Abstract
Sordaria macrospora is a homothallic ascomycete which is able to form fertile fruiting bodies without a mating partner. To analyze the molecular basis of homothallism and the role of mating products during fruiting body development, we have deleted the mating type gene Smta-1 encoding a high-mobility group domain (HMG) protein. The ΔSmta-1 deletion strain is morphologically wild type during vegetative growth, but it is unable to produce perithecia or ascospores. To identify genes expressed under control of Smta-1, we performed a cross-species microarray analysis using Neurospora crassa cDNA microarrays hybridized with S. macrospora targets. We identified 107 genes that are more than twofold up- or down-regulated in the mutant. Functional classification revealed that 81 genes have homologues with known or putative functions. Comparison of array data from ΔSmta-1 with those from three phenotypically similar mutants revealed that only a limited set of ten genes is deregulated in all mutants. Remarkably, the ppg2 gene encoding a putative lipopeptide pheromone is 500-fold down-regulated in the ΔSmta-1 mutant while in all other sterile mutants this gene is up-regulated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The regulation of sexual reproduction is one of the central processes in the life cycle of most eukaryotes. Like sex chromosomes in animals and plants, the allele type at the fungal mating type locus determines sexual compatibility between haploid individuals in fungi (Fraser and Heitman 2004, 2005). In ascomycetous fungi, alternative versions of the mating type locus on homologous chromosomes are completely dissimilar and encode unrelated proteins (Coppin et al. 1997; Shiu and Glass 2000). They have thus been termed idiomorphs instead of alleles (Metzenberg and Glass 1990). The genetic breeding mechanism of fungi in which sexual reproduction occurs only between strains of opposite mating type is called heterothallism.
The mating system of Neurospora crassa is the best characterized of any heterothallic filamentous ascomycete. The idiomorphs conferring mating behavior of the partners are designated mat A and mat a (Glass et al. 1988). The A-idiomorph contains three genes: mat A-1, mat A-2 and mat A-3 (Ferreira et al. 1996; Glass et al. 1990a). The MAT A-1-polypeptide, which is the major regulator of sexual development in A strains, accommodates a DNA-binding motif that shows similarity to the Saccharomyces cerevisiae mat α1 mating-type protein. The N. crassa genes mat A-2 and mat A-3 encode a protein without characteristic DNA-binding motif and a protein with a high mobility group (HMG) domain, respectively (Ferreira et al. 1998). The HMG motif is found in several DNA-binding proteins (Grosschedl et al. 1994).
The a-idiomorph contains two genes, mat a-1 and mat a-2. The mat a-1 gene encodes a HMG domain protein that is the major regulator of mating in a strains, whereas mat a-2 codes for a protein of unknown function (Pöggeler and Kück 2000; Staben and Yanofsky 1990).
In addition to heterothallism, a second mating system, designated homothallism, can be observed in ascomycetes. Homothallic species have no genetically definable mating-type and are self-fertile. The mycelium derived from a uninucleate ascospore or a vegetative spore of a homothallic fungus is able to complete the sexual cycle without the interaction with a mating partner (Pöggeler 2001). Sordaria macrospora is a homothallic ascomycete that is closely related to the heterothallic N. crassa. An analysis of the mating-type locus of S. macrospora revealed that this locus contains sequences homologous to both the mat a and mat A idiomorphs of N. crassa (Fig. 1) (Pöggeler et al. 1997). In the mating-type locus of S. macrospora, four different ORFs were identified, Smt a-1, Smt A-3, Smt A-2, and SmtA-1, all of which are transcribed (Pöggeler and Kück 2000). Two of these genes (SmtA-1 and Smta-1) contain domains typical for eukaryotic transcription factors (Fig. 1). When expressed in haploid strains of both mating types of the related heterothallic Podospora anserina, the mating-type locus of S. macrospora can induce fruiting body formation (Pöggeler et al. 1997). Furthermore, we previously demonstrated that SMTA-1 and SMTa-1 are able to form a heterodimer and are able to activate transcription of reporter genes in yeast. The mating-type proteins, SMTa-1 and SMTA-1, thus contain properties characteristic for mating-type-encoded transcriptional regulators of other ascomycetes (Jacobsen et al. 2002).
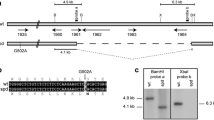
Deletion of the S. macrospora Smta-1 gene. a Structure of the Smta-1 genomic region and construction of a ΔSmta-1 deletion strain. Positions of primers used to amplify the deletion construct and verify the homologous recombination at the mating type locus are indicated. P, A. nidulans trpC promoter; hph, E. coli hygromycin resistance gene; B, BamHI. b Southern analysis. Genomic DNA from S. macrospora wild type (wt) and seven transformants (T1, T2, T3, T5, T20, T28 and T34) was digested with BamHI, separated on a 1% agarose gel, blotted and hybridized with the 840 bp-32P probe indicated in (a). c. PCR analysis for verification of homologous recombination from a single spore isolate SpT2 (S59403) of primary transformant T2 and the wild type. Positions of primers are indicated in (a)
In the yeast S. cerevisiae, pheromone signaling was shown to enable cells of opposite mating type to detect each other. Here, the unique proteins encoded by the two different mating-type loci directly act as transcriptional regulators on mating-type specific expression of pheromone and pheromone receptor genes (Bardwell 2005; Herskowitz 1989). Interestingly, genes encoding two different pheromone precursors and two pheromone receptors have not only been found in the heterothallic N. crassa, but also in the homothallic S. macrospora (Bobrowicz et al. 2002; Pöggeler 2000; Pöggeler and Kück 2001). The pheromone-precursor gene ppg1 is predicted to encode an α-factor-like peptide pheromone and the ppg2 gene an a-factor-like lipopeptide pheromone (Pöggeler 2000). The deduced gene products of pre1 and pre2 show structural similarities to the yeast a-factor receptor Ste3p and α-factor receptor Ste2p, respectively (Pöggeler and Kück 2001). As in the yeast S. cerevisiae, the expression of pheromone genes and pheromone-receptor genes in filamentous ascomycetes is supposed to be controlled directly by transcription factors encoded by the mating-type genes (Debuchy 1999). In the heterothallic N. crassa, mat A or mat a mating-type specific transcription factors are present in mat A or mat a strains, respectively, and it was demonstrated that pheromone-encoding genes of N. crassa are predominantly expressed in a mating-type-specific manner (Bobrowicz et al. 2002). On the contrary, both types of mating-type regulators are expressed in the same individual of the homothallic ascomycete S. macrospora (Pöggeler et al. 1997a). It is thus not surprising that both pheromone-precursor genes and pheromone-receptor genes of S. macrospora were shown to be transcriptionally expressed during sexual development in the same individual (Pöggeler 2000; Pöggeler and Kück 2001).
To further determine the functionality of the S. macrospora mating-type genes, we deleted the Smta-1 gene encoding a HMG protein, analyzed the phenotype of the ΔSmta-1 mutant, and examined the effect of the deletion on the expression of pheromone genes. The high degree of sequence identity between S. macrospora and N. crassa allowed us to perform cross-species microarray hybridizations using N. crassa cDNA microarrays for hybridization with targets prepared from S. macrospora (Nowrousian et al. 2004, 2005). Using this cross-species microarray technology, we have identified 107 genes that are at least twofold up- or down-regulated in the ΔSmta-1 mutant.
Materials and methods
Strains and culture conditions
Cloning and propagation of recombinant plasmids was done under standard conditions (Sambrook et al. 2001). E. coli strain SURE was used as the host for plasmid amplification (Greener 1990). The S. macrospora strain K (S17736) has a wild-type phenotype and was derived from our laboratory collection (Department of General and Molecular Botany, Bochum, Germany). The wild-type strain and the S. macrospora knockout mutant ΔSmta-1 and the complemented strain ΔSmta-1 + Smta-1 were cultivated on corn-meal medium or CM medium (Esser 1982; Nowrousian et al. 1999). S. macrospora strains used for the isolation of RNA were grown as described previously (Nowrousian et al. 2005).
Construction of the knockout plasmid
Deletion of the S. macrospora mating type gene Smta-1 was achieved as follows. The 5′- and 3′- regions of the Smta-1 gene were amplified by PCR from genomic DNA of a wild-type S. macrospora strain using oligonucleotide pairs 780/781 and 1200/1201 as primers (Fig. 1). The sequences of all PCR primers used in this work are summarized in Table 1. Amplification with 1200 and 1201 generated ApaI sites at the ends. The Smta-1-fragment (1,448 bp) containing the 5′-region of the Smta-1 gene was inserted into vector pDrive (Qiagen, Hilden, Germany) resulting in plasmid psa1prom1. Plasmid psa1term was generated by cloning the 1,523 bp 3′-Smta-1-fragment into pGEM-T (Promega, Mannheim, Germany). The 1.5 kb-ApaI fragment encoding the 3′-Smta-1-region was then inserted into the ApaI site of plasmid psa1prom1. The resulting plasmid psa1-1 contains the 5′-region of the Smta-1 gene adjacent to the 3′-region of the gene. The two sequences are separated by a single SalI restriction site. Next, the 1.4 kb SalI hph cassette of pCB1003 (Carroll et al. 1994) was inserted into the single SalI site of plasmid psa1-1. The resulting plasmid psa1hyg-2 was used as a template to amplify the Smta-1-hph cassette with oligonucleotides 781/1201 as primers. The 4.4 kb PCR fragment obtained was transformed into S. macrospora wild-type strain to generate a knockout of the Smta-1 gene by homologous recombination (Fig. 1).
Transformation of S. macrospora and crosses
Transformation of S. macrospora was performed according to Nowrousian et al. (1999). Fungal protoplasts were transformed with linear PCR fragments amplified from plasmid psa-1hyg-2. Transformants were selected on hygromycin B-containing medium (110 U/ml).
Genomic DNA was isolated as described previously (Pöggeler et al. 1997). Transformants were screened for homologous integration (Fig. 1) by Southern blot analysis. Genomic DNA from 45 hygromycin-resistant transformants was subjected to Southern blot analysis after digestion with BamHI. Among these, one strain lacking wild-type nuclei (corresponding to a 3.6 kb hybridizing fragment), but carrying ΔSmta-1 nuclei (8.8 kb hybridizing fragment) was identified (strain T2, Fig. 1b). Successful homologous recombination was confirmed by PCR amplification. Primers 766 and h3 were used for the 5′-flanking region and primers tC1 and 1203 for the 3′-flanking region of the Smta-1 gene. Fungal transformants are often heterokaryotic, i.e. mycelia carry transformed and non-transformed nuclei. Therefore, homokaryotic single spore isolates of primary transformants carrying the Smta-1 knockout were created by crossing the primary transformant and a spore color mutant fus1-1 and selecting the hygromycin-resistant progeny. The single spore isolate S59403 was used for further analyses.
For complementation of mutant defects, strain S59403 was transformed with cosmid vector D9 containing the mating-type locus of S. macrospora (Pöggeler et al. 1997). Since strain S59403 is already hygromycin-resistant, cosmid D9 was used in cotransformation with plasmid pNat1 containing the nourseothricin-coding gene nat1. The nat1 gene provides a selectable marker for fungal protoplasts in transformation experiments (Krugel et al. 1993). In plasmid pNat1, nat1 is expressed under the control of the gpd promotor and trpC terminator of A. nidulans (Kück and Pöggeler, unpublished). Transformants were selected on nourseothricin-containing medium (50 μg/ml).
Preparation of nucleic acids and hybridization protocols
DNA isolation was performed as described by Pöggeler et al. (1997). RNA and poly(A) RNA was prepared as described previously (Nowrousian et al. 2005). The integrity of RNAs was verified by agarose gel electrophoresis and Northern blot analysis prior to extraction of poly(A) RNA. Southern and Northern blotting were performed and hybridized according to standard techniques (Sambrook et al. 2001) using 32P-labeled DNA probes.
Microarray hybridization
The N. crassa microarrays used in these experiments were prepared from two unigene libraries described previously (Dvorachek et al. 2001; Nowrousian et al. 2003). Microarray hybridization with Cy3- and Cy5-labeled probes were performed as described by Nowrousian et al. (2005). For analysis of the ΔSmta-1 mutant, two independent experiments were carried out; in the first, the mutant cDNA was labeled with Cy3 and the wild type with Cy5; in the second experiment, the dyes were switched. Analyses of TIFF-files from arrays and normalization were performed as described previously (Nowrousian et al. 2005). Clones were defined as regulated differentially in the mutant if they fulfilled one of the following criteria: (1) mutant/wild type expression ratio <0.5 or >2 in the two independent experiments; (2) the mean ratio of transcript levels of the mutant versus the wild type from two independent experiments was <0.5 or >2 with a coefficient of variance <0.5.
Quantitative real-time PCR
For quantitative real-time PCR (RT-PCR), reverse transcription of 1 μg of total RNA was performed as described previously (Nowrousian et al. 2005). Real-time PCR was performed in a DNA Engine Opticon 2 (MJ Research) with qPCR MasterMix for SybrGreen (Eurogentec) in a volume of 20 μl. Each reaction was carried out in triplicate with each primer at 0.3 μM (Table 1). Primer pairs were selected to have melting temperatures of 61–62°C and to yield amplicons of 120–150 bp. PCR conditions were as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, followed by a melting curve analysis. Amplicon size for each primer pair was verified by gel electrophoresis. Mean Ct values (threshold cycles) were calculated from triplicates and used for calculations of expression ratios according to (Pfaffl 2001) with primer-specific efficiencies. The Ct values for an amplicon of the SSUrRNA with primer pair SSU-f/SSU-r were used as a reference for normalization. For each primer pair, real-time experiments were carried out at least twice with biologically independent samples, and the significance of differential expression was verified using REST (Pfaffl et al. 2002).
Microscopy
A Zeiss Axiophot microscope was used for light microscopy. Pictures were captured with an AxioCam camera. Recorded images were edited using Adobe Photoshop 6.0™.
Results
Generation and phenotype analysis of the S. macrospora ΔSmta-1 mutant strain
The S. macrospora mating-type gene Smta-1 is predicted to encode HMG domain DNA binding protein of 288 amino acids (Pöggeler et al. 1997). To determine the role of the putative transcription factor SMTa-1 during vegetative and sexual development of the homothallic ascomycete S. macrospora, we generated a ΔSmta-1 mutant strain. The knockout strain was generated as described in Materials and methods and was confirmed by Southern blot and PCR analysis (Fig. 1). To be sure that the deletion strain bears no traces of wild-type nuclei, a primary transformant (T2) carrying the expected deletion was crossed with the S. macrospora spore-color mutant fus1-1 that produces brown ascospores. From this cross, we selected the hygromycin-resistant homokayotic ΔSmta-1 single spore isolate S59403.
Mutant strain S59403 was examined for defects in vegetative and sexual development. No irregularities in vegetative growth and mycelial morphology were seen when compared to S. macrospora wild type. The development of sexual reproductive structures was analyzed on corn-meal medium. Although the mutant strain showed the formation of ascogonia and protoperithecia, it was unable to perform the transition to mature fruiting bodies (Fig. 2). When grown on fructification medium, ΔSmta-1 produced 240 protoperithecia/cm2, similar to the wild-type strain (231 protoperithecia/cm2). The mean diameter of protoperithecia in mutant and wild type is 55 and 46 μm, respectively. However, fruiting bodies and ascospores that are formed in the wt (39 perithecia/cm2) were never observed in the ΔSmta-1 mutant even after extended incubation time. Thus, the mutant is completely sterile and resembles phenotypically previously described pro mutants which also do not produce any perithecia (Masloff et al. 1999; Nowrousian et al. 2005; Pöggeler and Kück 2004).
Sexual developmental stages of the wild type (wt) and ΔSmta-1 mutant strain. a Strains of S. macrospora were grown on fructification medium for 9 days. Note the complete absence of perithecia in the mutant strain when compared with the wt strain. b Differential interference contrast microscopy identifies ascogonia (wt, ΔSmta-1), protoperithecia (wt, ΔSmta-1), young perithecia (only wt strain) and perithecia (only wt strain). Strains were grown on fructification medium and examined after growth at 25°C for days as indicated
Conventional genetic analysis of more than ten ordered tetrads from a cross of the ΔSmta-1 mutant with the fus1-1 mutant revealed a Mendelian 1:1 segregation of fus and ΔSmta-1 mutant phenotype. The ΔSmta-1 mutant phenotype was always linked with the hygromycin resistance, thus indicating that deletion of the Smta-1 gene is responsible for the mutant phenotype. To further demonstrate that the phenotype was due to mutation of Smta-1, we complemeted the mutant phenotype of ΔSmta-1 by transformation with the wild-type Smta-1 gene. Complemented mutants were able to produce perithecia and ascospores (data not shown).
Transcriptional expression of pheromone and pheromone receptor genes
It was shown previously that the pheromone precursor genes in the heterothallic filamentous ascomycete N. crassa are expressed in a mating type-specific manner. The homologue of the ppg2 gene, mfa-1, is expressed in mat a strains, whereas ccg-4, a homologue of the S. macrospora ppg1 gene, is expressed in mat A strains (Bobrowicz et al. 2002). To examine the effect of the HMG-domain protein SMTa-1 on the expression of the pheromone precursor genes in a homothallic ascomycete, we compared mRNA expression of ppg1 and ppg2 in the wild type versus the ΔSmta-1 strain by means of quantitative RT-PCR experiments.
Real-time PCR experiments were performed three times with RNA prepared from independently grown mycelia. In all experiments, ppg2 transcript levels were strongly down-regulated in the ΔSmta-1 strain compared to the wild type (Fig. 3). On average, mRNA amounts in the mutant strain were reduced about 500-fold. These results indicate that the mating type protein SMTa-1 acts as a positive regulator on the expression of the pheromone precursor gene ppg2. Unlike ppg2, transcript levels of ppg1, pre1 and pre2 varied across RT-PCR experiments, which were up-regulated in some experiments and down-regulated in some. Mean expression ratios for three experiments are given in Fig. 3, but significance analysis with REST (Pfaffl et al. 2002) indicated that none of the three genes is differentially regulated in ΔSmta-1 compared to wild type. Thus, no clear effect of the deletion of Smta-1 on ppg1, pre1 and pre2 expression can be seen; if anything, the genes seem to be deregulated in the absence of Smta-1.
Quantitative real-time PCR analysis of ppg1, ppg2, pre1, and pre2 in ΔSmta-1 and the wild type. Ratios are given as logarithmic values (base 2) of means of three independent experiments, means were calculated with REST. Only ppg2 (white bar) is differentially expressed (P-value 0.001, determined with REST)
Microarray analysis of the ΔSmta-1 mutant
In a previous study, we have found that cross-species microarray hybridizations with S. macrospora targets on N. crassa cDNA microarrays can be used to identify developmentally regulated genes in S. macrospora (Nowrousian et al. 2005). We, therefore, performed cross-species microarray hybridizations with targets derived from the S. macrospora wild type and the ΔSmta-1 mutant on N. crassa cDNA arrays to identify genes that are differentially regulated in the ΔSmta-1 strain. Two hybridization experiments were performed with dye-switching in the second experiment as described previously (Nowrousian et al. 2005).
Of the 2,880 cDNA clones on the arrays, 1,063 gave hybridization signals significantly above background in all hybridizations. For these 1,063 clones, the ratio of mutant- versus wild-type expression could be determined in both independent experiments, and only these clones were used for further analysis. Among the spots that gave signals in all hybridizations, we identified 107 genes (10%) that were up- or down-regulated more than twofold in the ΔSmta-1 compared to the wild type. Of these, 74 genes were down-regulated, and 33 were up-regulated in the mutant strain (see Tables S1 and S2 in the supplementary material).
Among the genes that are down-regulated in ΔSmta-1 is SMU4533, the homologue of NCU04533.2. This gene had already been identified as down-regulated in other developmental mutants of S. macrospora (Nowrousian et al. 2005). Transcriptional expression of SMU4533 in the ΔSmta-1 strain was verified by Northern blot analysis and was found to be strongly reduced (data not shown), thereby confirming that the cross-species microarray data correctly reflect expression patterns as was found before (Nowrousian et al. 2005).
In a previous analysis, we had already identified genes that are differentially regulated in the three developmental mutants pro1, pro11, and pro22 using cross-species microarray hybridizations (Nowrousian et al. 2005). These mutants only produce protoperithecia, and thus phenotypically resemble the ΔSmta-1 strain. Therefore, we analyzed whether any gene is regulated in a similar manner in the pro mutants and the ΔSmta-1 mutant. A combined analysis of regulated genes revealed a total of 176 genes that gave hybridization signals with all four mutant strains as well as the wild type and were regulated differentially in at least one of the mutant strains. Of these, 56 genes were down-regulated and 24 genes up-regulated only in the ΔSmta-1 mutant strain, whereas 33 genes were up-regulated and 36 genes down-regulated in one or more of the pro mutants, but not in the ΔSmta-1 mutant (Table 2 and Table S1–S4 in the supplementary material). However, nine genes were down-regulated and one was up-regulated in all four mutants strains (Table 3). Thus, it seems that there is only a limited overlap among the genes regulated by Smta-1 and the pro genes, respectively (Table 2). This is also reflected in the fact that the pheromone precursor genes ppg1 and ppg2 are up-regulated in all three pro mutants (Nowrousian et al. 2005), whereas ppg2 is down-regulated in the ΔSmta-1 strain (Fig. 3).
The EST sequences corresponding to the genes regulated differentially in ΔSmta-1 were compared to the Neurospora genome database (http://www.broad.mit.edu/annotation/fungi/neurospora/) and to GenBank (http://www.ncbi.nlm.nih.gov/BLAST/) using BLASTX (Altschul et al. 1997). Seventy-five percent of the regulated genes have homologues with known or putative function and were sorted into 10 putative functional categories (Fig. 4). Among these genes are a number of genes that are involved in differentiation processes in other fungi or have homology to signal transduction components. Examples are a homologue to a putative G-protein coupled receptor (NCU04931.2), the G protein γ subunit GNG-1 (NCU00041.2), a putative rho1 GDP-GTP exchange protein (NCU02131.2), an adenylyl cyclase (NCU08377.2), and two serine threonine kinases (NCU06179.2; NCU00685.2). The gng-1 and the adenylyl cyclase encoding gene cr-1 (NCU08377.2) are up-regulated, whereas all other genes are down-regulated in the ΔSmta-1 mutant. Implications of the findings are discussed below.
Functional classification of differentially regulated genes. In microarray experiments with targets prepared from wild type and the S. macrospora ΔSmta-1 mutant, we found 74 to be down- and 33 to be up-regulated in the mutant strain. BLASTX analysis of the 107 regulated genes yielded 80 genes (75%) with a homologue with known or putative function. Twenty-seven genes encode unknown but conserved fungal proteins
Discussion
The S. macrospora ΔSmta-1 mutant strain is unable to form fertile fruiting-bodies
In heterothallic filamentous ascomycetes, two important steps in sexual reproduction are regulated by the mating-type encoded transcription factors: (1) the initial fertilization event mediated by chemo-attraction between reproductive structures of two compatible partners and (2) before karyogamy, the paired migration of nuclei of opposite mating types into the ascogenous hyphae (Coppin et al. 1997; Shiu and Glass 2000). Homothallic fungi have no genetically defined mating type and typically carry one mating-type locus that incorporates homologues from genes of both mating types of related heterothallic species (Pöggeler et al. 1997, Yun et al. 1999). In contrast to the heterothallic ascomycetes, fruiting body development is an apandrous process in the homothallic S. macrospora and lacks the cooperative interaction of two opposite mating-type strains. Sexual development starts with the formation of female gametangia called ascogonia. The ascogenous cells are enveloped by sterile hyphae to form protoperithecia as fruiting body precursors; conidiospores, spermatia, and trichogynes are lacking. After autogamous fertilization (i.e. pairwise fusion of nuclei present within the ascogonium, without cell fusion having taken place), the protoperithecia differentiate inner ascus initials, embedded in sterile paraphyses, and an outer-pigmented peridial tissue. Meiosis and a postmeiotic division lead to eight meiotically derived ascospores in each ascus (Esser 1982; Esser and Straub 1958). Thus, although the fertilization process does not involve two genetically distinct nuclei, the homothallic S. macrospora must ensure that karyogamy occurs exactly between two nuclei. Therefore, it has been proposed that individual nuclei of homothallic fungi could be functionally heterothallic in order to allow nucleus recognition (Coppin et al. 1997; Glass et al. 1990b). This might be achieved through differential expression of mating-type genes (Pöggeler and Kück 2000).
Furthermore, it was not clear whether mata genes are essential for sexual reproduction in homothallic members of the family Sordariaceae, as some homothallic Neurospora species carry only matA specific genes but no mata genes (Glass et al. 1990b; Glass and Smith 1994) and matA strains of the heterothallic Sordaria brevicollis have been shown to produce fruiting bodies with ascospores under certain cultural conditions (Robertson et al. 1998). This study demonstrates that the Smta-1 gene is required for fruiting body development and sexual reproduction in the homothallic S. macrospora. Deletion of Smta-1 encoding a putative HMG-domain transcription factor converts the self-fertile S. macrospora to a self-sterile fungus, no longer able to produce fruiting bodies and ascospores. Similarly, it has been previously demonstrated that deletion of the HMG domain protein encoding MAT1-2 gene, a putative orthologue of the S. macrospora Smta-1, in the homothallic Nectriaceae Gibberella zeae resulted in a self-sterile strain. In G. zeae, the MAT1-2 deletion strain was still able to outcross with a homothallic tester strain (Lee et al. 2003). Likewise, the S. macrospora ΔSmta-1 strain displayed a 1:1 segregation of the hph marker gene when outcrossed with the fus1-1 mutant, thus indicating that the ΔSmta-1 strain has retained the ability to be sexual.
Expression of the pheromone precursor gene ppg2 is controlled by the mating type protein SMTa-1
In ascomycetes, two types of pheromone precursor genes are present in the same genome. For the yeast S. cerevisiae, it has been shown that expression of pheromone genes is directly regulated by transcription factors encoded by the mating-type locus (Herskowitz 1989).
While in heterothallic filamentous ascomycetes such as N. crassa, Magnaporthe grisea, and Podospora anserina, pheromone genes are expressed in a mating-type dependent manner, both genes are expressed in the homothallic S. macrospora having a - and A-specific transcription factors in its mating-type locus (Bobrowicz et al. 2002; Coppin et al. 2005; Pöggeler et al. 1997, 2000; Shen et al. 1999). It was demonstrated for the heterothallic P. anserina that mating-type genes activate their specific pheromone gene and repress the complementary pheromone gene (Coppin et al. 2005).
In our study, the ΔSmta-1 mutant that lacks the a-specific HMG transcription factor SMTa-1 displayed a 500-fold reduction of lipopeptide ppg2 gene expression; thus indicating that SMTa-1 has a direct or indirect impact on the activation of ppg2 gene expression. Similarly, it was demonstrated for the heterothallic N. crassa that the a-specific pheromone gene mfa-1 was not expressed in a mat am1 mutant strain that has a frameshift mutation in the mat a1 coding region (Bobrowicz et al. 2002; Staben and Yanofsky 1990). Previously, it was shown that the N. crassa MATa-1 polypeptide produced in E. coli binds to specific DNA sequences whose core is CTTTG or CTTCG and that DNA binding is a function of the HMG domain (Philley and Staben 1994). The CTTTG motif corresponds to the pheromone response element recognized by the PRF1 HMG transcription factor in the basidiomycete Ustilago maydis (Hartmann et al. 1996). The S. macrospora SMTa-1 protein shows 86.0% identity with the corresponding Mata-1 protein of N. crassa, and the 80-aa HMG domain of both proteins display even 96.0% of identity, suggesting that the cellular functions and the specific DNA-binding recognition sites for both proteins are highly similar, possibly identical. Analysis of the 5′ non-coding region of the S. macrospora ppg2 gene revealed the presence of the above-mentioned HMG-domain protein binding sequences: two CTTTG and four CTTCG sequences. These sequences, similar to the N. crassa binding sites, might thus be binding sites for the SMTa-1 protein, which might then directly activate transcription of ppg2. However, the action of SMTa1 on ppg2 transcription might also be an indirect activation of a transcriptional activator that again might bind the 5′ noncoding sequence of ppg2.
In contrast, no clear effect on the expression of the complementary pheromone gene ppg1 gene has been observed in the S. macrospora ΔSmta-1 strain. Similarly, the deletion of Smta-1 had no clear effect on the transcriptional expression of the pheromone receptor genes pre1 and pre2. This result does not come as a surprise, since transcription of pheromone receptor genes was previously shown to be mating-type independent in the heterothallic N. crassa (Pöggeler and Kück 2001). However, in our study we only analyzed transcriptional expression of ppg1, pre1 and pre2; therefore, a posttranscriptional effect of SMTa-1 on the expression of these genes cannot be ruled out. In P. anserina, it was recently shown that the orthologue of SMTa-1, FPR1, not only activates the expression of the mat- specific pheromone gene but also represses the expression of the complementary mat+ specific pheromone gene posttranscriptionally (Coppin et al. 2005).
The mating type protein SMTa-1 affects the expression of several genes including a common set of developmental genes deregulated in sterile pro mutants
Since a Δppg2 pheromone knockout mutant is still able to form fruiting-bodies and ascospores, the 500-fold down-regulation of ppg2 gene expression cannot be responsible for the sterile phenotype of ΔSmta-1 (Mayrhofer et al. 2006). To examine the role of SMTa-1 in the regulation of other genes that might be target genes of SMTa-1, we performed a cross-species microarray analysis. Following signal intensity quantification and normalization, 107 genes showed significant alterations in mRNA abundance in the ΔSmta-1 mutant. Among these, a BLAST search identified 80 genes with known or putative functions (Fig. 4).
The analysis of three other independent mutants affected in fruiting body formation showed that only a limited set of genes are deregulated in a similar way. Mutants, pro1, pro11 and pro22, carrying mutations in different genes have a developmental block just after protoperithecia formation and, therefore, resemble the phenotype of ΔSmta-1. The pro1 gene encodes a zinc finger transcription factor, pro11 encodes a WD40 repeat protein and pro22 a putative membrane protein (Masloff et al. 1999; Nowrousian et al. 2005; Pöggeler and Kück 2004). Only ten genes were regulated similarly in all mutants (Table 2). Thus, this set of deregulated genes seems to be fruiting body development specific rather than mating-type specific regulated. Among the genes down-regulated in all mutants is NCU04931.2 (Table 3 and Tables S1 and S2 of the supplementary material). The protein encoded by NCU04931.2 is predicted to have seven transmembrane spanning domains (http://www.enzim.hu/hmmtop/) and is a putative homologue of the recently described seven-helix transmembrane protein BTP1 of the asexual plant pathogen Botrytis cinerea. In B. cinerea, this putative G-protein coupled receptor was suggested to play a role in mediation of plant signals and pathogenicity. However, targeted gene replacement of the B. cinerea bpt1 did not result in a phenotype affecting either pathogenicity or sensitivity to chemical stress (Schulze Gronover et al. 2005). Since B. cinerea reproduces only asexually, influences of this receptor on the development of fruiting bodies have not been described yet. The putative receptor may play a role in targeting signals relevant for sexual development in S. macrospora. Down-regulated expression in all mutants was also found for two genes encoding putative transcription factors. A putative bZIP transcription factor (NCU08055.2) and a putative fork head domain transcription factor (NCU06173.2) showed a fourfold to fivefold decrease in mRNA level in ΔSmta-1 and are also down-regulated in the pro mutants (Table 3, supplementary material). NCU08055.2 is a putative homologue of IDI-4, a bZIP transcription factor inducing autophagy and cell death in P. anserina when overexpressed (Dementhon et al. 2004). The second transcription factor gene down-regulated in ΔSmta-1 and the pro mutants, NCU06173.2, is a homologue of the S. pombe forkhead transcription factor Sep1. The S. pombe Sep1 protein regulates mitotic gene transcription and is involved in the cell separation at the end of mitosis. (Buck et al. 2004; Ribar et al. 1999). Another gene that is down-regulated in all mutants is a homologue of NCU02532.2 encoding a putative NEDD8 activating enzyme (Liakopoulos et al. 1998). NEDD8 is an ubiquitin-like molecule that covalently ligates to members of the cullin-family proteins through an enzymatic cascade analogous to ubiquitylation. Previously, it has been demonstrated that the COP9 signalosome (CSN) which interacts with cullin-containing proteins is a regulator of fruiting body development in Aspergillus nidulans (Busch et al. 2003). Among the genes down-regulated in all mutants are, in addition, genes involved in cell wall biogenesis and structure (NCU04533.2 and the trihydroxynaphtalene reductase homologue NCU09390.2, Table 3). The homologue of NCU04533.2 was demonstrated to be highly expressed during sexual development of S. macrospora (Nowrousian, unpublished), while trihydroxynaphthalene reductase is most probably necessary for ascospore and perithecial cell wall pigmentation (Wang and Breuil 2002).
Of the 107 genes that were regulated differentially in the ΔSmta-1 mutant, 56 and 24 genes were only down- and up-regulated, respectively, in the ΔSmta-1 mutant and not in the pro mutants (Table 2, see also Tables S1 and S2 supplementary material). Thus, the regulation of these genes seems to be Smta-1-specific rather than fruiting body specific. These data indicate that Smta-1 is involved in the regulation of pathways distinct from pathways regulated by the pro genes. Of the 80 genes regulated differentially in ΔSmta-1 alone, several encode components of different signal transduction pathways. The gene homologous to NCU00041.2 encodes the γ-subunit GNG-1 of a heterotrimeric G-protein. In the ΔSmta-1 mutant, the gng-1 gene is up-regulated. Recently it was demonstrated that deletion of gng-1 negatively influences levels of Gα proteins (GNA-1, GNA-2, and GNA-3) in plasma membrane fractions isolated from various tissues of N. crassa and leads to a significant reduction in the amount of intracellular cyclic AMP (Krystofova and Borkovich 2005). Thus, up-regulation of gng-1 in the ΔSmta-1 might lead to an up-regulation of genes encoding Gα subunits and adenylate cyclases. Interestingly, our study revealed that indeed the transcript level of adenylate cyclase gene cr-1 (NCU08377.2) was elevated in the ΔSmta-1 mutant. In N. crassa it has been shown previously that the adenylate cyclase together with the Gα subunit GNA-1 contributes to proper fruiting body and ascospore development (Ivey et al. 2002). Similarly, cAMP signaling might be involved in sexual development of S. macrospora.
Another component of a signal transduction pathway exhibiting a more than five-fold decreased mRNA level in the ΔSmta-1 mutant is the homologue to NCU02131.2. The N. crassa gene encodes a putative homologue of the S. pombe guanine nucleotide exchange factor Rgfp3. In S. pombe, Rgfp3 regulates the activity of the small GTPase Rho1p which is involved in β-1,3-glucan biosynthesis and cell integrity during septation (Tajadura et al. 2004). In this context, it is worth noting that in ΔSmta-1 the NCU04431.2 homologue encoding a putative endo-1,3-β-glucanase is one of the most strongly down-regulated genes. In addition to this gene, the expression of further six genes involved in cell wall biosynthesis is deregulated in ΔSmta-1.
The genes affected by SMTa-1 are numerous and include the pheromone gene ppg2 and several genes for components of signaling cascades. Deletion of Smta-1 results in sterility of S. macrospora and thus, similar to other sterile mutants, genes up-regulated during fruiting body development are down-regulated in the ΔSmta-1 mutant. Taken together, the mating-type proteins of homothallic filamentous ascomycetes seem to control sexual reproduction by regulating a variety of essential cellular processes. Further studies are necessary to elucidate the molecular mechanism underlying this complex regulatory network.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bardwell L (2005) A walk-through of the yeast mating pheromone response pathway. Peptides 26:339–350
Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ (2002) The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and circadian clock. Mol Microbiol 45:795–804
Buck V, Ng SS, Ruiz-Garcia AB, Papadopoulou K, Bhatti S, Samuel JM, Anderson M, Millar JBA, McInerny CJ (2004) Fkh2p and Sep1p regulate mitotic gene transcription in fission yeast. J Cell Sci 117:5623–5632
Busch S, Eckert SE, Krappmann S, Braus GH (2003) The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol Microbiol 49:717–730
Carroll AM, Sweigard JA, Valent B (1994) Improved vectors for selecting resistance to hygromycin. Fungal Genet Newslett 41:22
Coppin E, Debuchy R, Arnaise S, Picard M (1997) Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61:411–428
Coppin E, de Renty C, Debuchy R (2005) The function of the coding sequences for the putative pheromone precursors in Podospora anserina is restricted to fertilization. Eukaryotic Cell 4:407–420
Debuchy R (1999) Internuclear recognition:A possible connection between euascomycetes and homobasidiomycetes. Fungal Genet Biol 27:218–223
Dementhon K, Saupe SJ, Clave C (2004) Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Mol Microbiol 53:1625–1640
Dvorachek WHJ, Dolan PL, Nelson MA, Natvig DO (2001) A provisional UniGene clone set based on ESTs from Neurospora crassa. Fungal Genet. Newslett 48:12
Esser K (1982) Cryptogams—Cyanobacteria, algae, fungi, lichens. Cambridge University Press, London
Esser K, Straub J (1958) Genetische Untersuchungen an Sordaria macrospora Auersw.: Kompensation und Induktion bei genbedingten Entwicklungsdefekten. Z Vererbungslehre 89:729–746
Ferreira AV, Saupe S, Glass NL (1996) Transcriptional analysis of the mtA idiomorph of Neurospora crassa identifies two genes in addition to mtA-1. Mol Gen Genet 250:767–774
Ferreira AV, An Z, Metzenberg RL, Glass NL (1998) Characterization of mat A-2, mat A-3 and deltamatA mating-type mutants of Neurospora crassa. Genetics 148:1069–1079
Fraser JA, Heitman J (2004) Evolution of fungal sex chromosomes. Mol. Microbiol 51:299–306
Fraser JA, Heitman J (2005) Chromosomal sex-determining regions in animals, plants and fungi. Curr Opin Genet Dev 15:645–651
Glass NL, Smith ML (1994) Structure and function of a mating-type gene from the homothallic species Neurospora africana. Mol Gen Genet 244:401–409
Glass NL, Vollmer SJ, Staben C, Grotelueschen J, Metzenberg RL, Yanofsky C (1988) DNAs of the two mating-type alleles of Neurospora crassa are highly dissimilar. Science 241:570–573
Glass NL, Grotelueschen J, Metzenberg RL (1990a) Neurospora crassa A mating-type region. Proc Natl Acad Sci USA 87:4912–4916
Glass NL, Metzenberg RL, Raju NB (1990b) Homothallic Sordariaceae from Nature: the absence of strains containing only the a mating type sequence. Exp Mycol 14:274–289
Greener A (1990) New competent cells for highest transformation efficiencies. Strategies 3:5–6
Grosschedl R, Giese K, Pagel J (1994) HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10:94–100
Hartmann HA, Kahmann R, Bölker M (1996) The pheromone response factor coordinates filamentous growth and pathogenicity in Ustilago maydis. EMBO J 15:1632–1641
Herskowitz I (1989) A regulatory hierarchy for cell specialization in yeast. Nature 342:749–757
Ivey FD, Kays AM, Borkovich KA (2002) Shared and independent roles for a Gαi protein and Adenylyl Cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot Cell 1:634–642
Jacobsen S, Wittig M, Pöggeler S (2002) Interaction between mating-type proteins from the homothallic fungus Sordaria macrospora. Curr Genet 41:150–158
Krugel H, Fiedler G, Smith C, Baumberg S (1993) Sequence and transcriptional analysis of the nourseothricin acetyltransferase-encoding gene nat1 from Streptomyces noursei. Gene 15:1127–1131
Krystofova S, Borkovich KA (2005) The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot Cell 4:365–378
Lee J, Lee T, Lee YW, Yun SH, Turgeon BG (2003) Shifting fungal reproductive mode by manipulation of mating type genes:obligatory heterothallism of Gibberella zeae. Mol Microbiol 50:145–152
Liakopoulos D, Doenges G, Matuschewski K, Jentsch S (1998) A novel protein modification pathway related to the ubiquitin system. EMBO J. 17:2208–2214
Masloff S, Pöggeler S, Kück U (1999) The pro1 + gene from Sordaria macrospora encodes a C6 zinc finger transcription factor required for fruiting body development. Genetics 152:191–199
Mayrhofer S, Weber JM, Pöggeler S (2006) Pheromones and pheromone receptors are required for proper sexual development in the homothallic ascomycete Sordaria macrospora. Genetics (in press)
Metzenberg RL, Glass NL (1990) Mating type and mating strategies in Neurospora. Bioessays 12:53–59
Nowrousian M, Masloff S, Pöggeler S, Kück U (1999) Cell differentiation during sexual development of the fungus Sordaria macrospora requires ATP Citrate lyase activity. Mol Cell Biol 19:450–460
Nowrousian M, Duffield GE, Loros JJ, Dunlap JC (2003) The frequency gene is required for temperature-dependent regulation of many clock-controlled genes in Neurospora crassa. Genetics 164:923–933
Nowrousian M, Würtz C, Pöggeler S, Kück U (2004) Comparative sequence analysis of Sordaria macrospora and Neurospora crassa as a means to improve genome annotation. Fungal Genet Biol 41:285–292
Nowrousian M, Ringelberg C, Dunlap JC, Loros JJ, Kück U (2005) Cross-species microarray hybridization to identify developmentally regulated genes in the filamentous fungus Sordaria macrospora. Mol Gen Genomics 273:137–149
Pfaffl MW (2001) A new mathematical model for relative quantification in RT-PCR. Nucleic Acids Res 29:e45
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36
Philley ML, Staben C (1994) Functional analyses of the Neurospora crassa MT a-1 mating type polypeptide. Genetics 137:715–722
Pöggeler S, Risch S, Kück U, Osiewacz HD (1997) Mating-type genes from the homothallic fungus Sordaria macrospora are functionally expressed in a heterothallic ascomycete. Genetics 147:567–580
Pöggeler S (2000) Two pheromone precursor genes are transcriptionally expressed in the homothallic ascomycete Sordaria macrospora. Curr Genet 37:403–411
Pöggeler S (2001) Mating-type genes for classical strain improvements of ascomycetes. Appl Microbiol Biotechnol 56:589–601
Pöggeler S, Kück U (2000) Comparative analysis of the mating-type loci from Neurospora crassa and Sordaria macrospora: identification of novel transcribed ORFs. Mol Gen Genet 263:292–301
Pöggeler S, Kück U (2001) Identification of transcriptionally expressed pheromone receptor genes in filamentous ascomycetes. Gene 280:9–17
Pöggeler S, Kück U (2004) A WD40 repeat protein regulates fungal cell differentiation and can functionally be replaced by the mammalian homologue striatin. Eukaryot Cell 3:232–240
Ribar B, Grallert A, Olah E, Szallasi Z (1999) Deletion of the sep1(+) forkhead transcription factor homologue is not lethal but causes hyphal growth in Schizosaccharomyces pombe. Biochem Biophys Res Commun 263:465–474
Robertson SJ, Bond DJ, Read ND (1998) Homothallism and heterothallism in Sordaria brevicollis. Mycol Res 102:1215–1223
Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schulze Gronover C, Schumacher J, Hantsch P, Tudzynski B (2005) A novel seven-helix transmembrane protein BTP1 of Botrytis cinerea controls the expression of GST-encoding genes, but is not essential for pathogenicity. Mol Plant Pathol 6:243–256
Shen WC, Bobrowicz P, Ebbole DJ (1999) Isolation of pheromone precursor genes of Magnaporthe grisea. Fungal Genet Biol 27:253–263
Shiu PK, Glass NL (2000) Cell and nuclear recognition mechanisms mediated by mating type in filamentous ascomycetes. Curr Opin Microbiol 3:183–188
Staben C, Yanofsky C (1990) Neurospora crassa a mating-type region. Proc Natl Acad Sci USA 87:4917–4921
Tajadura V, Garcia B, Garcia I, Garcia P, Sanchez Y (2004) Schizosaccharomyces pombe Rgf3p is a specific Rho1 GEF that regulates cell wall beta-glucan biosynthesis through the GTPase Rho1p. J Cell Sci 117:6163–6174
Wang HL, Breuil C (2002) A second reductase gene involved in melanin biosynthesis in the sap-staining fungus Ophiostoma floccosum. Mol Genet Genomics 267:557–563
Yun SH, Berbee ML, Yoder OC, Turgeon BG (1999) Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci USA 96:5592–5597
Acknowledgment
The authors wish to thank Silke Nimtz, Gisela Isowitz, Susanne Schlewinski and Swenja Ellßel for excellent technical assistance. This work was funded by the Deutsche Forschungsgemeinschaft PO523/3-1 and SFB480-TPA1 (Bonn, Germany) and the Ruhr-University of Bochum.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Perez-Martin
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Pöggeler, S., Nowrousian, M., Ringelberg, C. et al. Microarray and real-time PCR analyses reveal mating type-dependent gene expression in a homothallic fungus. Mol Genet Genomics 275, 492–503 (2006). https://doi.org/10.1007/s00438-006-0107-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-006-0107-y