Abstract
Cold-induced sweetening (CIS) is a crucial factor influencing the processing quality of potato tubers. To better understand the molecular events of potato CIS and different CIS-sensitivity among various potato species, a suppression subtractive hybridization library and cDNA microarray gene filters were developed. A total of 188 genes were found to be differentially expressed (DE) in Solanum berthaultii (ber) upon cold stimulation. These functional genes were mostly related to cell rescue, defense and virulence, metabolism, energy and protein fate, included in various processes of plant defense against abiotic stresses. Four expression patterns of these DE genes were profiled by qRT-PCR using the cold-stored tubers of both CIS-resistant (ber) and CIS-sensitive (E-potato 3, a variety of S. tuberosum) potatoes. The expression pattern and abundance of many DE genes encoding proteins involved in metabolism were different in these two potato tubers, especially genes associated with amylolysis, sucrose decomposition and glycolysis pathways, indicating distinct regulatory mechanisms between ber and E3 in response to cold stress, which may be crucial for potato CIS. Further investigation of these cold-regulated genes will deepen our understanding of the regulatory mechanisms of potato CIS and direct approaches for the genetic improvement of potato processing quality.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Low temperature is beneficial for preventing from sprout growth, water loss and microorganism infection of potato tubers and so prolonging their storage after harvest. However, temperatures <10 °C stimulate cold-induced sweetening (CIS), which is characterized by accumulation of reducing sugars (RS) in the tubers. CIS is a widespread phenomenon first described in 1882 (Menéndez et al. 2002). As potato tubers are processed into chips and fries, RS react with amino acids to generate unacceptable dark-colored, bitter-tasting products in the non-enzymatic Maillard reaction, as well as accumulating acrylamide—a known neurotoxin and suspected carcinogen (Friedman 2003). CIS has posed a significant challenge to the potato industry and is of commercial interest (Xin and Browse 2000).
Physiology and biochemistry of CIS have been investigated extensively and the biological processes have been partially elucidated. Sugars that accumulate in the cold are derived from starch. The starch degradation occurs via phosphorolytic and hydrolytic reaction and probably requires concerted action of several enzymes (Beck and Ziegler 1989). This process starts primarily through the action of starch phosphorylase (Davies and Viola 1992; Smith et al. 2005), although amylases may have an important facilitating role (Cottrell et al. 1993). Hexosephosphates are then exported from the amyloplasts via a modified phosphate carrier, converted to sucrose via sucrose–phosphate synthase, and then sucrose can be cleaved into RS (i.e., glucose and fructose) by invertase (Sowokinos 2001). Other pathways such as glycolysis, mitochondrial respiration and hexogenesis are also causal processes influencing RS content (Sowokinos 2001).
CIS inheritance is considered to be controlled by multiple genes and seems impossible to understand without molecular information indicating a genome-wide response in potato tubers. By differential screening of a cDNA library, 16 cold-induced fragments were identified from nearly 25,000 clones of the library (van Berkel et al. 1994). With a diploid potato, the first molecular-function map with 85 loci was constructed in potato, based on 69 carbohydrate metabolism and transport associated genes (Chen et al. 2001). Another 26 quantitative trait locus (QTLs) were identified to control CIS in two diploid potato species (Menéndez et al. 2002). It is obvious that more events associated with CIS remain unknown. Heterologous GeneChips (made from tomato) were used to discover genes responding to cold stimulation at an early stage when potato tubers were exposed to low temperature, as many as 1,854 differentially expressed (DE) probe-sets were identified (Bagnaresi et al. 2008). However, this research mainly focuses on cold responses in a single CIS-sensitive genotype, and the genetic basis for the variation in CIS sensitivity among different genotypes has yet to be explained.
In the present study, more genes involved in potato CIS were identified through construction of the forward and reverse suppression subtractive hybridization (SSH) libraries and the transcriptional analysis in different genotypes with distinct CIS sensitivity. Possible functions of DE genes in response to cold stimuli are discussed by gathering the information available.
Materials and methods
Plant materials and bacterial strains
Mature tubers of diploid wild potato species Solanum berthaultii accession CW2-1 (ber, CIS-resistant) and tetraploid S. tuberosum variety E-potato 3 (E3, CIS-sensitive) grown in the greenhouse of Huazhong Agricultural University were used for the experiments. Freshly harvested tubers were kept first at 20 °C for 10 days in darkness for skin set, and then divided into two groups. One group was moved into 4 °C in darkness as the low temperature treatment, while another was maintained at 20 °C in darkness as the control. Tubers were sampled at specific time-points, immediately frozen in liquid nitrogen and stored at −70 °C until use. Escherichia coli DH5α used for propagation of the library was cultured in Luria–Bertani (LB) broth, supplemented with appropriate antibiotics (ampicillin, 50 μg/mL; Sigma, St. Louis, USA).
Sugar measurement
Reducing sugars of sampled tubers were quantified by a colorimetric estimation method described previously (Lindsay 1973). For the estimation of the total soluble sugars (TS), the ethanol extract from tubers was hydrolyzed by hydrochloric acid and then neutralized by sodium hydroxide. The content of TS was calculated according to the reducing power of the resulting solution, which was tested by the colorimetric estimation method mentioned above. The content of sucrose was determined as TS−RS. Each sample was tested in triplicate to ensure reproducibility and stability.
SSH library construction
The SSH libraries were constructed using a procedure described by Chen et al. (2010). In short, for construction of the forward subtractive library (FSL), tubers of ber incubated at 20 and 4 °C for 5 days were employed as Driver and Tester, respectively. By turns, tubers exposed to 4 and 20 °C for 5 days were, respectively, used as Driver and Tester for construction of the reverse subtractive library (RSL). Total RNA was extracted from the tubers according to Yang et al. (2006), then the mRNA was purified from the total RNA using an Oligotex mRNA isolation column (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. SSH was performed using the PCR-Select cDNA Subtraction Kit (Clontech, New Jersey, USA) by following the manufacturer’s instructions. The enriched specific PCR products were purified using the instructions of a DNA recovery kit (Takara, Dalian, China), and restricted by SmaI/NotI, ligated into plasmid pBluescript SK (−), and then transformed into E. coli DH5α. Single white colonies grown on LB agar containing Amp/X-gal/IPTG were propagated in 384-well plates (Greiner, Frickenhausen, Germany). These clones were primarily screened by PCR with primers specific for plasmid pBluescript SK (−) (T3 and T7 primers) and those with a single amplicon had glycerol added to 15 % (v/v) and stored at −70 °C.
cDNA microarray analysis
A cDNA microarray chip containing 3,696 probes obtained from the SSH libraries was designed according to the methods described previously (Wang et al. 2005). Briefly, inserts of the libraries were amplified and purified, then qualified spectrophotometrically to ensure adequate and equal PCR amplification prior to being robotically printed onto glass slides. The microarray slides were prepared by Takara. Human transferrin receptor (TFR) gene (DTX806; Takara) and pUC19 (D3219; Takara) were used as negative controls to assess non-specific hybridization. Potato genes encoding actin (GenBank accession no. X55747), β-tubulin (GenBank accession no. Z33382) and elongation factor (GenBank accession no. AB061263) were used as internal controls. An amplicon obtained from λ phage DNA (DTX803; Takara) was used as an external control to equalize hybridization signals generated from different samples. Three independent biological replicates were applied for microarray analysis. For each replicate, the total RNA samples (30 μg each) were reverse transcribed to cDNA in presence of fluorescently labeled dUTP (Cy5 or Cy3). The hybridization and subsequent scanning, visualization and quantitation were carried out, and DE genes (determined as >2-fold alteration of signals in the microarray) were selected for further analysis.
DNA sequencing and analysis
Sequencing of DE clones identified by the microarray analysis was performed on an ABI3730 machine (Applied Biosystems, California, USA). Vector and adaptor sequences were trimmed prior to further analysis. Sequence homology was assessed with the BLASTn (nucleotide homology) and BLASTx (translation homology) algorithm programs at the NCBI network service (http://www.ncbi.nlm.nih.gov/BLAST). To classify the DE genes into the Arabidopsis thaliana Gene Ontology (GO) categories, the Arabidopsis database at the MIPS website (http://mips.gsf.de/proj/plant/jsf/athal/index.jsp) was searched for each sequence.
Real-time quantitative RT-PCR (qRT-PCR) analysis
The total RNA (1 μg) was extracted as described above and converted into cDNAs using an oligo (dT) primer (20-mer) and reverse transcriptase according to the manufacturer’s instructions (Takara). Gene-specific primers used for qRT-PCR assays are listed in Appendix S1 (primers were synthesized by Sangon Biotechnology Inc, Shanghai, China).
Real-time qRT-PCRs were carried out in a 20-μL system containing 1× SYBR Green real-time PCR Master Mix (Toyobo, Osaka, Japan), 1 μL of cDNA and 0.5 μL forward and reverse primers (10 μmol/L). The gene expression levels were analyzed by an ABI 7500 Sequence Detection System and normalized with the results of elongation factor (AB061263). Real-time qRT-PCR was performed in triplicate for each sample and the standard deviation was calculated for individual means.
Results
Sugar accumulation in ber and E3 tubers stored at different temperatures
ber and E3 tubers were stored at 20 and 4 °C and sampled at 0, 3, 5, 10, 15, 20, 30, 45, 60, 75 and 90 days. The RS, sucrose and TS concentrations were determined (Fig. 1). When stored at 20 °C, both E3 and ber showed low levels of RS, sucrose and TS accumulation. During cold storage, the RS content of both genotypes did not change during 0–5 days, then the RS content of E3 tubers increased quickly to a high level, from 3.5 to 20.7 mg/g fresh weight (FW) during 5–30 days, and was maintained at high levels with only slight fluctuation until the end of the storage. However, there was only a slight increase in ber (the maximum RS content was 3.4 mg/g FW) (Fig. 1a). Therefore, ber was designated as a CIS-resistant genotype and E3 as CIS-sensitive. Both ber and E3 showed increased sucrose content 5 days after commencing the cold storage but it was always higher in ber than in E3 (Fig. 1b). There was a gradual decrease after 30 days in E3 tubers but a high level of sucrose was maintained until the end of storage in ber tubers, presumably a slow degradation of sucrose in ber might be a major factor in the low RS accumulation. Although there were higher levels of sucrose in ber tubers at 4 °C compared to E3, the TS level of the former was lower than the latter (Fig. 1c).
Identification of cold-responsive genes in tubers of ber
To conduct the comparative transcriptomic studies for screening DE genes in potato tubers under low temperature, we used PCR-based SSH to enrich DE cDNAs between low (4 °C) and room temperatures (20 °C). The clones in the subtracted cDNA libraries were cloned and printed onto chips for rapidly screening out non-DE noise. A total of 3,696 clones were printed on the array: 2,112 clones from the FSL and 1,584 from the RSL. RNA samples from tubers stored at 4 and 20 °C were reverse transcribed in presence of Cy5- and Cy3-labeled dUTP, respectively. The ratio of the two fluorescent signal intensities for each cDNA spot on the microarray was used as a relative measure to determine changes of the DE gene. Our results showed high reproducibility of microarray hybridization, which was expressed by the significant correlation coefficients of different replications (R 2 > 0.84). Data were filtered to exclude genes with a normalized expression ratio <2-fold. Thus, a total of 736 cold-related clones were identified, of which 663 were putatively up-regulated and 73 down-regulated.
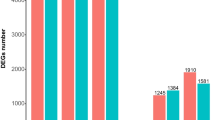
All 736 DE clones were sequenced and 719 (98 %) produced good results. BLAST analyses against the GenBank database revealed that most of these genes shared homology with diverse classes of genes from plants such as potato, Arabidopsis, tomato, tobacco and cucumber. A total of 188 non-redundant sequences were identified and their putative functions were classified by the BLASTx or BLASTn homology search (Appendix S2). The genes of known function were classified into 14 functional categories according to the gene annotations from the MIPS Arabidopsis database (Fig. 2). The largest set of genes (18.1 %) was assigned to cell rescue, defense and virulence. The genes contained in categories of metabolism (11.4 %) and energy (11.4 %) were tied as the second largest groups, followed by categories of protein fate (10.1 %), protein with binding function (8.7 %) and protein activity regulation (7.4 %). The remaining functional categories, with range of 0.7–5.4 %, were composed of a small number of transcripts. The DE genes are listed in Appendix S2.
Expression analysis by qRT-PCR
Of the 188 identified ESTs, most showed similarity to proteins associated with cell rescue, metabolism and energy, protein fate and unknown proteins. To further verify the results of the cDNA microarray hybridization and analyze the expression patterns of DE genes in tubers upon cold stimulation, qRT-PCR was conducted. RNA samples were extracted from ber and E3 tubers at four time-points in the early stage of cold storage (0, 5, 15 and 30 days), since the RS content dramatically changed during this period (Fig. 1a). Expression profiles at 20 °C were also characterized as the control. A total of 185 sequences could be amplified in both ber and E3. The relative transcription values were expressed as accumulation levels at different time-points normalized to that of 0 day for individual genes, then the logarithms of the relative transcription values were calculated and imported into the Gene Cluster Program 3.0 for clustering analysis. Hierarchical clustering was performed based on a Euclidean distance matrix and the complete linkage rule. According to the expression profiles, the differentially regulated genes were primarily grouped into four clusters (clusters A–D) (Fig. 3). The detailed information is listed in Appendix S1.
Expression patterns of differentially expressed (DE) genes in ber and E3 tubers under 20 and 4 °C. Tubers were sampled at 0, 5, 15 and 30 days. Total RNA was analyzed by qRT-PCR. Hierarchical clustering analysis of 185 DE genes isolated from ber tubers is shown. For clustering, the average logarithms of relative values of three biological replicates were used for each sample after normalization of the raw data. Clusters (A–D) are marked at the bottom. The color scale for logarithm signal values is shown on the left
Cluster A included 43 genes which were transiently induced in ber (5 days) but continuously induced in E3 at 4 °C. There were 12 transcripts related to defense in this cluster. C20-2-P21, C20-3-B04 and C20-3-B12 encode proteins belong to the heat shock proteins (HSPs) family, and were reported to be induced in almost all stresses and considered as stress tolerance factors in plants (Al-Whaibi 2011). Genes involved in the ubiquitination/proteasome pathway were identified, e.g., C20-5-C03 represented a 20S proteasome, which was required to control plant development and stress tolerance (Kurepa et al. 2009). These HSPs and the 20S proteasome have been reported to share common roles in mediating the degradation of denatured proteins under stress conditions. Differential regulation of these genes might indicate different strategies against cold-induced damaged proteins.
Eighty-two genes included in cluster B were up-regulated in both ber and E3 tubers in cold storage. The genes related to energy and metabolism were the first redundant subgroup in this cluster, such as β-amylase gene (C20-2-D03) for amylolysis (Fig. 4) and the genes (C20-1-A21, C20-6-F06 and C20-3-A14) involved in glycolysis (Fig. 5). The second subgroup in cluster B was composed of the genes related to protein synthesis, fate and activity regulation, and noticeably also a putative invertase inhibitor (InvInh) gene (C20-4-C15) (Fig. 6). Although these genes were induced in both ber and E3, most exhibited higher accumulation levels in ber at 4 °C, e.g., C20-1-E21 representing dicer-like protein (DCL) and C20-2-B07 encoding a putative calcineurin B-like (CBL)-interacting protein kinase (CIPK) (Fig. 7).
qRT-PCR validation of the genes involved in amylolysis in ber and E3 tubers stored at 20 and 4 °C. Values represent mean ± SD (n = 3 biological replicates with three technical repeats) normalized with elongation factor (AB061263) at each time point. Details of gene names and their putative function refer to Appendix S2. a DE genes involved in the amylolysis pathway and their predicted products are marked in the diagram, b expression level of C20-3-O14 (GWD) and c expression level of C20-2-D03 (β-amylase)
Expression of the genes involved in glycolysis pathway in ber and E3 tubers. a DE genes involved in glycolysis pathway and their predicted products are marked in the diagram. Transcript accumulation was validated by qRT-PCR for genes: b C20-6-F06 encoding a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and c C20-3-A14 encoding a pyruvate kinase
qRT-PCR validation of the genes associated with sucrose decomposition in ber and E3 tubers stored at 20 and 4 °C. Values represent mean ± SD (n = 3 biological replicates with three technical repeats) normalized with elongation factor (AB061263) at each time point. Details of gene names and their putative functions refer to Appendix S2. a A diagram of starch decomposition and DE genes involved in this pathway, b relative expression levels of C20-4-C15 (invertase inhibitor) and c relative expression levels of C20-6-K02 (xylose isomerase)
qRT-PCR validation of some DE genes related to signal transduction and transcription in ber and E3 tubers. Values represent mean ± SD (n = 3 biological replicates with three technical repeats) normalized with elongation factor (AB061263) at each time point. Details of gene names and their putative function refer to Appendix S2. a Expression levels of C20-4-D09 that represents an ethylene response factor 24 (ER24), b expression levels of C20-1-E21 that represents a dicer-like protein (DCL) and c expression levels of C20-2-B07 that encodes a calcineurin B-like (CBL)-interacting protein kinase (CIPK)
Cluster C consisted of 32 genes with a down-regulation pattern in cold storage. Genes encoding proteins functioned as ‘protein fate, synthesis or activity regulation’ were the most significant subgroups in this cluster, e.g., proteinase inhibitor homolog coding sequences (C4-1-B11, C4-3-J11 and C20-4-J09) (Fig. 8). We presumed that the suppression of these genes would lead to more liberation of proteinases for degradation of inappropriate proteins.
Expression changes of DE genes related to protein activity regulation were determined by qRT-PCR. Values represent mean ± SD (n = 3 biological replicates with three technical repeats) normalized with elongation factor (AB061263) at each time point. Details of gene names and their putative function refer to Appendix S2. a C4-1-B11 encodes a cystatin, b C4-3-J11 encodes a proteinase inhibitor II and c C20-4-J09 encodes a Kunitz-type proteinase inhibitor A6
The last cluster D had 28 genes down-regulated in cold treated ber but up-regulated in E3 tubers. For example, alpha-glucan, water dikinase gene (C20-3-O14) decreased at 5 days of cold storage in both ber and E3 tubers (Fig. 4), but significantly increased in E3 thereafter and remained suppressed in ber.
Discussion
A major drawback of potato tubers stored at low temperature is CIS, which limits the commercial value of the processed products. Considerable effort has therefore been directed toward understanding how potato tubers respond and adapt to low temperature. The process of CIS is a multigenic and quantitative trait associated with complex physiological and biochemical changes (Menéndez et al. 2002). It was noted that the sensitivity to low temperature differed among various potato species (McCann et al. 2010), and a S. berthaultii accession was found somewhat resistant to CIS in our laboratory (Liu et al. 2010). In the present study, we developed a ber-specific SSH/cDNA microarray gene filter and at least 188 genes were found associated with CIS.
Genes related to amylolysis
Previous studies have shown a complex carbohydrate metabolism network during occurrence of CIS, including starch degradation and resynthesis, conversion of sucrose to RS, as well as respiration (Menéndez et al. 2002). The degradation of starch was considered the source of RS. Our data also showed that there were two DE genes encoding proteins involved in amylolysis (Fig. 4). Of them, C20-3-O14—which is similar to Arabidopsis glucan, water dikinase (GWD)—is a key enzyme controlling the phosphorylation degree of starch and susceptibility of starch to degradation. It shares a synergistic effect in β-amylase mediated starch degradation (Mikkelsen et al. 2005; Edner et al. 2007). Yano et al. (2005) revealed that Arabidopsis GWD was cold induced, and GWD deficiency resulted in a starch over-accumulation phenotype and reduced freezing tolerance. Lower transcripts accumulation of this GWD gene in ber than in E3 might contribute to low RS content in the former. This speculation is supported by an antisense transformation of GWD which could reduce CIS in potato tubers (Lorberth et al. 1998).
The other gene, C20-2-D03, showed maximum homology (76 %) to Arabidopsis β-amylase BMY7. Its expression was induced by cold storage but was much higher in E3 than in ber (Fig. 4b). β-amylase is an exoamylase that hydrolyzes α-1,4 glycosidic linkages of polyglucan chains at the non-reducing end to produce reducing maltose. The primary function of β-amylase is starch breakdown in plants (Kossmann and Lloyd 2003). Interference of chloroplast β-amylase by antisense methods resulted in reduced ability to degrade starch and a starch-excess phenotype in potato leaves (Scheidig et al. 2002). The product of β-amylase is translocated to the cytosol by maltose translocator (Niittylä et al. 2004) to be metabolized to glucose units by cytosolic glucosyltransferases during transitory starch degradation (Lu and Sharkey 2004; Weise et al. 2004). The BMY7 was found to be a temperature-stress specifically induced starch-degrading enzyme in Arabidopsis (Kaplan and Guy 2004). Maltose accumulation resulting from the enzymolysis of BMY7 was demonstrated to be a stabilizing factor in the chloroplast stroma in response to acute temperature stress (Kaplan and Guy 2004). The present results suggest that this alteration might be an important source of RS accumulation in potato tubers under cold conditions. Higher transcripts abundance of C20-2-D03 in E3 than in ber during cold storage might contribute to a high RS accumulation in E3 tubers.
Sucrose decomposition
Incubation of potato tubers at <10 °C causes accumulation of soluble sugars (mainly sucrose, glucose and fructose) at the expense of starch, which is likely to increase the resistance of tissues to cold shock. However, sugar accumulation can negatively affect the quality of processed products of tubers (Sowokinos 2001). Data suggested that the ability to form sucrose, and the hydrolysis of sucrose to RS (glucose and fructose) were two critical determinants of potato tubers’ sugar accumulation in cold storage (Sowokinos 2001). Invertase (EC 3.2.1.26) catalyzes irreversible hydrolysis of sucrose into glucose and fructose, and may have an important role in regulating sugar content and composition in potato tubers (Zrenner et al. 1996; Liu et al. 2011). In the present study, two sucrose hydrolysis-related genes were identified: C20-4-C15 having a moderate identity (57 %) to InvInh from Arabidopsis and C20-6-K02 annotating a putative xylose isomerase (Fig. 6a). InvInh proteins were suggested to play a critical role in post-translational modification of invertase activity through protein–protein interactions (Rausch and Greiner 2004), and hence they may have great influence on CIS resistance (Liu et al. 2010; Brummell et al. 2011). In the present study, expression of C20-4-C15 was sharply induced by low temperature (Fig. 6b). Interestingly, the expression was much higher in ber than in E3 at the early stage of cold storage with a diminishing rate toward 15 days, which could partially explain a higher sucrose accumulation in the former compared to the latter (Fig. 1b). More transcripts of C20-4-C15 were detected in E3 by 30 days, possibly due to a contrary balance in plant cells to keep a relatively high concentration of soluble sugars to cope with cold stress. A putative xylose isomerase (C20-6-K02) was found to be differentially regulated (Fig. 6c). Xylose isomerase (EC 5.3.1.5) plays an essential role in sugar metabolism in many organisms, catalyzing reversible isomerization of d-xylose to d-xylulose, and is often referred to as ‘glucose isomerase’ since xylose isomerase can also catalyze the interconversion of d-glucose to d-fructose (Bhosale et al. 1996). Previous data suggested that the expression of bacterial xylose isomerase in potato tubers could result in a shift in the glucose to fructose ratio (Urbanczyk-Wochniak et al. 2003). The time course for C20-6-K02 transcripts accumulation revealed that C20-6-K02 was down-regulated in ber and up-regulated in E3 upon cold stimulation (Fig. 6c). However, the engagement of C20-6-K02 in regulation of CIS resistance and cold adaptation requires further investigation.
Genes involved in glycolysis pathway
It has been reported previously that the anaerobic pathway has a general function in aerobic metabolism under stress conditions, such as water deficit, SO2 fumigation, ozone exposure and low temperature, which would damage the intricate mitochondrial ATP-generating machinery (Kimmerer and Kozlowski 1982). Utilization of the anaerobic respiratory pathway was observed as one of the targets responsible for CIS resistance of potato tubers (Blenkinsop et al. 2003; Pinhero et al. 2007). Transgenic potato tubers expressing Arabidopsis pyruvate decarboxylase, which was characterized as a key enzyme in the glycolysis pathway, showed lower RS levels and higher resistance to CIS compared with wild type tubers (Pinhero et al. 2011). In the present study, two glycolysis-associated genes with higher transcripts in ber than in E3 at 4 °C were identified (Fig. 5a). The first one is C20-6-F06, encoding a putative glyceraldehyde 3-phosphate dehydrogenase (GAPDH, EC 1.21.12) which catalyzes the conversion of glyceraldehyde 3-phosphate to d-glycerate 1,3-biphosphate in the pathway of energy and carbon molecule supply (Fig. 5b). The second one is C20-3-A14, which encodes a protein similar to pyruvate kinase (EC 2.7.1.40) (Fig. 5c). Pyruvate kinase is a key rate-limiting enzyme in glycolysis and catalyzes the transfer of a phosphate group from phosphoenolpyruvate to ADP. Previous study suggested that pyruvate kinase might be involved in tolerance to hypoxic and oxidative stresses in animal cells (Shimizu et al. 2004; Aisaki et al. 2007). Taken together, our results suggest that up-regulation of the genes participating in the glycolytic pathway is important in cold adaptation of potato tubers, and would result in accelerated glycolysis and might contribute to the regulation of CIS.
Signal transduction and transcription associated genes
In the present study, several signal transduction and transcription-related genes were identified as regulated by low temperature. For example, ethylene response factor 24 (ER24) (C20-4-D09) was up-regulated in ber (Fig. 7a). C20-4-D09 showed a great increase at 5 days in ber and then sharply decreased, while it was induced in E3 tubers at a relatively lower level. Bagnaresi et al. (2008) previously observed a similar expression pattern of ER24. Ethylene response factors were demonstrated to play important roles in abiotic-stress responses together with abscisic acid in plants, including salinity, cold and dehydration stresses (Zhang et al. 2010). DCL proteins form a small protein family function as processors of small RNAs and are important for stress tolerance (Boyko et al. 2010). A DCL homolog (C20-1-E21) was isolated by microarray and confirmed by qRT-PCR (Fig. 7b). In addition, C20-2-B07 encoding a putative CIPK was up-regulated (Fig. 7c). CIPKs were functionally regulated by CBL proteins, the CBL–CIPK complexes have been demonstrated to relay calcium signals and mediate plant responses to a variety of external stresses (Huang et al. 2011). Further study is needed to clarify the roles of these transcription factors in the CIS process.
Protein fate
Misfolded or damaged proteins would accumulate in plant cells when growth conditions alter, and these undesirable proteins could then be eliminated by refolding or degradation to keep homeostasis (Wickner et al. 1999). A large number of protein fate-related genes were identified as DE in tubers at 4 °C, including HSPs and ubiquitination/proteasome and proteinase inhibitors. Here, we would like to highlight the down-regulation of proteinase inhibitors (Fig. 8), such as cystatin (C4-1-B11), proteinase inhibitor II (C4-3-J11) and Kunitz-type proteinase inhibitor A6 (C20-4-J09), which were reported to be up-regulated under stress conditions and considered as stress-resistance related factors in plants (Pernas et al. 2000; Van der Vyver et al. 2003). It was hypothesized that the significance of down-regulation of proteinase inhibitors might be the impact of liberating more proteinases for elimination of damaged proteins.
The potential roles of novel genes
Nearly one-third of the 188 genes identified were novel and have not been functionally characterized. Genes containing an evolutionarily conserved C-terminal region (ECT), such as ECT4 (C20-5-A13) and ECT11 (C4-3-D24) were down-regulated, while ECT7 (C20-1-F11) was up-regulated during cold storage (Fig. 9). ECT proteins exist in various eukaryotic organisms, although functions of these proteins have not been precisely stated, data suggest that they may be candidate signaling molecules and modulate calcium signal pathways (Ok et al. 2005). Certainly, further studies are warranted to elucidate the roles of these novel genes in low-temperature response of potato tubers.
Expression levels of some DE genes of unknown function. Values represent mean ± SD (n = 3 biological replicates with three technical repeats) normalized with elongation factor (AB061263) at each time point. a–c represent expression patterns of three differentially regulated genes containing an evolutionarily conserved C-terminal region
In summary, the present study identified 188 genes regulated by cold stress in CIS-resistant ber tubers. These DE genes were also expressed in the CIS-sensitive E3 tubers. However, the expression pattern and abundance of DE genes differed between these two tubers, especially genes encoding proteins involved in amylolysis, sucrose decomposition and glycolysis, indicating distinct regulatory mechanisms between ber and E3 in response to cold stimuli. Although further studies are needed to fully elucidate the mechanisms of CIS, our results present an extensive investigation of the gene expression changes of tubers in response to cold stress, providing information for understanding the complex network of gene expression regulation in tubers and the possible mechanisms of CIS resistance of the wild potato species ber.
References
Aisaki K, Aizawa S, Fujii H, Kanno J, Kanno H (2007) Glycolytic inhibition by mutation of pyruvate kinase gene increases oxidative stress and causes apoptosis of a pyruvate kinase deficient cell line. Exp Hematol 35(8):1190–1200
Al-Whaibi MH (2011) Plant heat-shock proteins: a mini review. J King Saud Univ Sci 23(2):139–150
Bagnaresi P, Moschella A, Beretta O, Vitulli F, Ranalli P, Perata P (2008) Heterologous microarray experiments allow the identification of the early events associated with potato tuber cold sweetening. BMC Genomics 9:176
Beck E, Ziegler P (1989) Biosynthesis and degradation of starch in higher plants. Annu Rev Plant Physiol Plant Mol Biol 40(1):95–117
Bhosale SH, Rao MB, Deshpande VV (1996) Molecular and industrial aspects of glucose isomerase. Microbiol Rev 60(2):280–300
Blenkinsop RW, Copp LJ, Yada RY, Marangoni AG (2003) A proposed role for the anaerobic pathway during low-temperature sweetening in tubers of Solanum tuberosum. Physiol Plant 118(2):206–212
Boyko A, Blevins T, Yao Y, Golubov A, Bilichak A, Ilnytskyy Y, Hollunder J, Meins F Jr, Kovalchuk I (2010) Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5(3):e9514
Brummell DA, Chen RK, Harris JC, Zhang H, Hamiaux C, Kralicek AV, McKenzie MJ (2011) Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J Exp Bot 62(10):3519–3534
Chen X, Salamini F, Gebhardt C (2001) A potato molecular-function map for carbohydrate metabolism and transport. Theor Appl Genet 102(2–3):284–295
Chen X, Song B, Yang J, Xie C, Liu J (2010) Construction and identification of potato tuber SSH library induced by low temperature. J Agric Sci Technol 12(6):68–74 (in Chinese)
Cottrell JE, Duffus CM, Paterson L, Mackay GR, Allison MJ, Main H (1993) The effect of storage temperature on reducing sugar concentration and the activities of three amylolytic enzymes in tubers of the cultivated potato, Solanum tuberosum L. Potato Res 36(2):107–117
Davies HV, Viola R (1992) Regulation of sugar accumulation in stored potato tubers. Post-harvest News Inf 3(5):97N–100N
Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, Ritte G (2007) Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol 145(1):17–28
Friedman M (2003) Chemistry, biochemistry, and safety of acrylamide. A review. J Agric Food Chem 51(16):4504–4526
Huang C, Ding S, Zhang H, Du H, An L (2011) CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci 181(1):57–64
Kaplan F, Guy CL (2004) Beta-amylase induction and the protective role of maltose during temperature shock. Plant Physiol 135(3):1674–1684
Kimmerer TW, Kozlowski TT (1982) Ethylene, ethane, acetaldehyde, and ethanol production by plants under stress. Plant Physiol 69(4):840–847
Kossmann J, Lloyd J (2003) Understanding and influencing starch biochemistry. Crit Rev Biochem Mol Biol 35(3):141–196
Kurepa J, Wang S, Li Y, Smalle J (2009) Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav 4(10):924–927
Lindsay H (1973) A colorimetric estimation of reducing sugars in potatoes with 3,5-dinitrosalicylic acid. Potato Res 16(3):176–179
Liu X, Song B, Zhang H, Li XQ, Xie C, Liu J (2010) Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol Genet Genomics 284(3):147–159
Liu X, Zhang C, Ou Y, Lin Y, Song B, Xie C, Liu J, Li XQ (2011) Systematic analysis of potato acid invertase genes reveals that a cold-responsive member, StvacINV1, regulates cold-induced sweetening of tubers. Mol Genet Genomics 286(2):109–118
Lorberth R, Ritte G, Willmitzer L, Kossmann J (1998) Isolation of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol 16(5):473–477
Lu Y, Sharkey TD (2004) The role of amylomaltase in maltose metabolism in the cytosol of photosynthetic cells. Planta 218(3):466–473
McCann LC, Bethke PC, Simon PW (2010) Extensive variation in fried chip color and tuber composition in cold-stored tubers of wild potato (solanum) germplasm. J Agric Food Chem 58(4):2368–2376
Menéndez CM, Ritter E, Schäfer-Pregl R, Walkemeier B, Kalde A, Salamini F, Gebhardt C (2002) Cold sweetening in diploid potato: mapping quantitative trait loci and candidate genes. Genetics 163(3):1423–1434
Mikkelsen R, Mutenda KE, Mant A, Schürmann P, Blennow A (2005) Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc Natl Acad Sci USA 102(5):1785–1790
Niittylä T, Messerli G, Trevisan M, Chen J, Smith AM, Zeeman SC (2004) A previously unknown maltose transporter essential for starch degradation in leaves. Science 303(5654):87–89
Ok SH, Jeong HJ, Bae JM, Shin JS, Luan S, Kim KN (2005) Novel CIPK1-associated proteins in Arabidopsis contain an evolutionarily conserved C-terminal region that mediates nuclear localization. Plant Physiol 139(1):138–150
Pernas M, Snchez-Monge R, Salcedo G (2000) Biotic and abiotic stress can induce cystatin expression in chestnut. FEBS Lett 467(2–3):206–210
Pinhero RG, Copp LJ, Amaya C, Marangoni AG, Yada RY (2007) Roles of alcohol dehydrogenase, lactate dehydrogenase and pyruvate decarboxylase in low-temperature sweetening in tolerant and susceptible varieties of potato (Solanum tuberosum). Physiol Plant 130(2):230–239
Pinhero R, Pazhekattu R, Marangoni AG, Liu Q, Yada RY (2011) Alleviation of low temperature sweetening in potato by expressing Arabidopsis pyruvate decarboxylase gene and stress-inducible rd29A: a preliminary study. Physiol Mol Biol Plants 17(2):105–114
Rausch T, Greiner S (2004) Plant protein inhibitors of invertases. Biochim Biophys Acta 1696(2):253–261
Scheidig A, Fröhlich A, Schulze S, Kossmann J, Lloyd JR (2002) Downregulation of a chloroplast-targeted beta-amylase leads to a starch-excess phenotype in leaves. Plant J 30(5):581–591
Shimizu T, Uehara T, Nomura Y (2004) Possible involvement of pyruvate kinase in acquisition of tolerance to hypoxic stress in glial cells. J Neurochem 91(1):167–175
Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Annu Rev Plant Biol 2005(56):73–98
Sowokinos JR (2001) Biochemical and molecular control of cold-induced sweetening in potatoes. Am J Potato Res 8(3):221–236
Urbanczyk-Wochniak E, Leisse A, Roessner-Tunali U, Lytovchenko A, Reismeier J, Willmitzer L, Fernie AR (2003) Expression of a bacterial xylose isomerase in potato tubers results in an altered hexose composition and a consequent induction of metabolism. Plant Cell Physiol 44(12):1359–1367
van Berkel J, Salamini F, Gebhardt C (1994) Transcripts accumulating during cold storage of potato (Solanum tuberosum L.) tubers are sequence related to stress-responsive genes. Plant Physiol 104(2):445–452
Van der Vyver C, Schneidereit J, Driscoll S, Turner J, Kunert K, Foyer CH (2003) Oryzacystatin I expression in transformed tobacco produces a conditional growth phenotype and enhances chilling tolerance. Plant Biotechnol J 1(2):101–112
Wang B, Liu J, Tian Z, Song B, Xie C (2005) Monitoring the expression patterns of potato genes associated with quantitative resistance to late blight during Phytophthora infestans infection using cDNA microarrays. Plant Sci 169(6):1155–1167
Weise SE, Weber AP, Sharkey TD (2004) Maltose is the major form of carbon exported from the chloroplast at night. Planta 218(3):474–482
Wickner S, Maurizi MR, Gottesman S (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286(5446):1888–1893
Xin Z, Browse J (2000) Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ 23(9):893–902
Yang J, Song B, Li Y, Liu J (2006) A simple and efficient method for RNA extraction from potato tuber. J Agric Biotechnol 14(2):297–298 (in Chinese)
Yano R, Nakamura M, Yoneyama T, Nishida I (2005) Starch-related alpha-glucan/water dikinase is involved in the cold-induced development of freezing tolerance in Arabidopsis. Plant Physiol 138(2):837–846
Zhang Z, Li F, Li D, Zhang H, Huang R (2010) Expression of ethylene response factor JERF1 in rice improves tolerance to drought. Planta 232(3):765–774
Zrenner R, Schüler K, Sonnewald U (1996) Soluble acid invertase determines the hexose-to-sucrose ratio in cold-stored potato tubers. Planta 198(2):246–252
Acknowledgments
We thank Dr. Xun Liu for designing primers for qRT-PCR analysis, Hui Fang and Yongbin Ou for technical assistance in qRT-PCR and sugar measurements. We also thank Mr. Andy Z. Jiang and International Science Editing for language editing. This research was supported by grants from the National Science Foundation of China (30571181 and 31171602) and Chenguang Project of Wuhan City (201050231068).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Appendix S1: Lists of DE genes in four groups classified according to expression profiles.
Appendix S2: Genes differentially expressed in ber under cold conditions.
Rights and permissions
About this article
Cite this article
Chen, X., Song, B., Liu, J. et al. Modulation of gene expression in cold-induced sweetening resistant potato species Solanum berthaultii exposed to low temperature. Mol Genet Genomics 287, 411–421 (2012). https://doi.org/10.1007/s00438-012-0688-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-012-0688-6