NtEXLA.
Each of the tobacco expansin proteins detected carried five to eight other motifs besides the two well-known conserved domains, DPPB_1 and Pollen_allerg_1. In total, eleven motifs were identified in the NtEXPs (Fig. 1), and the schematic diagrams of these motifs were listed in Additional file 6. The types and distribution of motifs for expansins belonging to the same subfamily were conserved, and were more conserved within the subgroups. The numbers and types of protein motifs were the same among members within NtEXLA and NtEXLB subfamilies, respectively, while within NtEXPA and NtEXPB subfamilies the types and distribution of protein motifs varied among subgroups. In most cases, members of NtEXPA subfamily contained seven motifs in the same order, except NtEXPA28 and NtEXPA30 which lacked motif 3, and nine genes which harbored motif 8 in addition to the seven motifs. Each of the two subgroups of NtEXPB contained one or two additional motifs which were different from each other. They had motifs 3, 4 and 6 which were only found in NtEXPA, as well as motifs 9 and 11 which were only found in NtEXLA and NtEXLB. Motifs 3, 4 and 5 were mainly detected in NtEXPA subfamily as well as in several NtEXPB genes, whereas motifs 9, 10 and 11 were only identified in NtEXPB, NtEXLA and NtEXLB subfamilies. Motifs 1, 2 and 7 existed in all the NtEXPs except NtEXLA genes which lacked motif 1.
The above analysis showed that NtEXPA was apparently different from the other three subfamilies in gene structure, motif type and distribution, whereas NtEXPB and NtEXLB were like transitional type either in motif composition or exon–intron organizations, suggesting a progressively closer evolutionary and phylogenetic relationship between NtEXLA, NtEXLB, NtEXPB and NtEXPA.
Gene duplication events are considered to be one of the evolutionary forces in genome evolution, providing materials for generation of new genes and development of new functions (Kong et al. 2007). Segmental and tandem duplications are considered to be the two main causes of gene family expansion in plants (Cannon et al. 2004; Sampedro et al. 2005; Xue et al. 2008), the impacts of which on the expansion of tobacco expansin gene family were focused in our study. The genome of allotetraploid tobacco (sstt, 2n = 48) is believed to have undergone polyploidization from a crossed progeny of Nicotiana tomentosiformis (tt, 2n = 24) and Nicotiana sylvestris (ss, 2n = 24) during evolution (Sierro et al. 2014). The tobacco expansin genes must have experienced duplication along with the whole genome duplication event. Although the physical positions on chromosomes of the tobacco expansin genes are still unclear, some genes were found closely adjacent to each other on the same contigs, such as NtEXPA9/NtEXPA10, NtEXPA12/NtEXPA13 and NtEXLB5/NtEXLB6 (Table 1). These three pairs of genes might be tandem repeats, accounting for 11.5 % of the tobacco expansin gene family. However, because the tobacco chromosomes have not yet been built and its genomic evolution is less understood, we could not identify genes involved in segmental duplication.
GO analysis
The GO figures were drawn using BGI WEGO with the InterproScan output file for each tobacco and Arabidopsis expansin gene based on their GO category annotations (Additional file 7, Additional file 8). The results showed that all the NtEXPs and AtEXPs were annotated with GO:0005576, which in the figures was displayed in “cellular component” and defined as “extracellular region”, suggesting that all the mature expansin proteins are located outside of the plasma membrane, which is the cell wall in plants. All the tobacco and Arabidopsis expansins mainly participated in three kinds of “biological process” as was shown in Additional file 7. However, the processes they participated in varied from different subfamilies. In both tobacco and Arabidopsis, the EXPA subfamily was annotated with biological process GO term “plant-type cell wall organization” (GO:0009664), indicating their involvement in a process, carried out at the cellular level, that results in the assembly and arrangement of constituent parts of the cellulose and pectin-containing cell wall, or in the disassembly of the cellulose and pectin-containing cell wall. This is in line with the suggested roles played by expansins on cell walls. As it was proved that EXPAs are cell wall loosening proteins with their enigmatic effects on cell wall rheology, which can rapidly induce creep and stress relaxation of primary cell walls (Cosgrove 2000a). The biological process GO term “sexual reproduction” (GO:0019953) was also enriched for the expansin genes. In tobacco, all the NtEXPB and NtEXLA subfamily members were annotated with GO:0019953, which suggested that they might participate in the tobacco sexual reproduction process. However, in Arabidopsis, all the members of AtEXPB subfamily, as well as AtEXLA2 and AtEXLB1 might participate in the sexual reproduction process. The group-1 grass pollen allergens of EXPB subfamily are known to facilitate pollen tube invasion of the stigma by dissolving the middle lamella (Sampedro et al. 2015). Here, we conjectured that some EXLA and EXLB subfamily members might also participate in this process in cell wall-dependent manners. The other two AtEXLA subfamily members, AtEXLA1 and AtEXLA3, and all the NtEXLB subfamily members were not annotated with any other GO terms except “cellular component”, suggesting that they might have unknown new functions in consideration of the relatively larger EXLB subfamily size in tobacco.
Cis-Acting elements detection in the promoter regions of tobacco expansin genes
Cis-Acting elements in the upstream region of genes play important roles in regulating gene expression in response to the changing environment as well as during different developmental stages. By employing PlantCARE, we found that in the 1.5 kb upstream regions of the NtEXPs seven kinds of cis-acting elements were apparently abundant, which may provide useful information about the regulatory mechanism of these expansin genes. They were light responsive elements, hormone responsive elements, environmental stress-related elements, development related elements, promoter related elements, site-binding related elements and other elements of unknown function (Additional file 9). A large number of these cis-elements have two or more copies with some being located at adjacent sites in the 1.5 kb upstream region of one tobacco expansin gene, which might enhance their binding effects to their corresponding trans-acting factors (Fig. 2).
The distribution of main cis-acting elements and their putative regulating factors in the 1.5 kb upstream promoter regions of tobacco expansin genes. The same types of cis-acting elements or their putative regulating factors are represented using the same shape but in different colors, e.g. 14 colored ellipses represents 14 types of cis-acting elements responsive to 6 kinds of plant hormones which were displayed in cross stars with six different colors, which in the figure were put together. See legend for details. The scale above was to measure nucleotides length (color figure online)
The most abundant cis-acting elements in the upstream regions of tobacco expansin genes are light-responsive elements which have up to 40 members. Eight of the 40 elements, AE-box, Box 4, Box I, G-box, G-Box, GT1-motif, I-box and Sp1, are more abundant than others and distribute in the promoter regions of over half of the 52 expansin genes. Box 4 and G-box appear to be the most abundant light-responsive elements, locating in the promoter regions of 49 and 41 tobacco expansin genes, respectively. Box 4 element is part of a conserved DNA module involved in light responsiveness in which it needs to form a complex with other light-responsive elements to perform its function. In fact, it is always located closely to other light-responsive elements, such as Box I, GT1-motif and AE-box (Fig. 2). In most cases, G-box and G-Box are adjacent to each other, and sometimes they share the same positions (Fig. 2). Moreover, both G-box and G-Box are less abundant in NtEXPB and NtEXLB subfamilies. About half of the 40 light-responsive elements are rarely appearing cis-acting regulatory elements (appearing in less than five genes), such as 4cl-CMA2b (only one copy in NtEXPA17), H-box (only one copy in NtEXPA10), CG-motif (only one copy in NtEXLA3) and chs-CMA2a (only one copy in NtEXLB2), etc. NtEXPA subfamily has the largest number of light-responsive element types of 37, and 9 elements are NtEXPA-specific, whereas the number of light-responsive element types contained in NtEXPB, NtEXLA and NtEXLB subfamilies is 23, 18 and 22, respectively.
Plant hormone responsive elements, including ABRE, GARE-motif, TCA-element and TGA-element, are another group of cis-acting regulatory elements enriched in the upstream promoter regions of tobacco expansin genes. A total of 14 types of hormone responsive related cis-acting elements were found (Additional file 9). TCA-element is the most abundant cis-acting hormone responsive element in the promoter regions of 31 tobacco expansin genes which is involved in salicylic acid (SA) responsiveness. However, it is not present in NtEXLA subfamily, indicating that the expression of most tobacco expansin genes except NtEXLAs can be regulated by SA. The hormone responsive elements harbored in the promoter regions of each tobacco expansin gene reflect the possibility that it could be regulated by the corresponding plant hormones, such as ABRE and motif IIb involved in abscisic acid (ABA) responsiveness, GARE, P-box and TATC-box in gibberellins (GA) responsiveness, TGA-element, TGA-box and AuxRR-core in auxin responsiveness, TGACG-motif and CGTCA-motif in the jasmonic acid (MeJA) responsiveness, etc. (Fig 2). NtEXPB subfamily does not contain any cis-acting elements responsive to ABA. We also found ERE, GCC-motif and GCC box in the promoter regions of 24 expansin genes, which are ethylene-responsive elements, indicating their involvement in plant maturity and senility. Similarly, two or more copies of one cis-element, and different types of cis-elements responsive to the same plant hormones were found located at adjacent sites, such as TGACG-motif and CGTCA-motif, GARE and P-box, TGA-element and AuxRR-core, indicating that they might be concerted in response to the same plant hormones.
The third important type of cis-acting elements in the upstream regions of tobacco expansin genes are environmental stress-related elements. In total, 12 types of elements were found in charge of about six kinds of external environment stresses, which are HSE in heat stress responsiveness, LTR in low-temperature responsiveness, MBS and C-repeat/DRE in drought responsiveness, ARE and GC-motif in flooding or anaerobic responsiveness, box S, W box and WUN-motif in wounding and pathogen responsiveness, and box E and Box-W1 in fungal elicitor responsiveness (Additional file 9). In addition, Box-W1 was always found beside W-box (Fig. 2). Over half of the 52 tobacco expansin genes appear to contain HSE, MBS, ARE, TC-rich repeats (in defense and stress responsiveness) and W box elements. The low temperature responsive element LTR and fungal elicitor responsive elements Box-W1were not detected in the NtEXLA and NtEXLB subfamilies, respectively. Again, two or more copies of one cis-element and cis-elements responsive to the same stress were found in the promoter region of one expansin gene, which might facilitate their responsiveness to environmental stresses. We conjectured that external environmental stresses could induce the expression of expansin genes containing their responsive cis-acting elements, further contributing to heightening plants’ resistances to environmental stresses.
The forth class of abundant cis-acting element type contains those responsive to development. Skn-1_motif is the most widespread regulatory element required for endosperm expression, which was located in the upstream regions of 44 tobacco expansin genes. 38 genes contain the circadian element, which is involved in circadian control, suggesting that these tobacco expansins might have a distinct diurnal expression pattern. What’s more, as-2-box was found in the promoter regions of 13 tobacco expansin genes which is involved in shoot-specific expression and light responsiveness, AC-I in 4 genes and AC-II in 2 genes which are involved in phloem and xylem vascular development, and HD-Zip 1 and HD-Zip 2 in the same 3 genes which are involved in the differentiation of palisade mesophyll cells and leaf morphology development.
Differential expression profiles of the tobacco expansin genes
TobEA, an atlas of tobacco gene expression from seed to senescence, provides us the opportunity to acquaint the expression profiles of tobacco expansin genes (Edwards et al. 2010). The microarray data of expansin gene expression in tobacco were downloaded from EMBL-EBI ArrayExpress with accession number E-MTAB-176. A heat map showing the hierarchical clustering of expression level of tobacco expansin genes in 19 different tissues from different developmental stages, corresponding to the periods from seed to flower and senescent leaf throughout the tobacco lifecycle, was built to analyze the expression profiles of each tobacco expansin gene (Fig. 3). The BLASTN search results showed that a total of 35 Unigene entries representing 52 expansin genes were identified. With Cluster 3.0 and TreeView softwares, the expression patterns of the tobacco expansin genes were divided into five groups (A, B, C, D and E), as it was shown in Fig. 3.
Heat map showing the hierarchical clustering of expression level of tobacco expansin genes in 19 different tissues across the tobacco lifecycle. Expression data were normalized based on the mean expression value of each gene in all tissues/organs. Different organs/tissues are displayed vertically above each column. Gene names are displayed to the right of each row. Samples were hierarchically clustered based on the average Pearson’s distance. For expression levels, see color legend (color figure online)
The expression patterns of the five groups were different, and that of members within each group were similar. Groups A and B only contain genes from NtEXLA and NtEXLB subfamilies, while genes of the other three groups all belonged to NtEXPA subfamily except NtEXPB3 and NtEXLB5 which was from NtEXPB and NtEXLB subfamilies, respectively. NtEXLB2 and NtEXLB4 from Group A were mainly expressed in senescent leaf, as well as in mature leaf and flower, and they had similar expression patterns, suggesting that they might have similar functions in the same space. The expression levels of the two NtEXLA genes from Group A were low, but were relatively higher in root, flower and mature leaf. All genes from group B had much higher expression levels in root and flower. NtEXLB6 also exhibited high expression values in lower stem and late senescent leaf, while NtEXLB7 showed high expression values in stem, seed, cotyledons and mid/late senescent leaf as well. In contrast, Groups C and D genes were seldom expressed in older organs such as mature and senescent leaf, but they showed higher expression levels in young tissues, including young leaf, bud, shoot apex and cotyledons. Genes such as NtEXPA9 and 19, 7 and 18, 28, 30 and NtEXPB3 which were clustered together showed similar expression patterns, indicating their involvement in the same biological functions. Being different from other NtEXLB members, NtEXLB5 was mainly expressed in bud, root and flower. Besides showing a relatively higher expression level in bud and stem, Group E genes particularly had high expression values in seed, indicating their important roles in seed development or germination. These results showed that the NtEXPs are expressed during the whole lifecycle of tobacco, with some genes having high expression levels at different developmental stages, suggesting their involvement in a certain developmental period directly or indirectly.
Expression analysis of 10 expansin genes in six different tobacco tissues with qRT-PCR
Based on the analysis above, it is clear that the tobacco expansin genes may be expressed differentially in different tissues and/or at different developmental stages. On account of this consideration, qRT-PCR was conducted for ten genes randomly selected from the five groups in tobacco K326 under normal growth conditions to detect their expression pattern in six different tissues, including germinating seeds (GS), root (RT), stem (ST) and leaf (LF) of fast growing period, flower (FL), and mid/late senescent leaf (SL). Primers for the ten selected genes as well as the actin house-keeping gene were listed in Additional file 10.
The expression levels of the ten genes were different in the six tobacco tissues, suggesting their importance in one or several developmental processes (Fig. 4). Our qRT-PCR results showed that NtEXPA4 was dominantly expressed in GS. This concurs with the results from microarray analysis where it was shown that NtEXPA4 from Group E was highly expressed in seed thus suggesting the involvement of this gene in the seed development and/or germination processes. NtEXPA21 and NtEXPA28 were mainly expressed in ST, which was again in line with results from microarray analysis, indicating their involvement in stem elongation and/or diameter enlargement. According to the qRT-PCR analysis, it is likely that NtEXPA28 might participate in the seed germination process and root development as well. In contrast, the results for NtEXPA11 from the two analyses were different. In our qRT-PCR analysis NtEXPA11 was highly expressed in LF while the microarray analysis showed that it had higher expression levels in seed and bud. This might suggest the involvement of NtEXPA11 in plant developmental processes such as seed and leaf development rather than seed germination. In addition, NtEXPB3 was also highly expressed in LF, GS and ST, indicating that this gene might belong to the group of EXPB that we do not know much rather than the group-1 grass pollen allergens which are known to facilitate pollen tube invasion (Cosgrove 2015). NtEXPA20, NtEXPA23, NtEXLA1 and NtEXLB1 were expressed in RT with relatively higher levels, indicating the possibility of their involvement in root growth and development. NtEXPA23 was expressed highly in LF and FL as well. Furthermore, it was noted that NtEXLA1 and NtEXLB2 were primarily expressed in SL in both qRT-PCR and microarray analysis, hence we can speculate that these genes play important roles during tobacco leaf senescence.
qRT-PCR analysis for ten expansin genes in six different tobacco tissues, GS (germinating seeds), RT (root), ST (stem), LF (leaf), FL (flower)and SL (senescent leaf). Data shown represent relative expression quantity (RQ) of an average of three independent experiments + SD. The error bars show the standard error. The one with lowest RQ was set as 1 for each gene in the six tissues
Discussion
Tobacco is one of the world’s most important economic crops, as well as an ideal model plant in scientific research. The tobacco industry collects tobacco leaves for cigarette production after a series of processes, such as flue-curing. The characteristics of flue-cured tobacco leaves, such as sponginess, flammability and harmfulness, are conjectured to be connected with the cell wall. Expansin is a kind of cell wall loosening protein, which might play important role in this process (Cosgrove 2000a). What’s more, expansins have been proved to be involved in a lot of plant developmental stages, as well as in biotic and abiotic stress response (Zhao et al. 2011; Li et al. 2011; Guo et al. 2011; Lü et al. 2013). To date, a large number of expansin gene families has been identified and characterized in both model and crop plants, including Arabidopsis, rice and maize, to mention but a few. However, no such kind of analysis has been done in tobacco, except expression studies for a few NtEXPs genes (Kuluev et al. 2013, 2014). Therefore, it is of great importance to identify the expansin gene family in the tobacco genome and explore the characteristics and functions of each expansin gene. The draft genome of cultivated tobacco was published recently (Sierro et al. 2014), facilitating a genome-wide analysis of this huge gene family in tobacco.
In this study, a total of 52 expansin genes containing full length ORFs and the two conserved domains, DPBB_1 and Pollen_allerg_1, were identified in the tobacco genome through genome-wide analysis. The length, molecular weight, isoelectric point and cleavage site of signal peptide for their corresponding proteins were also predicted with web-based tools and were listed in Table 1. Similar to other plants, the 52 tobacco expansins were divided into four subfamilies, NtEXPA, NtEXPB, NtEXLA and NtEXLB, and they clustered together with genes of the same subfamily from other plants rather than genes of the same species from different subfamilies, suggesting that their ancestors differentiated before the divergence of different plant species. Based on phylogenetic and syntenic analyses, Seader et al. (2016) estimated that there were 12–13 EXPA genes, 2 EXPB genes, 1 EXLA gene, and 2 EXLB genes in the last common ancestor of all angiosperms. In our study the four tobacco expansin subfamilies NtEXPA, NtEXPB, NtEXLA and NtEXLB were accordingly divided into 8, 2, 1 and 2 subgroups with each group of genes deriving from a common ancestor by frequent gene duplication. Attributing to the quality of the draft tobacco genome sequences, which represents about 80 % of the tobacco genome, the missing genes belonged to the other four subgroups in the NtEXPA subfamily might span different genome sequences that have not been scaffolded and therefore cannot be identified or be removed in our sequence analysis (Sierro et al. 2014). The disappearance of descendents from these ancestors could also attribute to gene deaths which were observed in Arabidopsis and rice as well (Sampedro et al. 2005). Expansin gene family from seven species, Arabidopsis, soybean, rice, maize, tobacco, tomato and potato were compared for the size of subfamily, results of which showed an uneven distribution of genes for each subfamily, as well as for different species. The size of EXPB subfamily in monocot plants, rice and maize, is much larger than dicot plants, which is in accordance with the observation that EXPBs are particularly numerous and abundantly expressed in grasses (Cosgrove 2000b). We also found that soybean, tobacco, tomato and potato have a much larger EXLB subfamily size compared to Arabidopsis, rice and maize. Different from other species, the size of EXPB and EXLB subfamilies was almost equal in Solanaceae. However, the EXPA subfamily has the largest size in all species except in maize. Previous studies reported that tandem and segmental duplication events are the main causes for gene family expansion and distribution (Cannon et al. 2004). A relatively much larger EXPB subfamily size in rice and maize, and a relatively much larger EXLB subfamily size in soybean and Solanaceae might be a result of higher degree of expansion and retention, whereas subfamilies of relatively smaller size, such as EXPB in soybean, might be due to large-scale gene loss after duplication events (Zhu et al. 2014). We can also conjecture that plants retaining certain kind of genes with larger subfamily size during the long evolutionary time was to increase its adaption to certain functions and environments. Therefore, the additional NtEXLB genes might have special functions in tobacco, which need to be validated.
Gene expression pattern can provide important clues for gene function, which are believed to be associated with the divergence of the promoter regions (Xue et al. 2008; Li et al. 2014). Cis-Acting regulatory elements contained in the promoter region of genes play key roles in conferring developmental and/or environmental regulation of gene expression. Using PlantCARE, a plant cis-acting regulatory element database, we could identify in the 1.5 kb DNA sequence upstream of the translation initiation codon the potential cis-acting elements for each tobacco expansin gene. Four types of cis-acting elements, light responsive elements, hormone responsive elements, environmental stress-related elements, and development related elements, were found to be apparently abundant for the NtEXPs. In plants, the expression of expansins are regulated by both internal and external factors, resulting in their involvement in a variety of developmental processes, such as seed germination (Yan et al. 2014), fruit growth and/or ripening (Rose et al. 1997; Brummell et al. 1999; Ishimaru et al. 2007), as well as in nutrient uptake and efficiency (Zhou et al. 2014), and stress tolerance (Li et al. 2011; Zhao et al. 2011; Lü et al. 2013), which are conjured to be regulated by the corresponding cis-acting elements they contain. Plant hormones can regulate the expression of expansin genes and the regulation of expansin activity by plant hormones is well documented (Zhao et al. 2012; Li et al. 2014; Lu et al. 2016). For example, cytokinin and auxin act synergistically to induce the accumulation and proteolytic processing of Cim1/GmEXPB1, a cytokinin-inducible β-expansin from soybean (Downes et al. 2001; Li et al. 2014); the Arabidopsis LBD18 up-regulates a subset of expansin genes, including EXPA4, EXPA14 and EXPA17, to enhance cell separation thus promoting lateral root emergence, the effects of which were promoted by exogenous application of auxin (Lee and Kim 2013); the rice OsMPS, whose expression is induced by ABA and cytokinin, but is repressed by auxin, GA and brassinolide (BR), is a direct negative upstream regulator of OsEXPA4/8 and OsEXPB2/3/6, illustrating an indirect regulation of these plant hormones to EXPs (Schmidt et al. 2013). Two tobacco expansin genes, NtEXPA1 and NtEXPA4, were induced by auxin in young leaves located near the terminal bud, whereas in the lower leaves BR could inhibit NtEXPA1 but up-regulate NtEXPA4 expression (Kuluev et al. 2014). The regulation of plant architecture by expansins to adapt to certain external environmental changes, such as drought, heat, low phosphorus (P), is also well studied (Cosgrove 2015; Lu et al. 2016). Some examples include the wheat (Triticum aestivum) TaEXPB23, Poa pratens PpEXPA1, soybean GmEXPB2 and rose (Rosa hybrida) RhEXPA4, the transgenic plants with which gained tolerance to drought (Li et al. 2011, 2013), tolerance to heat (Xu et al. 2014), improved P efficiency (Guo et al. 2011; Zhou et al. 2014) and tolerance to drought and salt (Lü et al. 2013; Yan et al. 2014), respectively. In addition, we found that for many cis-elements two or more copies located at adjacent sites, which might enhance the effects of their binding to trans-acting factors such as transcription factors. What’s more, cis-elements in response to the same factors such as plant hormones and environmental stresses also tended to be neighbors, which might promote the efficiency of gene transcription regulation and might be beneficial for plants to adapt to environmental changes. However, we also found that some cis-elements in response to different factors appeared closely to each other, such as Box I (light responsiveness) and ERE (ethylene responsiveness), G-box (light responsiveness) and ABRE (ABA responsiveness), indicating that the transcription regulation of these genes might be both light- and plant hormone-dependent. For example, ABA plays a major role in plant responses to abiotic environmental stresses, such as drought and salt (Leung and Giraudat 1998; Lü et al. 2013). ABRE and G-box are cis-elements that have been proved to be involved in the activation of gene expression by ABA (Leung and Giraudat 1998), and G-box is also involved in response to blue light (Sun et al. 2015). The close positions of G-box and ABRE in the promoter regions of some tobacco expansin genes suggested that these expansin genes might be components of blue light- and abiotic stress-responsive pathways. Overall, promoter analysis demonstrated the presence of a diversity of cis-acting regulatory elements in the upstream regions of tobacco expansin genes. This finding indicates that the 52 NtEXPs might be regulated by different internal and/or external factors separately or synergistically, and further supports the various functional roles of expansins in a wide range of developmental processes related to cell wall modification.
TobEA, an atlas of tobacco gene expression from seed to senescence, allows the characterization and comparison of the expansin gene expression patterns in tobacco (Edwards et al. 2010). In this study, the tobacco expansin genes were identified expressed differently in the 19 tobacco tissues, indicating their different roles in tobacco development. However, about twenty expansin genes were not identified in the custom microarray design, which might attribute to the specificity of probes (about 81 % of the mapped SGN unigenes represented by a unique probe set, and 99 % by four probe sets or less. Edwards et al. 2010). We have revealed that the expression levels of the tobacco expansin genes were closely associated with different tissues or organs as well as different developmental stages, and that the expression patterns differed among most expansin gene members in each group, which might attribute to the functional differentiation after gene duplication events. Groups A and B only contain genes from NtEXLA and NtEXLB subfamilies, with Group A genes being mainly expressed in senescent leaf and flower, while Group B genes in root and flower, as well as in senescent leaf. The cis-acting elements W-box and G-box were reported to play important roles in early senescence of rice flag leaf (Liu et al. 2016). In our study, in the promoter regions of both NtEXLA1 and NtEXLA2 were laden with these two cis-elements (Fig. 2). However, the NtEXLB subfamily members lacked the W-box and G-box, especially for NtEXLB2 and NtEXLB4, indicating that the involvement of these genes in tobacco leaf senescence was not W-box and/or G-box dependent. In contrast, group C and D genes, which contained mainly NtEXPA subfamily members, seldom expressed in older organs, but they showed higher expression in young tissues, such as young leaf, bud, shoot apex and cotyledons. Group E genes particularly had high expression values in seed, indicating their involvement in seed development or germination. There were also genes clustered together in the heat map that had similar expression patterns, such as NtEXLB2/NtEXLB4 and NtEXPA28/NtEXPA30, indicating their similarity in gene function in the same tissues. To verify the expression profiles of the tobacco expansin genes, qRT-PCR analysis was performed in six different tissues for ten expansin genes uniformly distributed in the five groups. The results showed that the expression pattern for each gene detected by qRT-PCR was roughly in consistency with the microarray analysis, which further confirm their preferential expression. However, there existed inconsistency, such as the expression patterns of NtEXPA11 and NtEXPA28. In the microarray analysis, NtEXPA11 had higher expression levels in seed and bud. Whereas NtEXPA28 was mainly expressed in upper stem, bud, young leaf and flower, and seldom expressed in seed. However, in our qRT-PCR analysis, NtEXPA11 and NtEXPA28 were highly expressed in leaf, flower and stem and in stem and germinating seeds, respectively. We conjectured that the “seed” in the custom microarray was different from “geminating seeds” in our study. Therefore, NtEXPA11 might participate in other seed developmental processes but not in seed germination. Similarly, NtEXPA28 might be involved in the seed germination process rather than seed development. To sum up, all the findings in this study indicate that expansin genes are expressed during the whole life of tobacco, most of which exhibited preferential expression in different tissues and/or at different developmental stages. However, the factual temporal and spatial expression patterns and the functions of the tobacco expansin genes need further experimental verifications.
In conclusion, in this study we presented a genome-wide analysis of expansin gene family in the tobacco genome and identified 36 NtEXPAs, 6 NtEXPBs, 3 NtEXLAs and 7 NtEXLBs, respectively. The four subfamilies were further grouped into 8, 2, 1 and 2 clades which were derived from 13 common ancestors by gene duplications. The NtEXPs exhibited specific characteristics in terms of exon–intron organization, amino acid sequences, or protein motif composition within subfamilies or subgroups. Whole-genome duplication and tandem duplication events might have contributed to the expansion of the NtEXPs. Each expansin gene contained a number of cis-acting elements in its 1.5 kb upstream region before the start codon for transcription, suggesting that its expression was regulated by various internal or environmental factors, such as light, plant hormones and stresses, thus participating in tobacco development and its resistance to stresses. The expression patterns of the NtEXPs were also studied using a custom microarray design and qRT-PCR, which revealed that most NtEXPAs had higher expression levels in young organs, while some NtEXLAs and NtEXLBs were preferentially expressed in mature or senescent tissues. Taken together, our expansin gene family data analysis presents a comprehensive analysis which could benefit researchers in investigating the important roles that expansins play in tobacco development. The results presented here may potentially advance our understanding of the biological roles of expansins in many agronomically important traits of tobacco, such as leaf development and senescence and other physiological processes.
References
Boron AK, Van Loock B, Suslov D, Markakis MN, Verbelen JP, Vissenberg K (2015) Over-expression of AtEXLA2 alters etiolated arabidopsis hypocotyl growth. Ann Bot 115:67–80
Brummell DA, Harpster MH, Dunsmuir P (1999) Differential expression of expansin gene family members during growth and ripening of tomato fruit. Plant Mol Biol 39:161–169
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Cheng Y, Li X, Jiang H, Ma W, Miao W, Yamada T, Zhang M (2012) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in maize. FEBS J 279:2431–2443
Cho HT, Kende H (1997) Expansins in deepwater rice internodes. Plant Physiol 113:1137–1143
Civello PM, Powell AL, Sabehat A, Bennett AB (1999) An expansin gene expressed in ripening strawberry fruit. Plant Physiol 121:1273–1280
Cosgrove DJ (2000a) Loosening of plant cell walls by expansins. Nature 407:321–326
Cosgrove DJ (2000b) New genes and new biological roles for expansins. Curr Opin Plant Biol 3:73–78
Cosgrove DJ (2015) Plant expansins: diversity and interactions with plant cell walls. Curr Opin Plant Biol 25:162–172
Downes BP, Steinbaker CR, Crowell DN (2001) Expression and processing of a hormonally regulated beta-expansin from soybean. Plant Physiol 126:244–252
Edwards KD, Bombarely A, Story GW, Allen F, Mueller LA, Coates SA, Jones L (2010) TobEA: an atlas of tobacco gene expression from seed to senescence. BMC Genom 11:142
Fleming AJ, McQueen-Mason S, Mandel T, Kuhlemeier C (1997) Induction of leaf primordia by the cell-wall protein expansion. Science 276:1415–1418
Georgelis N, Yennawar NH, Cosgrove DJ (2012) Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin. Proc Natl Acad Sci USA 109:14830–14835
Georgelis N, Nikolaidis N, Cosgrove DJ (2015) Bacterial expansins and related proteins from the world of microbes. Appl Microbiol Biotechnol 99:3807–3823
Guo W, Zhao J, Li X, Qin L, Yan X, Liao H (2011) A soybean β-xpansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J 66:541–552
Ishimaru M, Smith DL, Gross KC, Kobayashi S (2007) Expression of three expansin genes during development and maturation of Kyoho grape berries. J Plant Physiol 164:1675–1682
Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, Voesenek LA (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55:311–314
Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, dePamphilis CW (2007) Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J 50:873–885
Kuluev BR, Knyazev AB, YaP Lebede, Chemeris AV (2012) Morphological and physiological characteristics of transgenic tobacco plants expressing expansin Genes: AtEXP10 from Arabidopsis and PnEXPA1 from poplar. Russ J Plant Physiol 59:97–104
Kuluev BR, Safiullina MG, Kniazev AV, Chemeris AV (2013) Effect of ectopic expression of NtEXPA5 gene on cell size and growth of organs of transgenic tobacco plants. Russ J Plant Physiol 44:28–34
Kuluev BR, Knyazev AV, Nikonorov luM, Chemeris AV (2014) Role of the expansin genes NtEXPA1 and NtEXPA4 in the regulation of cell extension during tobacco leaf growth. Genetika 50:560–569
Lee HW, Kim J (2013) EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol 54:1600–1611
Lee Y, Choi DS, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4:527–532
Leung J, Giraudat J (1998) Abscisic acid signal transduction. Annu Rev Plant Physiol Plant Mol Biol 49:199–222
Li LC, Cosgrove DJ (2001) Grass group I pollen allergens (beta-expansins) lack proteinase activity and do not cause wall loosening via proteolysis. Eur J Biochem 268:4217–4226
Li F, Xing SC, Guo QF, Zhao MR, Zhang J, Gao Q, Wang GP, Wang W (2011) Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J Plant Physiol 168:960–966
Li F, Han Y, Feng Y, Xing S, Zhao M, Chen Y, Wang W (2013) Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J Biotechnol 163:281–291
Li X, Zhao J, Walk TC, Liao H (2014) Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl Microbiol Biotechnol 98:2805–2817
Lin YX, Jiang HY, Chu ZX, Tang XL, Zhu SW, Cheng BJ (2011) Genome-wide identification, classification and analysis of heat shock transcription factor family in maize. BMC Genom 12:76
Liu L, Xu W, Hu X, Liu H, Lin Y (2016) W-box and G-box elements play important roles in early senescence of rice flag leaf. Sci Rep 6:20881
Lu YG, Liu LF, Wang X, Han ZH, Ouyang B, Zhang JH, Li HX (2016) Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol Genet Genomics 291:597–608
Lü PT, Kang M, Jiang XQ, Dai FW, Gao JP, Zhang CQ (2013) RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237:1547–1559
Marowa P, Ding AM, Kong YZ (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:949–965
McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4:1425–1433
Rose JK, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA 9:5955–5960
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:242
Sampedro J, Lee Y, Carey RE, dePamphilis C, Cosgrove DJ (2005) Use of genomic history to improve phylogeny and understanding of births and deaths in a gene family. Plant J 44:409–419
Sampedro J, Guttman M, Li LC, Cosgrove DJ (2015) Evolutionary divergence of β-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits. Plant J 81:108–120
Schmidt R, Schippers JH, Mieulet D, Obata T, Fernie AR, Guiderdoni E, Mueller-Roeber B (2013) MULTIPASS, a rice R2R3-type MYB transcription factor, regulates adaptive growth by integrating multiple hormonal pathways. Plant J 76:258–273
Seader VH, Thornsberry JM, Carey RE (2016) Utility of the Amborella trichopoda expansin superfamily in elucidating the history of angiosperm expansins. J Plant Res 129:199–207
Sierro N, Battey JN, Ouadi S, Bakaher N, Bovet L, Willig A, Goepfert S, Peitsch MC, Ivanov NV (2014) The tobacco genome sequence and its comparison with those of tomato and potato. Nat Commun 5:3833
Sun Y, Xu W, Jia Y, Wang M, Xia G (2015) The wheat TaGBF1 gene is involved in the blue-light response and salt tolerance. Plant J 84:1219–1230
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Xu Q, Xu X, Shi Y, Xu J, Huang B (2014) Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress. PLoS One 9:e100792
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y (2008) Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genom 9:550
Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y (2014) AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS One 9:e85208
Yennawar NH, Li LC, Dudzinski DM, Tabuchi A, Cosgrove DJ (2006) Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize. Proc Natl Acad Sci USA 103:14664–14671
Yu ZM, Kang B, He XW, Lv SL, Bai YH, Ding WN, Chen M, Cho HT, Wu P (2011) Root hair-specific expansins modulate root hair elongation in rice. Plant J 66:725–734
Zhang SZ, Xu RR, Gao Z, Chen CT, Jiang ZH, Shu HR (2014a) A genome-wide analysis of the expansin genes in Malus × Domestica. Mol Genet Genomics 289:225–236
Zhang W, Yan HW, Chen WJ, Liu JY, Jiang CP, Jiang HY, Zhu SW (2014b) Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics 289:1061–1074
Zhao MR, Li F, Fang Y, Gao Q, Wang W (2011) Expansin-regulated cell elongation is involved in the drought tolerance in wheat. Protoplasma 248:313–323
Zhao MR, Han YY, Feng YN, Li F, Wang W (2012) Expansins are involved in cell growth mediated by abscisic acid and indole-3-acetic acid under drought stress in wheat. Plant Cell Rep 31:671–685
Zhou J, Xie JN, Liao H, Wang XR (2014) Overexpression of β-expansin gene GmEXPB2 improves phosphorus efficiency in soybean. Physiol Plant 150:194–204
Zhu Y, Wu NN, Song WL, Yin GJ, Qin YJ, YAN= YM, Hu YK (2014) Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14:93
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the National Key Technology R&D Program (No. 2015BAD15B03-05), the National Natural Science Foundation of China (Grant No. 31470291) and the Youth Elite Program (to YK).
Conflict of interest
All authors declare that she/he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
A. Ding and P. Marowa contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ding, A., Marowa, P. & Kong, Y. Genome-wide identification of the expansin gene family in tobacco (Nicotiana tabacum). Mol Genet Genomics 291, 1891–1907 (2016). https://doi.org/10.1007/s00438-016-1226-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1226-8