Abstract
Overexpression of ETS genes is involved in prostate cancer (PrCa), but there is little information on the non-ERG components of this family. We have investigated ETV1, ETV4, and ETV5 overexpression, with or without PTEN loss, and their association with grade group (GG), pathological stage, focality, and PSA recurrence in PrCa. ETS gene expression was analyzed by qPCR in 104 cases. ETV1 and PTEN immunohistochemistry was assessed in TMA sections from 194 additional cases (PSMAR-Biobank, Barcelona, Spain). ETS mRNA overexpression was found in 23.1%, being ETV1 the most frequently overexpressed (18.3%). ETV1 protein overexpression was detected in 30.4% cases (moderate in 19.6%, strong in 10.8%). PTEN protein expression loss was detected in 36.1% cases and was not associated with ETV1. Strong-moderate ETV1 protein overexpression reaches its highest values in GG3–4, whereas its negativity was significantly more common in GG1 tumors (p = 0.034). ETV1-overexpressing tumors were more often unifocal (p = 0.0007) and high stage (p = 0.032). PTEN loss was less common in GG1 (p = 0.012) and showed a trend to be less frequent in pT2 (p = 0.062) tumors. Strong ETV1 immunostaining (histoscore > 177) was associated with shorter time to PSA recurrence in the univariate (p = 0.002) and in the multivariate analysis (p = 0.018). Moreover, when strong ETV1 overexpression was not combined with PTEN loss, its association with PSA recurrence was even stronger (p = 0.0004). In conclusion, non-ERG ETS overexpression, particularly ETV1 overexpression, has a non-negligible role in PrCa. Strong ETV1 protein expression has a negative impact on prostate cancer outcome that seems to be independent of PTEN status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mutations in classic oncogenes and tumor suppressor genes are relatively uncommon in prostate carcinoma (PrCa) [1], while fusions of androgen-regulated promoters with members of the ETS family of transcription factors are a specially common event [2]. Seven PrCa subtypes defined by ETS fusions or mutations in driver genes were established by The Cancer Genome Atlas (TCGA) research network [3], and a remarkable molecular diversity has been reported among the ETS-fused prostate tumors.
TMPRSS2-ERG rearrangement has been extensively reported in about 50% of prostate tumors [4] and is the main cause of ERG overexpression [5, 6]. ERG has been shown to have oncogenic capacity [7] and to promote prostatic dedifferentiation [8], as well as tumor initiation and progression [9]. ERG has also SLC45A3 [10] and NDRG1 [11] as less frequent 5′ partners. There is evidence that SLC45A3-ERG fusion is added to some TMPRSS2-ERG rearranged tumors, resulting in higher ERG overexpression [12] and worse prognosis [13]. In addition, PTEN loss is considered a late event that cooperates with ERG overexpression to promote tumor progression [12, 14,15,16]. TMPRSS2 is also fused, less frequently, with other members of the ETS family, namely ETV1, ETV4, and ETV5. Chromosomal translocations affecting ETV1 or ETV4 have been involved in prostate cancer [17].
ETV1 is overexpressed in 5–10% of PrCa [4, 18]. Only in some of these cases overexpression is related to rearrangements involving TMPRSS2 and ETV1 [19]. The reported percentage of ETV4 overexpression ranges from 2% in an American series to 30% in a Chinese cohort [20, 21]. ETV1 and ETV4 are supposed to have overlapping functions in late prostatic carcinogenesis [22]. In this regard, they have been related with dissemination and metastasis [19, 23]. Moreover, ETV4 is needed for anchorage-independent growth, proliferation, and epithelial-mesenchymal transition [24]. Less information is available about ETV5 overexpression in PrCa, but a frequency of 1.5% is documented. Fusions of ETV5 with TMPRSS2 or SLC45A3 have been reported as the main mechanism for ETV5 overexpression [25]. The effect of ETV5 overexpression has been related to the invasive capacity of PrCa cells, but not to proliferation.
In summary, there are limited data on the role of the non-ERG ETS genes (ETV1, ETV4, and ETV5) in the pathogenesis of PrCa. The aim of the present work has been to further understand the role of ETV1, ETV4, and ETV5 in prostatic carcinogenesis and their relationship with PTEN loss, as well as with the clinical-pathological features of tumors and PSA recurrence.
Materials and methods
Patients and tumor samples
Two independent cohorts, including prostate tumors from laparoscopic radical prostatectomy, were selected retrospectively from the files of the Parc de Salut MAR Biobank (MARBiobanc, Barcelona, Spain): (1) The first one consisted of 104 frozen prostate tumors and 3 frozen non-tumor prostate samples from cases with nodular hyperplasia. (2) The second cohort that was used for the construction of the TMA blocks consisted of 194 formalin fixed, paraffin embedded (FFPE) prostate tumors. The distribution of the grade groups (GG), tumor stage (pT), uni- or multifocality, and pre-operative PSA are summarized in Table 1.
Total RNA extraction and retrotranscription
Representative tumor areas containing at least 50% and the vast majority of cases with 70 to 90% of tumor cells were selected and manually microdissected from frozen tissues. Standard hematoxylin and eosin (H&E) slides served as templates. About 10–15 sections of 10 μm from the tumor area were cryopreserved with Ultraspec (Biotecx Laboratories, Houston, TX, USA) at − 80 °C. Total RNA was extracted in tumor and non-tumor samples with the RNeasy Mini kit (Qiagen, Cathsworth, CA, USA). RNA purity and quality were assessed with the Nanodrop® ND-100 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and the Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA).
cDNA was synthesized using 1 μg of total RNA and Superscript IV Kit (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA, USA) according to the manufacturer’s instructions.
ETS genes mRNA expression analysis by qPCR
ETV1, ETV4, and ETV5 expression was analyzed from cDNA in the 104 frozen prostate tumors and the 3 non-tumor samples. qPCR reactions were done with ABI PRISM 7500 Sequence Detection System and Taqman® Gene Expression Assays (Applied Biosystems, Thermo Fisher Scientific, Carlsbad, CA, USA): ETV1 (Hs00951951_m1), ETV4 (Hs00383361_g1), ETV5 (Hs00927557_m1), and GAPDH (4310884E–NM_002046.3).
Samples were run in triplicate, and the mean value was calculated for each case. GAPDH gene was used as internal control to normalize levels of mRNA expression. The three non-tumor prostate samples were used to determine normal expression levels of the ETS genes, and overexpression was considered for values of 2^(− ∆∆Ct) ≥ 2.6. For the cutoff definition, we tried to perform ROC curves, but clear cutoff values for high and low levels were not found with this method. Thus, the median values were taken as reference points, several cutoff values were tested, and the cutoff showing the best discrimination power between the different clinical-pathological groups was selected.
Immunohistochemistry of ETV1 and PTEN
Nine tissue microarrays (TMAs) were constructed as previously reported [16]. The dominant tumor was sampled in all cases. Whenever possible, several cores were taken from different regions. In addition, when the size and thickness of the focus allowed it, secondary foci where also sampled. Thus, smaller foci (< 3 mm in diameter) were not included for technical reasons. For the present study, all the cores were analyzed, but only that with the highest grade was included in the evaluation. Likewise, to analyze the relationship between expression of ETV1 and PTEN and the clinical-pathological parameters, the core with the highest grade and containing the maximum number of altered proteins was selected. The immunohistochemical expression of ETV1 and PTEN was assessed in 469 cores from the 230 cases. Information on both proteins was available in 194 cases. Results of PTEN have been previously reported [16], and we have incorporated them in the present study for comparison with ETV1.
Immunohistochemical staining of PTEN was carried out as previously reported [16]. For ETV1 immunostaining, the primary rabbit anti-ETV1 polyclonal antibody (clone PA5-41484 Thermo Fisher Scientific, Life Technologies Corporation, Carlsbad, CA, USA) and the Dako Envision+ System-HRP (Dako, Glostrup, Denmark) were used. ETV1 expression was cytoplasmic or nuclear. Both were assessed and quantified with the histoscore system. ETV1 expression was graded as: negative = 0, weak = 1, moderate = 2, and strong = 3. The histoscore was the sum of the product of the staining intensity and the corresponding tumor percentage (histoscore = [1 × (%1+ cells)] + [2 × (%2+ cells)] + [3 × (%3+ cells)]). A final score of 0 was considered negative expression; there were no cases with values between 1 and 4; 5–99 was considered weak expression, 100–176 moderate overexpression, and ≥ 177 strong overexpression. Endothelial cells were used as internal positive control and stromal cells as internal negative control. In addition, a TMA containing many different tissues was used that included testes as a positive external control. In both the problem TMA blocks and the control TMA block, additional sections were immunostained, replacing the primary antibody with buffer solution as additional control for the specificity of the immunostaining.
Statistical analysis
Categorical variables were presented as frequencies and percentages. The Fisher or chi-square tests were used to compare these categorical variables between groups. For the statistical analysis, SPSS version 15.0 statistical software (SPSS Inc., Chicago, IL, USA) was used. p values less than 0.05 were considered statistically significant.
Survival analysis
Patients were followed clinically at regular intervals of 3 months for the first year and every 6 months the subsequent years, with PSA values available for each follow-up visit. Patients were censored at their last clinical follow-up or when an increase in serum PSA > 0.2 ng/ml was detected (i.e., two consecutive increases) [26].
PSA recurrence was analyzed using Kaplan-Meier (log rank) in two independent cohorts of patients. The first consisted of 94 cases (10 were lost for follow-up) with available ETV1, ETV4, and ETV5 quantitative mRNA expression analysis. In this group, the median follow-up was 49 months (1–130). The second cohort included 174 cases (20 were lost for follow-up) with available ETV1 and PTEN immunostaining analysis. In this group, the mean follow-up was 102.5 months (1–212). A multivariate Cox proportional hazard regression analysis was used to assess the association between strong ETV1 cytoplasmic overexpression and PSA-recurrence risk, with their corresponding hazard ratio (HR), 95% confidence intervals (CI), and p values, after adjusting for other prognostic variables (grade group, tumor stage, tumor focality, and pre-operative PSA). In this analysis, 37 patients were lost for follow-up.
Results
Analysis of ETS genes (ETV1, ETV4, and ETV5) mRNA quantitative expression
ETV1, ETV4, and ETV5 mRNA expression was analyzed by qPCR in all 104 frozen prostate tumor samples. In 24 tumors (23.1%), we found overexpression of one or more ETS genes, while the remaining 80 tumors had basal levels of all ETS genes. ETV1 was the most frequently overexpressed of them (19 of 104; 18.3%), followed by ETV4 (9 of 104; 8.6%), and ETV5 (3 of 104; 2.8%). Only one ETS gene was overexpressed in 79.2% (19 of 24) cases, while 20.8% (5 of 24) overexpressed more than one. Isolated ETV1 overexpression was the most frequent event (14.4%; 15 of 104), followed by isolated ETV4 (3.8%; 4 of 104), and ETV5 was always overexpressed in combination with other ETS genes.
Relationship of ETS mRNA overexpression with grade group and tumor stage
Regarding the grade group tumor classification, ETV1 overexpression was seen in 19.2% (5 of 26) GG1, 21.2% (7 of 33) GG2, 14.3% (3 of 21) GG3, 23.1% (3 of 13) GG4, and 9.1% (1 of 11) GG5 tumors (Pearson X2, p = 0.902). ETV4 overexpression was not seen in GG5, while it was detected in 15.4% (4 of 26) GG1, 3.1% (1 of 33) GG2, 14.3% (3 of 21) GG3, and in 7.7% (1 of 13) GG4 tumors (Pearson X2, p = 0.331). ETV5 overexpression was only detected in 7.7 (2 of 26) GG1 and 7.7% (1 of 13) GG4 tumors (Pearson X2, p = 0.199).
Finally, lack of ETS gene (ETV1, ETV4, or ETV5) overexpression was detected in 76.9% (20 of 26) GG1, 78.8% (26 of 33) GG2, 71.4% (15 of 21) GG3, 69.2% (9 of 13) GG4, and 90.9% (10 of 11) GG5 tumors (Fisher’s exact test, p = 0.751). Comparing the proportion of GG5 vs GG1–4 (70 of 93; 75.3%) cases lacking ETS gene overexpression, this was higher in the former, but without statistical differences (Fisher’s exact test, p = 0.450).
Regarding tumor stage classification, ETV1 overexpression was found in 18.7% (6 of 32) pT2 tumors and in 18.1% (13 of 72) pT3 tumors (Pearson X2, p = 0.933). ETV4 overexpression was found in 12.5% (4 of 32) pT2 tumors and in 6.9% (5 of 72) pT3 tumors (Fisher’s exact test, p = 0.452). ETV5 overexpression was found in 6.2% (2 of 32) pT2 tumors and in 1.4% (1 of 72) pT3 tumors (Fisher’s exact test, p = 0.223). Finally, lack of ETS gene overexpression was detected in 78.1% (25 of 32) pT2 tumors and in 76.4% (55 of 72) pT3 tumors (Pearson X2, p = 0.845).
Analysis of ETV1 and PTEN immunostaining in TMA
For ETV1, we have found almost exclusively cytoplasmic staining and only one case showing nuclear expression (Fig. 1b, e). Thus, only the cytoplasmic expression pattern of ETV1 was considered. The positive controls (endothelial cells as internal control and testes in the control TMA) showed nuclear and cytoplasmic staining. The fibroblast and muscle cells in the cases were negative. The negative control without ETV1 antibody was completely devoid of cytoplasmic or nuclear stain. Therefore, although it is not usually reported in the literature, these controls support the real presence of ETV1 in the cytoplasm of tumor cells and allow to exclude a non-specific reaction. 27.5% (55 of 194) of tumors were totally negative, and 41.2% (80 of 194) showed weak expression. Overexpression was classified as moderate or strong immunostaining: 19.6% (38 of 194) of tumors presented moderate overexpression and 10.8% (21 of 194) strong overexpression (Figs. 1 and 2).
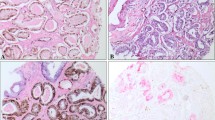
ETV1 immunostaining in overexpressing PrCa. a and d Case 1: prostate adenocarcinoma, GG4, with nuclear histoscore = 0 and cytoplasmic histoscore = 140 (moderate overexpression). b and e Case 2: prostate adenocarcinoma, GG1, with nuclear histoscore = 105 and cytoplasmic histoscore = 215 (strong overexpression). c and f Case 3: prostate adenocarcinoma, GG2, with nuclear histoscore = 0 and cytoplasmic histoscore = 185 (strong overexpression). Original magnification ×50 (a, b, and c) and ×200 (d, e, and f)
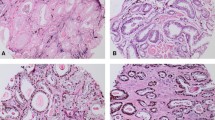
ETV1 immunostaining in non-overexpressing PrCa. a and d Case 4: prostate adenocarcinoma, GG5, with nuclear histoscore = 0 and cytoplasmic histoscore = 80 (weak expression). b and e Case 5: prostate adenocarcinoma, GG3, with nuclear and cytoplasmic histoscore = 0. c and f Case 6: prostate adenocarcinoma, GG5, with nuclear and cytoplasmic histoscore = 0. Endothelial cells were used as controls. Original magnification ×50 (a, b, and c) ×200 (d, e, and f)
PTEN expression loss was detected in 70 cases (36.1%). Eighteen of the 59 (30.5%) cases with moderate or strong ETV1 overexpression, and 52 of the 135 (38.5%) with negative or weak ETV1 showed PTEN loss (Pearson X2, p = 0.285). Thus, ETV1 overexpression was not associated with PTEN loss.
Relationship of ETV1 and PTEN immunostaining with grade group, tumor stage, and tumor focality
Regarding grade group tumor classification, ETV1 overexpression (moderate and strong) and weak or negative stain are shown in Fig. 3a. ETV1 overexpression was found in 23.3% (7 of 30) GG1, 29.7% (22 of 74) GG2, 40.9% (9 of 22) GG3, 44.8% (13 of 29) GG4, and 20.5% (8 of 39) GG5 tumors, with highest values in GG3 and GG4. Weak ETV1 expression was seen in 33.3% (10 of 30) GG1, 36.5% (27 of 74) GG2, 31.8% (7 of 22) GG3, 48.3% (14 of 29) GG4, and 56.4% (22 of 39) GG5 tumors. In contrast, lack of expression was found in 43.3% (13 of 30) GG1, 33.8% (25 of 74) GG2, 27.3% (6 of 22) GG3, 6.9% (2 of 29) GG4, and 23.1% (9 of 39) GG5 tumors, being significantly higher in GG1 (Pearson X2, p = 0.034).
Regarding stage, ETV1 overexpression (moderate or strong) was found in 26.5% (35 of 132) pT2 and in 39.6% (21 of 53) pT3, weak expression in similar percentages in pT2 (53 of 132; 40.1%) and pT3 (24 of 53; 45.3%) tumors, and lack of expression in 33.3% (44 of 132) pT2 and 15.1% (8 of 53) pT3 cases (Pearson X2, p = 0.032). Thus, ETV1 overexpression seems to be associated with high stage and negative expression with low stage (Fig. 3b).
Finally, regarding tumor focality, ETV1 overexpression (moderate or strong) was found in 22.6% (30 of 133) of the multifocal tumors and 50% (26 of 52) of cases with a single tumor focus; weak ETV1 expression was higher in multifocal (64 of 133; 48.1%) than in unifocal (13 of 52; 25%) PrCa, and lack of expression was present in similar proportion of multifocal (39 of 133; 29.3%) and unifocal tumors (13 of 52; 25%) (Pearson X2, p = 0.0007). Thus, ETV1 overexpression seems to be associated with cases with a single tumor focus and weak expression with multifocal cancers (Fig. 3c).
PTEN protein loss was detected in 13.3% (4 of 30) GG1, 39.2% (29 of 74) GG2, 40.9% (9 of 22) GG3, 34.5% (10 of 29) GG4, and 46.2% (18 of 39) GG5 tumors, thus being significantly lower in GG1 (Pearson X2, p = 0.012). Regarding stage, PTEN protein loss was detected in 32.6% (43 of 132) pT2 and in 47.2% (25 of 53) pT3 tumors (Pearson X2, p = 0.0623). Thus, PTEN protein loss tends to associate with high stage tumors. Finally, PTEN protein loss was similar in multifocal (50 of 133; 37.6%) and unifocal (19 of 52; 36.5%) cases (Pearson X2, p = 0.893).
ETS overexpression, PTEN loss, and PSA recurrence analysis in PrCa
Kaplan-Meier analysis for PSA recurrence was performed in the two independent cohorts of the study.
In the cohort with ETS quantitative mRNA data, PSA recurrence was detected in 24/94 cases (25.5%): 4 GG1 (16%), 8 GG2 (25.8%), 4 GG3 (19.1%), 3 GG4 (37.5%), and 5 GG5 (55.6%) tumors. Kaplan-Meier analysis was performed for ETV1 overexpression vs basal ETV1 levels (log rank test, p = 0.733), ETV4 overexpression vs basal ETV4 levels (log rank test, p = 0.905), ETV5 overexpression vs basal ETV5 levels (log rank test, p = 0.306), and overall ETS overexpression vs non-overexpression (log rank test, p = 0.274).
In the TMA protein expression cohort, PSA recurrence was detected in 37/174 cases (21.3%): 6 GG1 (23.1%), 13 GG2 (19.7%), 1 GG3 (4.8%), 7 GG4 (29.2%), and 10 GG5 (27%) tumors. Kaplan-Meier analysis was performed for moderate and strong ETV1 overexpression (≥ 100) vs negative or weak expression (log rank test, p = 0.460) and for strong ETV1 overexpression (≥177) vs negative, weak, or moderate expression (log rank test, p = 0.002) (Fig. 4a). Strong ETV1 overexpression with wt PTEN was associated with the worst outcome, followed by combined strong ETV1 overexpression and PTEN loss. Negative, weak, or moderate ETV1 expression, regardless of PTEN status, had the best survival (log rank test, p = 0.0004) (Fig. 4b).
PSA recurrence (Kaplan-Meier) plot for patients with strong cytoplasmic ETV1 overexpression vs patients with negative, weak, or moderate expression of ETV1 (log rank test, p = 0.002) (a) and for patients with strong cytoplasmic ETV1 overexpression/wt PTEN vs strong ETV1 overexpression/PTEN loss vs negative, weak, or moderate ETV1 expression/PTEN loss vs negative, weak, or moderate ETV1 expression/wt PTEN (log rank test, p = 0.0004) (b)
In addition, in the multivariate Cox proportional hazard regression analysis (Table 2), strong ETV1 cytoplasmic overexpression was significantly associated to a 2.714 times higher risk of PSA recurrence (HR 2.714, p = 0.018). None of the remaining parameters (GG, tumor stage, tumor focality, and pre-operative PSA values) were statistically associated with a higher risk of PSA recurrence.
Discussion
Fusions involving androgen-regulated genes and members of the ETS family are the most common molecular abnormality [1, 2] and are considered to drive carcinogenesis in many prostate tumors [7, 17]. Fusion-positive and fusion-negative PrCa have been proposed as two main different subtypes of the disease [3]. Fusion transcripts involving TMPRSS2 and ETS genes have been suggested to be good markers of PrCa aggressiveness, with variable results in different series [27,28,29,30].
Although the relevance of ERG rearrangements in PrCa is well known, the role of other ETS genes, as ETV1, ETV4, and ETV5 deserves further investigation. We designed this study to gain further insight on the potential involvement of these genes in prostate carcinogenesis.
In our series, ETS gene overexpression was a relatively frequent event, involving 23.1% of clinically localized prostate tumors. ETV1 was the most frequently overexpressed gene, ETV4 was the second, and ETV5 was uncommon. Several papers have proposed that rearrangements between TMPRSS2 and different ETS genes could be mutually exclusive [22, 31]. In this regard, our results show that ETS genes are overexpressed as an isolated event in most cases.
In the TMA cohort, we have analyzed the protein expression of ETV1 and PTEN in an independent series of 194 prostate tumors. The almost exclusively cytoplasmic pattern of ETV1 expression found in our cases needs a special comment. The Human Protein Atlas (https://www.proteinatlas.org) shows ETV1 nuclear sub-localization, but their data are not related to normal or neoplastic prostate tissue, as they are restricted to cell lines (PC-3 and RH-30). There are no previous reports on immunohistochemical expression of ETV1 in PrCa. RNA in situ hybridization has been used to detect ETV1 fusions in clinical specimens [32], but this cannot be equated to the protein expression pattern. In other tumor types, both nuclear immunostaining and cytoplasmic immunostaining have been reported [33]. In our series of tumors, ETV1 cytoplasmic proteins were detected moderately overexpressed in 19.6% of tumors and strongly overexpressed in 10.8%. By contrast, the positive controls showed consistent nuclear expression, occasional cytoplasmic expression, and background staining in some cases, while the negative controls with omission of the primary antibody were completely negative. Loss of PTEN expression has extensively been considered a crucial event in PrCa [14, 15], and it was found in 36.1% of our tumors. Many studies have reported it more often in cases with ERG overexpression, and both genes cooperate in PrCa progression. By contrast, our results seem to indicate that PTEN expression loss is not associated with ETV1 overexpression, suggesting that the interaction between PTEN and ETV1 would not be decisive for PrCa progression in this pathway.
The TMA results in the present study indicate that PTEN protein expression loss is rarely found in the lowest GG and in pT2 stage tumors, in accordance with many previous reports of a similar relationship between PTEN loss and advanced prostate cancer [34]. Conversely, PTEN protein loss was similarly found in both multifocal and unifocal prostate tumors.
To the best of our knowledge, our data show for the first time that cytoplasmic ETV1 overexpression is significantly more common in GG3–4, and interestingly, lack of expression is significantly more common in GG1. Also, that ETV1 overexpression seems to be associated with high stage, whereas negative expression is more frequent in low stage tumors. Finally, we also show that ETV1 overexpression seems to be much more common in unifocal cases, whereas weak expression is more common in multifocal tumors. A possible explanation for this difference could be that unifocal and multifocal PrCa evolved through different carcinogenetic pathways, but this conclusion cannot be taken from the results of the present study alone.
It is noteworthy that strong ETV1 immunostaining was associated with shorter time to PSA recurrence in the univariate analysis. In addition, strong ETV1 overexpression was also associated in the multivariate analysis with a higher risk of PSA-recurrence with an overall 2.714-fold increase compared with tumors in which ETV1 expression was negative, weak, or moderate. Moreover, strong ETV1 overexpression was even more strongly associated with this parameter in PTEN wt cases, compared with the combinations strong ETV1 overexpression/PTEN loss, negative, weak or moderate ETV1 expression/PTEN loss, and negative, weak, or moderate ETV1 expression/wt PTEN. In conclusion, strong ETV1 protein expression has a negative impact on prostate cancer outcome that seems to be independent of PTEN status. All these conclusions stress the relevant role of ETV1 in a subset of aggressive prostate tumors. Thus, ETV1 deserves further research, both to better understand its role in prostate carcinogenesis and also as a clinically helpful marker for prognostic and therapeutic stratification of prostate cancer patients.
References
Kaffenberger SD, Barbieri CE (2016) Molecular subtyping of prostate cancer. Curr Opin Urol 26:213–218
Arora K, Barbieri CE (2018) Molecular subtypes of prostate cancer. Curr Oncol Rep 20:55–58
Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, Annala M, Aprikian A, Armenia J, Arora A, Auman JT, Balasundaram M, Balu S, Barbieri CE, Bauer T, Benz CC, Bergeron A, Beroukhim R, Berrios M, Bivol A, Bodenheimer T, Boice L, Bootwalla MS, Borges dos Reis R, Boutros PC, Bowen J, Bowlby R, Boyd J, Bradley RK, Breggia A, Brimo F, Bristow CA, Brooks D, Broom BM, Bryce AH, Bubley G, Burks E, Butterfield YSN, Button M, Canes D, Carlotti CG, Carlsen R, Carmel M, Carroll PR, Carter SL, Cartun R, Carver BS, Chan JM, Chang MT, Chen Y, Cherniack AD, Chevalier S, Chin L, Cho J, Chu A, Chuah E, Chudamani S, Cibulskis K, Ciriello G, Clarke A, Cooperberg MR, Corcoran NM, Costello AJ, Cowan J, Crain D, Curley E, David K, Demchok JA, Demichelis F, Dhalla N, Dhir R, Doueik A, Drake B, Dvinge H, Dyakova N, Felau I, Ferguson ML, Frazer S, Freedland S, Fu Y, Gabriel SB, Gao J, Gardner J, Gastier-Foster JM, Gehlenborg N, Gerken M, Gerstein MB, Getz G, Godwin AK, Gopalan A, Graefen M, Graim K, Gribbin T, Guin R, Gupta M, Hadjipanayis A, Haider S, Hamel L, Hayes DN, Heiman DI, Hess J, Hoadley KA, Holbrook AH, Holt RA, Holway A, Hovens CM, Hoyle AP, Huang M, Hutter CM, Ittmann M, Iype L, Jefferys SR, Jones CD, Jones SJM, Juhl H, Kahles A, Kane CJ, Kasaian K, Kerger M, Khurana E, Kim J, Klein RJ, Kucherlapati R, Lacombe L, Ladanyi M, Lai PH, Laird PW, Lander ES, Latour M, Lawrence MS, Lau K, LeBien T, Lee D, Lee S, Lehmann KV, Leraas KM, Leshchiner I, Leung R, Libertino JA, Lichtenberg TM, Lin P, Linehan WM, Ling S, Lippman SM, Liu J, Liu W, Lochovsky L, Loda M, Logothetis C, Lolla L, Longacre T, Lu Y, Luo J, Ma Y, Mahadeshwar HS, Mallery D, Mariamidze A, Marra MA, Mayo M, McCall S, McKercher G, Meng S, Mes-Masson AM, Merino MJ, Meyerson M, Mieczkowski PA, Mills GB, Shaw KRM, Minner S, Moinzadeh A, Moore RA, Morris S, Morrison C, Mose LE, Mungall AJ, Murray BA, Myers JB, Naresh R, Nelson J, Nelson MA, Nelson PS, Newton Y, Noble MS, Noushmehr H, Nykter M, Pantazi A, Parfenov M, Park PJ, Parker JS, Paulauskis J, Penny R, Perou CM, Piché A, Pihl T, Pinto PA, Prandi D, Protopopov A, Ramirez NC, Rao A, Rathmell WK, Rätsch G, Ren X, Reuter VE, Reynolds SM, Rhie SK, Rieger-Christ K, Roach J, Robertson AG, Robinson B, Rubin MA, Saad F, Sadeghi S, Saksena G, Saller C, Salner A, Sanchez-Vega F, Sander C, Sandusky G, Sauter G, Sboner A, Scardino PT, Scarlata E, Schein JE, Schlomm T, Schmidt LS, Schultz N, Schumacher SE, Seidman J, Neder L, Seth S, Sharp A, Shelton C, Shelton T, Shen H, Shen R, Sherman M, Sheth M, Shi Y, Shih J, Shmulevich I, Simko J, Simon R, Simons JV, Sipahimalani P, Skelly T, Sofia HJ, Soloway MG, Song X, Sorcini A, Sougnez C, Stepa S, Stewart C, Stewart J, Stuart JM, Sullivan TB, Sun C, Sun H, Tam A, Tan D, Tang J, Tarnuzzer R, Tarvin K, Taylor BS, Teebagy P, Tenggara I, Têtu B, Tewari A, Thiessen N, Thompson T, Thorne LB, Tirapelli DP, Tomlins SA, Trevisan FA, Troncoso P, True LD, Tsourlakis MC, Tyekucheva S, van Allen E, van den Berg DJ, Veluvolu U, Verhaak R, Vocke CD, Voet D, Wan Y, Wang Q, Wang W, Wang Z, Weinhold N, Weinstein JN, Weisenberger DJ, Wilkerson MD, Wise L, Witte J, Wu CC, Wu J, Wu Y, Xu AW, Yadav SS, Yang L, Yang L, Yau C, Ye H, Yena P, Zeng T, Zenklusen JC, Zhang H, Zhang J, Zhang J, Zhang W, Zhong Y, Zhu K, Zmuda E (2015) The molecular taxonomy of primary prostate cancer. Cell 163:1011–1025
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM (2005) Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310:644–648
Clark J, Cooper C (2009) ETS gene fusions and prostate cancer. Nat Rev Cancer 6:429–439
Font-Tello A, Juanpere N, De Muga S, Lorenzo M, Lorente JA, Fumado L, Serrano L, Serrano S, Lloreta J, Hernández S (2015) Association of ERG and TMPRSS2-ERG with grade, stage, and prognosis of prostate cancer is dependent on their expression levels. Prostate 75:1216–1226
Sreenath TL, Dobi A, Petrovics G, Srivastava S (2011) Oncogenic activation of ERG: a predominant mechanism in prostate cancer. J Carcinog 10:37–49
Sun C, Dobi A, Mohamed A, Li H, Thanagapazham RL, Furusato B, Shaheduzzaman S, Tan SH, Vaidyanathan G, Whithman E, Hawksworth DJ, Chen Y, Nau M, Patel V, Vahey M, Gutkind JS, Sreenath T, Petrovics G, Sesterhenn IA, McLeod DG, Srivastava S (2008) TMPRSS2-ERG fusion, a common genomic alteration in prostate cancer activates C-MYC and abrogates prostate epithelial differentiation. Oncogene 27:5348–5353
Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON (2009) ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci U S A 106:12465–12470
Esgueva R, Perner S, J LaFargue C, Scheble V, Stephan C, Lein M, Fritzsche FR, Dietel M, Kristiansen G, Rubin MA (2010) Prevalence of TMPRSS2-ERG and SLC45A3-ERG gene fusions in a large prostatectomy cohort. Mod Pathol 23:539–546
Pflueger D, Rickman DS, Sboner A, Perner S, J Lafargue C, Svensson MA, Moss BJ, Kitabayashi N, Pan Y, de la Taille A, Kuefer R, Tewari AK, Demichelis F, Chee MS, Gerstein MB, Rubin MA (2009) N-myc downstream regulated gene 1 (NDRG1) is fused to ERG in prostate cancer. Neoplasia 11:804–811
Hernández S, Font-Tello A, Juanpere N, de Muga S, Lorenzo M, Salido M, Fumadó L, Serrano L, Cecchini L, Serrano S, Lloreta J (2016) Concurrent TMPRSS2-ERG and SLC45A3-ERG rearrangements plus PTEN loss are not found in low grade prostate cancer and define an aggressive tumor subset. Prostate 76:854–865
Perner S, Rupp NJ, Braun M, Rubin MA, Moch H, Dietel M, Wernert N, Jung K, Stephan C, Kristiansen G (2013) Loss of SLC45A3 protein (prostein) expression in prostate cancer is associated with SLC45A3-ERG gene rearrangement and an unfavorable clinical course. Int J Cancer 132:807–812
Han B, Mehra R, Lonigro RJ, Wang L, Suleman K, Menon A, Palanisamy N, Tomlins SA, Chinnaiyan AM, Shah RB (2009) Fluorescence in situ hybridization study shows association of PTEN deletion with ERG rearrangement during prostate cancer progression. Mod Pathol 22:1083–1093
King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, Gerald WL, Sawyers CL (2009) Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 41:524–526
Hernández-Llodrá S, Juanpere N, de Muga S, Lorenzo M, Gil J, Font-Tello A, Agell L, Albero-González R, Segalés L, Merino J, Serrano L, Fumadó L, Cecchini L, Lloreta-Trull J (2017) ERG overexpression plus SLC45A3 (prostein) and PTEN expression loss: strong association of the triple hit phenotype with an aggressive pathway of prostate cancer progression. Oncotarget 26:74106–74118
Oh S, Shin S, Janknecht R (2012) ETV1, 4, and 5: an oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 1826:1–12
Hermans KG, Van Der Korput HA, Van Marion R, Van de Wijngaart DJ, Ziel-van der Made A, Dits NF, Boormans JL, Van der Kwast TH, Van Dekken H, Bangma CH, Korsten H, Kraaij R, Jenster G, Trapman J (2008) Truncated ETV1, fused to novel tissue-specific genes, and full-length ETV1 in prostate cancer. Cancer Res 68:7541–7549
Cai C, Hsieh CL, Omwancha J, Zheng Z, Chen SY, Baert JL, Shemshedini L (2007) ETV1 is a novel androgen receptor-regulated gene that mediates prostate cancer cell invasion. Mol Endocrinol 21:1835–1846
Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM (2006) TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res 66:3396–3400
Qi M, Liu Z, Shen C, Wang L, Zeng J, Wang C, Li C, Fu W, Sun Y, Han B (2015) Overexpression of ETV4 is associated with poor prognosis in prostate cancer: involvement of uPA/uPAR and MMPs. Tumor Biol 36:3565–3572
Mesquita D, Barros-Silva JD, Santos J, Skotheim RI, Lothe RA, Paulo P, Teixeira MR (2015) Specific and redundant activities of ETV1 and ETV4 in prostate cancer aggressiveness revealed by co-overexpression cellular contexts. Oncotarget 6:5217–5236
Aytes A, Mitrofanova A, Kinkade CW, Lefebvre C, Lei M, Phelan V, LeKaye HC, Koutcher JA, Cardiff RD, Califano A, Shen MM, Abate-Shen C (2013) ETV4 promotes metastasis in response to activation of PI3-kinase and Ras signaling in a mouse model of advanced prostate cancer. Proc Natl Acad Sci U S A 110:3506–3515
Pellecchia A, Pescucci C, De Lorenzo E, Luceri C, Passaro N, Sica M, Notaro R, De Angioletti M (2012) Overexpression of ETV4 is oncogenic in prostate cells through promotion of both cell proliferation and epithelial to mesenchymal transition. Oncogenesis 1:1–11
Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, Mehra R, Chinnaiyan AM (2008) Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res 68:73–80
Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, Van Der KT, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N (2014) EAU guidelines on prostate cancer. Part II: treatment of advanced , relapsing , and castration-resistant prostate cancer. Eur Urol 65:467–479
Sanguedolce F, Cormio A, Brunelli M, D’Amuri A, Carrieri G, Bufo P, Cormio L (2016) Urine TMPRSS2: ERG fusion transcript as a biomarker for prostate cancer: literature review. Clin Genitourin Cancer 14:117–121
Rubin MA, Maher CA, Chinnaiyan AM (2011) Common gene rearrangements in prostate cancer. J Clin Oncol 29:3659–3668
Tandefelt DG, Boormans J, Hermans K, Trapman J (2014) ETS fusion genes in prostate cancer. Endocr Relat Cancer 21:143–152
Kumar-Sinha C, Tomlins SA, Chinnaiyan AM (2008) Recurrent gene fusions in prostate cancer. Nat Rev Cancer 8:479–511
Svensson MA, LaFargue CJ, MacDonald TY, Pflueger D, Kitabayashi N, Santa-Cruz AM, Garsha KE, Sathyanarayana UG, Riley JP, Yun CS, Nagy D, Kosmeder JW, Pestano GA, Tewari AK, Demichelis F, Rubin MA (2011) Testing mutual exclusivity of ETS rearranged prostate cancer. Lab Investig 91:404–412
Kunju LP, Carskadon S, Siddiqui J, Tomlins SA, Chinnaiyan AM, Palanisamy N (2014) Novel RNA hybridization method for the in situ detection of ETV1, ETV4, and ETV5 gene fusions in prostate cancer. Appl Immunohistochem Mol Morphol 22:32–40
Jomrich G, Maroske F, Stieger J, Preusser M, Ilhan-Mutlu A, Winkler D, Kristo I, Paireder M, Cshoppmann SF (2018) MK2 and ETV1 are prognostic factors in esophageal adenocarcinomas. J Cancer 9:460–468
Yoshimoto M, Ding K, Sweet JM, Ludkovski O, Trottier G, Song KS, Joshua AM, Fleshner NE, Squire JA, Evans AJ (2013) PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol 26:435–447
Funding
The study has been funded with FIS/Instituto Carlos III/PI15/FEDER PI15/00452 from the Spanish Ministry of Health. The authors thank the Parc de Salut MAR Biobank (MARBiobanc), Barcelona, Spain, for the archival support. The authors are grateful to Xavier Duran from the Consulting Service on Methodology for Biomedical Research, the Parc de Salut MAR Biobank (MARBiobanc).
Author information
Authors and Affiliations
Contributions
N.J., M.L., R.A.-G., L.C., L.F., and J.B. have contributed to specimen preparation and clinical data acquisition. J.L.-T., and S.H.-L. designed the research study. N.J., and J.L.-T. participated in the pathological data management and staining evaluation. M.L. constructed and stained the tissue microarrays. L.S. and S.H.-L. performed the qPCR experiments and analysis. L.S., J.L.-T., and S.H.-L. participated in the data preparation, statistical analysis, and wrote the paper. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
National and international guidelines (code of ethics, Declaration of Helsinki) and legal regulations on data confidentiality (Spanish Organic Law 15/1999 of December 13 on Protection of Personal Data [LOPD]) have been followed.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Segalés, L., Juanpere, N., Lorenzo, M. et al. Strong cytoplasmic ETV1 expression has a negative impact on prostate cancer outcome. Virchows Arch 475, 457–466 (2019). https://doi.org/10.1007/s00428-019-02573-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-019-02573-1