Abstract
Plant expansins are capable of inducing pH-dependent cell wall extension and stress relaxation. They may be useful as targets for crop improvement to enhance fruit development and stress resistance. Tomato is a major agricultural crop and a model plant for studying fruit development. Because only some tomato expansins have been studied, a genome-wide analysis of the tomato expansin family is necessary. In this study, we identified 25 SlEXPAs, eight SlEXPBs, one SlEXLA, four SlEXLBs, and five short homologs in the tomato genome. 25 of these genes were identified as being expressed. Bioinformatic analysis showed that although tomato expansins share similarities with those from other plants, they also exhibit specific features regarding genetic structure and amino acid sequences, which indicates a unique evolutionary process. Segmental and tandem duplication events have played important roles in expanding the tomato expansin family. Additionally, the 3-exon/2-intron structure may form the basic organization of expansin genes. We identified new expansin genes preferentially expressed in fruits (SlEXPA8, SlEXPB8, and SlEXLB1), roots (SlEXPA9, SlEXLB2, and SlEXLB4), and floral organs. Among the analyzed genes those that were inducible by hormone or stress treatments, including SlEXPA3, SlEXPA7, SlEXPB1–B2, SlEXPB8, SlEXLB1–LB2, and SlEXLB4. Our findings may further clarify the biological activities of tomato expansins, especially those related to fruit development and stress resistance, and contribute to the genetic modification of tomato plants to improve crop quality and yield.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Expansins are proteins that can induce pH-dependent cell wall extension and stress relaxation and are commonly found in plants (Cosgrove et al. 2002). These proteins have diverse biological roles, but most are related to cell wall modification, including leaf initiation and development (Goh et al. 2012), root hair growth (Cho and Cosgrove 2002), fiber development (Harmer et al. 2002), stem and pollen tube elongation (Cho and Kende 1997; Tabuchi et al. 2011), fruit development and ripening (Nardi et al. 2015; Brummell et al. 1999), seed germination (Yan et al. 2014; Chen et al. 2001), mycorrhiza and root nodule formation (Balestrini et al. 2005; Flemetakis et al. 2004), abscission (Tsuchiya et al. 2015; Belfield et al. 2005), and biotic and abiotic stress resistance (Abuqamar et al. 2013; Lü et al. 2013). Because of their important functions in plant development and stress resistance, researchers have considered expansins as potential targets for crop improvement (Choi et al. 2006).
Expansins are encoded by a large multigene family. A typical expansin contains 250–275 amino acids and is made up of two domains (domain 1 and domain 2) preceded by a signal peptide consisting of 20–30 amino acids (Li et al. 2003). Domain 1 shares some key structural features with the catalytic domain of glycoside hydrolase family 45 proteins, including a series of conserved cysteines and a histidine-phenylalanine-aspartate (HFD) motif. Domain 2 is homologous to group-2 grass pollen allergens, which have been hypothesized to form a binding strip for cell wall polysaccharides (Cosgrove et al. 1997). Plant expansins can be phylogenetically divided into four groups, designated as EXPANSIN A (EXPA), EXPANSIN B (EXPB), EXPANSIN-LIKE A (EXLA), and EXPANSIN-LIKE B (EXLB) (Kende et al. 2004). Expansin-like proteins (EXLA and EXLB) also possess domains 1 and 2 and a signal peptide, but the amino acid sequences are different from those of EXPAs and EXPBs (Choi et al. 2006). Some EXPAs and EXPBs are capable of inducing rapid cell wall extension or stress relaxation (Cosgrove et al. 1997), whereas no biological or biochemical function has been established for any EXLAs or EXLBs (Sampedro and Cosgrove 2005).
Proteins with homology to only one of the expansin domains also exist in plants, including grass secreted proteins, whose functions are unclear, and citrus tree p12 proteins, which are speculated to function as peptide hormones to maintain water and solute balance (Ludidi et al. 2002). Expansin orthologs were also identified in non-plant organisms such as bacteria and fungi. These proteins may function in plant cell wall digestion to facilitate pathogen invasion (Cosgrove et al. 2002).
Tomato (Solanum lycopersicum) is a major crop plant worldwide and biotic and abiotic stresses are seriously threatening its production. Expansins may play significant roles in different stress resistance processes. Tomato is also a model system for the study of fruit development. Expansins may cooperatively function in every fruit development stage. Studies on tomato expansins will benefit tomato production and provide valuable information regarding how fruits develop in other crops. The tomato expansin family is composed of many members. Although several related studies have been reported (Keller and Cosgrove 1995; Rose et al. 1997), to the best of our knowledge, there have been no systematic studies conducted on the whole family despite the fact the genome was sequenced over 2 years.
In this study, we identified 38 expansin family members and five short homologs in tomato. We systematically analyzed the phylogenetic relationships, gene structure, and protein architecture of the identified members. The transcription patterns of some expressed expansin genes in different organs, and their expression patterns following different hormone and abiotic stress treatments were investigated. We identified new expansin genes with organ-specific or preferential expression patterns, as well as those with stress- and/or hormone-responsive properties. Our findings may advance our understanding of the roles of different expansins in tomato growth, development, and responses to environmental stresses. This study may provide important information for future investigations of the biological functions of expansins.
Materials and methods
Identification of the tomato expansin gene family
To identify all tomato expansin genes, eight previously reported tomato expansin gene cDNA sequences were extracted from a National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/nucleotide/) and subjected to BLASTN searches using the SOL Genomics Network (SGN; http://solgenomics.net/) and KaFTom (http://www.pgb.kazusa.or.jp/kaftom/) databases (Brummell et al. 1999; Chen et al. 2001). We also searched the databases for homologs using ‘expansin’ as a keyword. Redundant sequences were removed after a similarity comparison. The open reading frames were identified by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Expansin protein sequences from other species were retrieved from the databases of the National Center for Biotechnology Information, the Arabidopsis Information Resource (www.arabidopsis.org/), the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), and the Expansin Central website (http://www.personal.psu.edu/fsl/ExpCentral/index.html).
Sequence analysis and phylogenetic tree construction
Deduced protein sequences were aligned using Clustal X 1.83 with the following parameters: gap opening, 4.0; gap extension, 0.2; delay divergent sequences, 30 %; DNA transition weight, 0.5; and use negative matrix, off (Larkin et al. 2007). Putative motifs were analyzed using the MEME program 4.10.2 with the following parameters: number of repetitions, any; maximum number of motifs, 20; and Motif width, 20–100 (http://meme-suite.org/tools/meme, Bailey et al. 2009). A phylogenetic tree was constructed using the maximum likelihood method and MEGA 6 software with default parameters (Tamura et al. 2013). The exon–intron structures were generated using GSDS software 2.0 (http://gsds.cbi.pku.edu.cn/, Hu et al. 2015).
Duplication analysis of tomato expansin genes
We completed duplication analysis of the tomato expansin gene family using MCScanX software (http://chibba.pgml.uga.edu/mcscan2/, Wang et al. 2012). All tomato expansin and expansin-like genes were compared with themselves and others using the BLASTp program, with an E value <1E−5. The BLAST search outputs were imported into MCScanX software using the default criterion. Diagrams were generated with Circos software, version 0.63 (http://circos.ca/, Krzywinski et al. 2009).
Plant growth during hormone and stress treatments
Tomato seeds (S. lycopersicum cv. Ailsa Craig) were germinated and grown in a greenhouse under a photosynthetic photon flux density of approximately 120 μmol/m2/s with a 12-h light/dark photoperiod. The temperature and relative humidity were maintained at 25 ± 2 °C and 60 %, respectively. Five-week-old seedlings were used to investigate the effects of hormones and abiotic stresses on expansin genes. Seedlings were sprayed with solutions containing 100 μM 6-benzylaminopurine (6-BA), indole-3-acetic acid (IAA), ethephon, abscisic acid (ABA), and gibberellic acid (GA). Control plants were treated with water. Seedling shoots were collected 0, 1.5, 3.0, 6.0, 12, and 24 h after spraying. Heat and cold stresses were applied by exposing the seedlings to 42 and 4 °C, respectively. Shoots were harvested after 1.5, 3.0, 6.0, 12, and 24 h. Drought stress was induced by placing the seedlings on a clean bench after removal from the soil. Shoots were collected after 0.5, 1.0, 1.5, 2.0, and 2.5 h. Untreated plants served as controls. To investigate organ-specific expression profiles, sixteen kinds of organ samples were collected from adult plants. All samples were quickly frozen in liquid nitrogen and stored at −70 °C. For all experiments, at least two biological replicates were used for further analysis.
Real-time quantitative reverse transcription (qRT)-PCR analysis
Total RNA was extracted using TRIzol reagent. First-strand cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen) with 3 μg RNA, the product was finally diluted to a volume of 90 µl. Real-time qRT-PCR was performed using the LightCycler 480 system (Roche Diagnostics). Each reaction contained 5 µl SYBR Premix Ex Taq (Takara), 0.5 µl cDNA, and 0.4 µl 10 µM gene-specific primers in a final volume of 10 µl. The PCR cycle was as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 25 s. Data were collected during the extension step. Three technical replicates were performed for each sample. The tomato actin gene (Solyc04g011500) was used as an internal control (Supplementary Table 1). Data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen 2001).
Results
Identification of tomato expansin genes
By removing redundant sequences from databases, 38 expansin genes and five short homologs were identified in the tomato genome. Among these, 25 were identified based on expressed sequence tag data (Table 1). Some of the genes were identified and named previously while others have never been reported. According to the nomenclature rules for expansin superfamily genes and proteins suggested by Kende et al. (2004), genes expressing proteins with only one expansin domain were not classified as expansins. For convenience, we first constructed a phylogenetic tree using the deduced sequences of proteins with both expansin domains (Supplementary Fig. 1). Phylogenetic analysis showed that these proteins can be divided into four subfamilies, EXPA, EXPB, EXLA, and EXLB. Newly identified expansins and expansin-like genes were named according to their locations in the phylogenetic tree and their relationships with Arabidopsis thaliana homologs. Three members that were homologous to only domain 1 were designated as SlPNPL1–L3. Two members that were homologous to only domain 2 were designated as SlG2A1 and SlG2A2 (Table 1).
The expansin genes were unevenly distributed on 11 chromosomes. Some of the genes may have evolved from gene duplications (Supplementary Fig. 2). On chromosome 3, six expansin genes (SlEXPA7 and SlEXPA19–A23) were clustered in a small 25.4 kb region. These genes were also classified in a small group in the phylogenetic tree, suggesting their close evolutionary relationship. Of these genes, only SlEXPA7 was identified as being expressed (Supplementary Fig. 1).
Gene structure and phylogenetic analysis
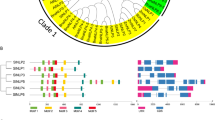
Similar to those of other plants (Lee et al. 2001), tomato expansin gene exon–intron organizations were diverse (Fig. 1). However, members of a subfamily showed a similar structure. Among the 25 SlEXPAs, 15 had a 3-exon/2-intron structure, nine had a 2-exon/1-intron structure, and only one member had a 4-exon/3-intron structure. As shown in Fig. 1, other structures likely originated from an intron loss or intron gain in a 3-exon/2-intron structure. The structures of SlEXPA7, SlEXPA19, and SlEXPA20–A22 may have resulted from the loss of the second intron (generally called intron B), while the structures of SlEXPA1, SlEXPA3, and SlEXPA24 may be due to the loss of the first intron (intron A). Conversely, the 4-exon/3-intron structure of SlEXPA13 may have been the result of an intron-gain (intron E) in the first exon. All SlEXPB family members consisted of the 4-exon/3-intron structure, which is likely because of an intron-gain in the second or third exon of a 3-exon/2-intron structure (intron C and F).
Exon–intron organization of tomato expansin family genes. Gene structures were generated with GSDS 2.0 (http://gsds.cbi.pku.edu.cn/)
To explore the phylogenetic relationship among plant expansins, we constructed a phylogenetic tree using all of the reported expansin and expansin-like protein sequences of tomato, A. thaliana, and rice (Fig. 2). These expansins were grouped into four major classes, namely EXPA, EXPB, EXLA and EXLB. In all species, EXPA was the largest subfamily. As previously reported, among the EXPB members, there are more monocots (e.g., rice) than eudicots (e.g., A. thaliana and tomato). The EXLA and EXLB groups are small subfamilies. There are three and four EXLA members in rice and A. thaliana, respectively, but only one in tomato. Four EXLB members were identified in tomato, while only one was found in rice and A. thaliana.
Phylogenetic analysis of the expansin family in tomato, A. thaliana, and rice. The phylogenetic tree was generated using the maximum likelihood method in MEGA 6. Rice and A. thaliana expansin sequences were retrieved from the databases of Arabidopsis Information Resource (www.arabidopsis.org/), the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), and the Expansin Central website (http://www.personal.psu.edu/fsl/ExpCentral/index.html)
All expansin and expansin-like genes are believed to have evolved from a common ancestor. This conclusion was also supported by our phylogenetic analysis, as some members, which come from different plant species, like SlEXPA24, AtEXPA13, and OsEXPA10, can be clustered in one small subfamily (Fig. 2). Differences in the expansins between monocots and eudicots, as well as those between two dicotyledonous species can also be distinguished in the phylogenetic tree, suggesting that the expansin gene family has evolved separately in each plant species.
Sequence alignment and putative motif identification
To obtain detailed information regarding the tomato expansin superfamily, we completed multiple sequence alignments of the deduced protein sequences (Fig. 3). Most of the expansins and expansin-like proteins consisted of three parts, a 20–30 amino acid signal peptide, domain 1, and domain 2. The amino acid sequence of domain 1 was more conserved than that of domain 2, especially among EXPB members. As reported by Li et al. (2002), the EXPA and EXPB subfamilies can be easily distinguished based on two types of insertions. The α-insertions existed only in the EXPA subfamily and consisted of about 14 residues, with most insertions containing four highly conserved residues at the 3′ end, WCNP (Trp-Cys-Asn-Pro). The β-insertions were present among EXPB, EXLA, and EXLB members. In EXPB members, the β-insertion was about seven residues long and contained a conserved G residue. The β-insertions among the EXLA and EXLB members consisted of 7–9 residues but were relatively diverse with no obvious conserved residues. The HFD motif was conserved among all EXPB and the majority of EXPA members, while members with either absent (i.e., SlEXPA23) or incompletely conserved (i.e., SlEXPA3, SlEXPA24, and all SlEXLA and SlEXLB members) HFD motif were also found. A series of conserved Cys (N-terminal) and Trp (C-terminal) residues reported in other plant expansins were also present in tomato expansins. The Cys residues were strictly conserved in all members, while the Trp residues were not as conserved, especially in SlEXLAs and SlEXLBs. These expansin properties were also reflected in the MEME analysis (Supplementary Figs. 3, 4).
Multiple sequence alignment of tomato expansins. Sequence alignments of tomato expansins and three A. thaliana expansins were prepared using the Clustal X software. Colored shading indicates identical and conversed amino acid residues. Conserved features such as signal peptides, expansin domains, HFD motif, and conserved Cys and Trp residues are also indicated
Organ-preferential expression of tomato expansin genes
For a more comprehensive understanding of tomato expansin gene expression, we analyzed the transcription profiles of 23 members, including 15 SlEXPAs, four SlEXPBs, one SlEXLA, and three SlEXLBs using real-time qRT-PCR. All selected members were identified as expressed genes in SGN database except for SlEXPA24. The results showed that the tested genes were expressed differently in tomato parts (Fig. 4).
Expression profiles of tomato expansin genes in various organs. The y-axis represents the expression levels of expansin or expansin-like genes relative to that of SlActin. The x-axis represents different organs. Rt root, YS young stem, MS mature stem, FA flower axis, YL young leaf, ML mature leaf, Al alabastrum, Se sepal, Pe petal, St stamen, SO stigma and ovary, IM pericarp of immature fruit, MG pericarp of mature green fruit, BR pericarp of breaker fruit, YR pericarp of yellow ripe fruit, RR pericarp of red ripe fruit. Error bars represent standard deviations for three replicates
First, we observed differences in expression levels. Three members (SlEXPA1, SlEXPA11, and SlEXPA12) were highly expressed in several organs with expression levels more than 4-fold higher than that of the reference gene. Six members (SlEXPA6, SlEXPA14, and four SlEXPB genes) were expressed at a very low level (<0.1 relative to the expression of the reference gene) in all examined organs. The other tested genes exhibited a relative expression level of 0.1–1.5 times the expression of the reference gene in at least one organ.
Second, there were differences in expression patterns. Most genes showed an organ-preferential expression pattern. Some genes were expressed more in the roots, including SlEXPA3, SlEXPA9, SlEXPA18, SlEXPB1, SlEXPB2, SlEXPB8, SlEXLB2, and SlEXLB4. Among these genes, SlEXPB2 and SlEXLB4 exhibited a root-specific expression pattern. Genes that were expressed more in fruits included SlEXPA1, SlEXPA3, SlEXPA5, SlEXPA6, SlEXPB8, and SlEXLB1. Among these, SlEXPA1 was expressed specifically in yellow and red ripe fruit, SlEXPB8 was expressed preferentially in yellow ripe fruit, SlEXLB1 was expressed preferentially during the breaker stage, and SlEXPA3, SlEXPA5, and SlEXPA6 were preferentially expressed in immature or mature green fruit.
Some genes were expressed more in floral organs, including SlEXPA2 (alabastrum, sepal, and petal), SlEXPA4 (petal), SlEXPA10 (alabastrum), SlEXPA8 (flower axis and alabastrum), SlEXPA11 (petal), SlEXPA14 (sepal), SlEXPA18 (alabastrum), SlEXPA24 (sepal), SlEXPB3 (alabastrum), SlEXLB1 (sepal), SlEXPA4 (stigma or ovary), and SlEXLB2 (stigma or ovary). Additionally, SlEXPA7 was preferentially expressed in mature leaves. We also observed that SlEXPA12 was constitutively expressed in every tested organ except for the stamen in which it was expressed at very low levels.
Differential expression of expansin genes after phytohormone treatment
The expression of some expansin genes can be regulated by phytohormones. We investigated the expression of selected expansin genes in shoots in response to the application of exogenous hormones (Fig. 5). The expression of most of the analyzed genes was up-regulated. Seven genes (SlEXPA2, SlEXPA3, SlEXPA5, SlEXPA8, SlEXPA11, SlEXPB2, and SlEXPB8) were up-regulated by 6-BA, ten (SlEXPA1, SlEXPA3, SlEXPA5–A9, SlEXPA12, SlEXPB8, and SlEXLB2) were up-regulated by IAA, four (SlEXPA2, SlEXPA10, SlEXPA11, and SlEXLB2) were up-regulated by GA, eight (SlEXPA4–A6, SlEXPA11, SlEXPA18, SlEXPB8, SlEXLB2, and SlEXLB4) were up-regulated by ethephon, and eight (SlEXPA1, SlEXPA3, SlEXPA5–A7, SlEXPA9, SlEXLB1, and SlEXLB2) were up-regulated by ABA. The expressions of some expansin genes were up-regulated by multiple hormones. For example, SlEXPA5 was up-regulated by all hormones except for GA, while SlEXPA3 was up-regulated by 6-BA, IAA, and ABA. However, SlEXPB2 and SlEXLB4 were up-regulated by only one hormone. Some members were markedly up-regulated by one or several hormones and exhibited higher expression levels than the control, including SlEXPB2, SlEXPB8, SlEXLB1, SlEXLB2, and SlEXLB4.
Expression patterns of tomato expansin genes following exogenous phytohormone treatments. The heatmap was constructed based on the expression level of each expansin gene in the shoot relative to that of SlActin. Blue and red boxes indicate lower and higher expression levels, respectively. The scale bar represents the fold change (log2 value). H2O, ABA, GA, SA, IAA, and ETH represent seedlings sprayed with water, or 100 μM ABA, GA, SA, IAA, or ethephon, respectively. The numbers 1.5, 3, 6, 12, and 24 represent the time (h) after treatment. CK represents untreated seedlings at the start of the experiment (i.e., time zero)
Induced expression of expansin genes during exposure to abiotic stress
Most of the analyzed genes were up- or down-regulated following exposure to stress (Fig. 6). Genes that were up-regulated in response to drought stress included SlEXPA4, SlEXPA10, SlEXPA12, SlEXPA14, SlEXPB2, SlEXPB8, SlEXLA1, SlEXLB1, and SlEXLB4. Of these genes, the transcription levels for SlEXPA10, SlEXPA14, SlEXPB8, SlEXLA1, SlEXLB1, and SlEXLB4 were 4-, 14-, 120-, 12-, 24-, and 20-fold higher than that of the control, respectively. Genes down-regulated by drought treatment included SlEXPA1, SlEXPA18, and SlEXPB3, and the SlEXPA1 and SlEXPB3 transcription levels were decreased by nearly 80 %.
Expression patterns of tomato expansin genes following exposure to different abiotic stresses. The heatmap was constructed based on the expression level of each expansin gene in the shoot relative to that of SlActin. Blue and red boxes indicate lower and higher expression levels, respectively. The scale bar represents the fold change (log2 value). Drought 0.5–2.5 represent samples collected 0.5, 1.0, 1.5, 2.0, and 2.5 h after exposure to drought stress. Cold and heat 1.5–24 represent samples collected 1.5, 3.0, 6.0, 12, and 24 h after cold (4 °C) or heat (42 °C) stress treatments. CK represents untreated seedlings at the start of the experiment (i.e., time zero)
Following cold stress, the transcription levels of SlEXPB2, SlEXLA1, and SlEXLB4 were nearly 30-, 12-, and 100-fold higher than that of the control, respectively. Conversely, the expression levels of SlEXPA4, SlEXPA5, SlEXPA10, and SlEXLB2 were significantly decreased. There were no significant expression level changes for the other genes following cold stress treatment.
The expression levels of more than half of the analyzed genes were affected by heat stress. Up-regulated genes included SlEXPA2, SlEXPA3, SlEXPA6–A9, SlEXPA12, SlEXPA14, SlEXPA24, SlEXPB1, and SlEXLB4. Down-regulated genes included SlEXPB3 and SlEXLB2. Among the up-regulated genes, SlEXLB4 exhibited the highest expression level change, with an 80-fold increase over the control.
Discussion
Previous studies have revealed that EXPAs and EXPBs existed when vascular plants and mosses were diverging, whereas EXLAs and EXLBs can only be traced back to the last angiosperm and gymnosperm ancestors (Sampedro and Cosgrove 2005). The expansin family has more recently continued to grow and diversify in different species. The last common ancestors of eudicots and monocots are believed to have 12 EXPAs, two EXPBs, one EXLA, and two EXLBs (Sampedro and Cosgrove 2005), and the number of genes has almost doubled or tripled in “modern” plants. In this study, we identified 38 tomato expansin and expansin-like genes, including 25 EXPAs, eight EXPBs, one EXLA, and four EXLBs (Table 1). These findings are more consistent with those of A. thaliana (26 EXPAs, six EXPBs, three EXLAs, and one EXLB) than with those of rice (34 EXPAs, 19 EXPBs, four EXLAs, and one EXLB). Rice and maize contain more EXPBs than eudicots (e.g., tomato and A. thaliana) (Zhang et al. 2014), which may be because of the differences in their cell wall composition and their evolutionary differences (Cosgrove et al. 2002).
The growth of the plant expansin superfamily is mostly due to gene duplications (Zhu et al. 2014). Our results indicate that segmental and tandem duplication play key roles in the growth of the tomato expansin gene family (Supplementary Fig. 2). A cluster of expansin genes that had evolved from tandem duplications was present in tomato and A. thaliana (Li et al. 2002), but not in rice. These expansins were grouped together in the phylogenetic tree (including SlEXPA7, SlEXPA19–A23, AtEXPA19, and AtEXPA21–A26) (Fig. 2), suggesting this duplication may have occurred after the eudicots and monocots diverged, but before the divergence of tomato and A. thaliana.
It has been assumed that EXPA and EXPB members have similar functions, but on different cell wall polymers (Cosgrove et al. 2002). Differences in substrate preferences may be mainly attributed to compositional diversity in domain 2 (Fig. 3). This domain was suggested as the polysaccharide (cellulose)-binding domain because several aromatic (e.g., Trp) and polar residues on the expansin surface are located in this region (Cosgrove 2000). It is reasonable that different sequences in this domain exhibit diverse substrate affinity. SlEXLA and SlEXLB members consist of the same motifs as SlEXPB members. However, there are sequence differences, especially related to the HFD motif (Fig. 3), which greatly decreased the possibility these proteins would have cell wall loosening activity in vivo.
In addition to the fruit-specific expansin gene SlEXPA1, five other SlEXPAs expressed in the fruit, SlEXPA3–A7, were identified by Brummell et al. (1999). Unlike SlEXPA1, these expansin genes are expressed more during cell division and/or cell expansion. SlEXPA3 is also highly expressed during the breaker stage. The previously described fruit expression patterns of SlEXPA1 and SlEXPA3–A6 were confirmed in this study, while our findings indicated that SlEXPA7 expression levels were higher only in mature leaves (Fig. 4). Additionally, SlEXPA8, SlEXPB8, SlEXLB1, and SlEXLB2 were determined as genes that were preferentially expressed in the fruit. Diverse fruit expression patterns of these expansin and expansin-like genes may mean they have diverse functions during fruit development. They may have coordinated functions in cell division and enlargement stages in which cell wall modifications occur. Fruit-specific or preferential expression patterns have also been reported in other plants such as strawberry (Nardi et al. 2015), pear (Hiwasa et al. 2003), and grapevine (Santo et al. 2013).
Expansins involved in root or root hair development have been widely reported in different plant species, including A. thaliana (Cho and Cosgrove 2002), maize (Wu et al. 2001), grapevine (Santo et al. 2013), and rice (Cho and Kende 1997). In this study, eight expansin genes preferentially expressed in the root were identified, including three EXPAs, three EXPBs, and two EXLBs (Fig. 4). Because the root is the main organ responsible for water and nutrient acquisition, it has always been speculated that expansins expressed in the root play important roles in water and nutrient absorption. Overexpression of the wheat expansin gene, TaEXPB23, enhanced drought tolerance in transgenic tobacco (Han et al. 2014). Overexpression of GmEXPB2 in soybean stimulated root growth and improved phosphate use efficiency in phosphate-limited conditions (Zhou et al. 2014). Therefore, the tomato expansin genes identified in this study as being specifically or preferentially expressed in the root may also have similar effects.
Phytohormones play critical roles in regulating plant growth, development, and adaptation to environmental changes. Expansins are regarded as the mediators of hormone activities. Actively expressed expansin genes are always observed in growing tissues with high levels of growth-promoting hormones, including GA and IAA (Lee and Kende 2002; Cho and Cosgrove 2004). We also identified expansin genes preferentially or specifically expressed in rapidly growing organs (Fig. 4), including young leaves (SlEXPA10), young shoots (SlEXPA2 and SlEXPA5), and immature fruits (SlEXPA3 and SlEXPA5). Preferential or specific expression of these expansin genes in these organs may be induced by endogenous hormones. The expression of most of these genes can be induced in shoots by exogenous cytokinin, GA, or IAA (Fig. 5).
Ethylene can induce fruit maturation and leaf senescence. The expression of the tomato ripening-specific expansin gene, SlEXPA1, can be considerably induced by exogenous ethylene (Rose et al. 1997), which can also stimulate root hair formation. Expression of two A. thaliana expansin genes, AtEXP7 and AtEXP18, which are tightly linked to root hair initiation, is also stimulated by exogenous ethylene (Cho and Cosgrove 2002). In our study, the expression of four expansin genes specifically or preferentially expressed in the root, SlEXPA3, SlEXPB8, SlEXLB2, and SlEXLB4, were also induced by exogenous ethylene applied to the shoots. Of these genes, SlEXLB4 was specifically expressed in roots and was induced only by exogenous ethylene (Fig. 5).
Various stresses, including salinity, drought, and extreme temperatures, can induce ABA production to activate genes needed for plant stress resistance. The relationship between expansins and ABA is still unclear. Maize can adapt to relatively low water potentials. This adaptation has been associated with the increased expression and activities of three expansins present in the root. Roots treated with the ABA biosynthesis inhibitor did not exhibit these adaptive features. However, the application of exogenous ABA to well-watered roots failed to increase expansin gene expression levels to those induced by drought stress (Wu et al. 2001). Cho and Cosgrove (2004) proposed that ABA may be required to induce root responses, but drought-modulated expansin gene expression may be regulated by an ABA-independent pathway. In our study, the expression of most expansin genes in shoots was induced by at least one stress treatment (Fig. 6). Some genes were also induced by the application of exogenous ABA, such as SlEXPA3, SlEXPA7, SlEXPA9, and SlEXLB1 (Fig. 5). This suggests their expression may depend on ABA. However, the expression of some genes was not induced by ABA, such as SlEXPA8, SlEXPA12, SlEXPA24, SlEXPB1, and SlEXPB8, which suggests their expression may be regulated by an ABA-independent pathway.
In conclusion, we identified 25 SlEXPAs, eight SlEXPBs, one SlEXLA, four SlEXLBs, and five short homolog sequences in the tomato genome. Twenty-five of these genes were identified being expressed. These expansins exhibited some specific features in terms of gene structure and amino acid sequences, suggesting that they underwent unique evolutionary processes. Segmental and tandem duplication events contributed to the expansion of the tomato expansin family. The 3-exon/2-intron pattern may represent the basic genetic structure, with other structural patterns arising from intron loss/gain events. Additionally, we identified three previously unreported expansin genes, SlEXPA8, SlEXPB8, and SlEXLB1, which were preferentially expressed in the fruit. We also identified expansin genes preferentially expressed in the root (SlEXPA9, SlEXLB2, and SlEXLB4), hormone and/or stress inducible expansin genes (SlEXPA3, SlEXPA7, SlEXPB1–B2, SlEXPB8, SlEXLB1–LB2, and SlEXLB4), and expansin genes preferentially expressed in floral parts. We believe our study findings may advance our understanding of the biological roles of tomato expansins. A more comprehensive understanding may be important for future crop improvement efforts involving genetic engineering.
References
Abuqamar S, Ajeb S, Sham A, Enan MR, Iratni R (2013) A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol Plant Pathol 14:813–827
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Balestrini R, Cosgrove DJ, Bonfante P (2005) Differential location of α-expansin proteins during the accommodation of root cells to an arbuscular mycorrhizal fungus. Planta 220:889–899
Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56:817–823
Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11:2203–2216
Chen F, Dahal P, Bradford KJ (2001) Two tomato expansin genes show divergent expression and localization in embryos during seed development and germination. Plant Physiol 127:928–936
Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14:3237–3253
Cho HT, Cosgrove DJ (2004) Expansins as agents in hormone action. In: Davies PJ (ed) Plant hormones, 3rd edn. Kluwer Academic, Dordrecht, pp 262–281
Cho HT, Kende H (1997) Expression of expansin genes is correlated with growth in deepwater rice. Plant Cell 9:1661–1671
Choi D, Cho HT, Lee Y (2006) Expansins: expanding importance in plant growth and development. Physiol Plant 126:511–518
Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407:321–326
Cosgrove DJ, Bedinger P, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA 94:6559–6564
Cosgrove DJ, Li LC, Cho HT, Hoffmann-Benning S, Moore RC, Blecker D (2002) The growing world of expansins. Plant Cell Physiol 43:1436–1444
Flemetakis E, Efrose RC, Desbrosses G, Dimou M, Delis C, Aivalakis G, Udvardi MK, Katinakis P (2004) Induction and spatial organization of polyamine biosynthesis during nodule development in Lotus japonicus. Mol Plant Microbe Interact 17:1283–1293
Goh HH, Sloan J, Dorca-Fornell C, Fleming A (2012) Inducible repression of multiple expansin genes leads to growth suppression during leaf development. Plant Physiol 159:1759–1770
Han Y, Chen Y, Yin S, Zhang M, Wang W (2014) Over-expression of TaEXPB23, a wheat expansin gene, improves oxidative stress tolerance in transgenic tobacco plants. J Plant Physiol 173:62–71
Harmer SE, Orford SJ, Timmis JN (2002) Characterisation of six alpha-expansin genes in Gossypium hirsutum (upland cotton). Mol Genet Genomics 268:1–9
Hiwasa K, Rose JK, Nakano R, Inaba A, Kubo Y (2003) Differential expression of seven alpha-expansin genes during growth and ripening of pear fruit. Physiol Plant 117:564–572
Hu B, Jin JP, Guo AY, Zhang H, Luo JH, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Keller E, Cosgrove DJ (1995) Expansins in growing tomato leaves. Plant J 8:795–802
Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, Voesenek LA (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55:311–314
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Lee Y, Choi D, Kende H (2001) Expansins: ever-expanding numbers and functions. Curr Opin Plant Biol 4:527–532
Lee Y, Kende H (2002) Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiol 130:1396–1405
Li Y, Darley CP, Ongaro V, Fleming A, Schipper O, Baldauf SL, McQueen-Mason SJ (2002) Plant expansins are a complex multigene family with an ancient evolutionary origin. Plant Physiol 128:854–864
Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6:603–610
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lü P, Kang M, Jiang X, Dai F, Gao J, Zhang C (2013) RhEXPA4, a rose expansin gene, modulates leaf growth and confers drought and salt tolerance to Arabidopsis. Planta 237:1547–1559
Ludidi NN, Heazlewood JL, Seoighe C, Irving HR, Gehring CA (2002) Expansin-like molecules: novel functions derived from common domains. J Mol Evol 54:587–594
Nardi CF, Villarreal NM, Rossi FR, Martínez S, Martínez GA, Civello PM (2015) Overexpression of the carbohydrate binding module of strawberry expansin2 in Arabidopsis thaliana modifies plant growth and cell wall metabolism. Plant Mol Biol 88:101–117
Rose JK, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA 94:5955–5960
Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6:242
Santo SD, Vannozzi A, Tornielli GB, Fasoli M, Venturini L, Pezzotti M, Zenoni S (2013) Genome-wide analysis of the expansin gene superfamily reveals grapevine-specific structural and functional characteristics. PLoS One 8:e62206
Tabuchi A, Li LC, Cosgrove DJ (2011) Matrix solubilization and cell wall weakening by β-expansin (group-1 allergen) from maize pollen. Plant J 68:546–559
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tsuchiya M, Satoh S, Iwai H (2015) Distribution of XTH, expansin, and secondary-wall-related CesA in floral and fruit abscission zones during fruit development in tomato (Solanum lycopersicum). Front Plant Sci 6:323
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49
Wu Y, Thorne ET, Sharp RE, Cosgrove DJ (2001) Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol 126:1471–1479
Yan A, Wu M, Yan L, Hu R, Ali I, Gan Y (2014) AtEXP2 is involved in seed germination and abiotic stress response in Arabidopsis. PLoS One 9:e85208
Zhang W, Yan H, Chen W, Liu J, Jiang C, Jiang H, Zhu S, Cheng B (2014) Genome-wide identification and characterization of maize expansin genes expressed in endosperm. Mol Genet Genomics 289:1061–1074
Zhou J, Xie J, Liao H, Wang X (2014) Overexpression of β-expansin gene GmEXPB2 improves phosphorus efficiency in soybean. Physiol Plant 150:194–204
Zhu Y, Wu N, Song W, Yin G, Qin Y, Yan Y, Hu Y (2014) Soybean (Glycine max) expansin gene superfamily origins: segmental and tandem duplication events followed by divergent selection among subfamilies. BMC Plant Biol 14:93
Acknowledgments
This work was supported by the National High Technology Research and Development Program for of China (863 Program, No. 2012AA100105-4) and the National Natural Science Foundation of China (Grant No. 31171960 and Grant No. 31572133).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Y. Lu and L. Liu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lu, Y., Liu, L., Wang, X. et al. Genome-wide identification and expression analysis of the expansin gene family in tomato. Mol Genet Genomics 291, 597–608 (2016). https://doi.org/10.1007/s00438-015-1133-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1133-4