Abstract
The measurement of microvessel density (MVD) is a widely accepted method for assessing the neoangiogenetic activity in neoplasia. The aim of the present study was to compare MVD with single nucleotide polymorphisms (SNPs) in the vascular endothelial growth factor receptor (VEGFR)-1 and VEGFR-2 genes and, furthermore, with quantitative measurements of the receptors in colorectal cancer (CRC) tissue. Prognosis was also assessed. Blood and tissue were collected from 110 patients surgically resected for CRC. SNPs were analysed from genomic DNA by polymerase chain reaction. MVD was assessed by immunohistochemistry using CD34 and CD105 combined with caldesmon in order to identify also immature vessels. Microvessels were counted in three fields of vision, and the mean MVD was used for statistical analysis. The VEGFR-2 1192 C/T and −604 T/C SNPs were associated with the MVD assessed by CD105. The median MVD score for the 1192 CC genotype was significantly lower compared to the CT + TT genotypes (p = 0.002). The median MVD score for the −604 CC genotype was significantly higher compared to the TT + TC genotypes (p = 0.009). A possible association, although non-significant, was demonstrated for the CD34-positive microvessels. The 1192 CC genotype and the −604 TT + TC genotypes correlated with improved survival. This is the first report on correlations between SNPs in the VEGF receptor genes and MVD in patients with CRC. Associations were shown between two SNPs in the VEGFR-2 gene and the CD105-positive microvessels indicating an impact on neoangiogenesis. Moreover, an association between the SNPs and survival was demonstrated. The clinical implications of these findings need further investigations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant tumours are dependent on proliferating capillaries in order to supply oxygen and nutrients for their continuous growth. This knowledge has been used to develop molecular targeted agents against the vascular endothelial growth factor (VEGF) system, which is one of the most important growth factor systems involved in the angiogenetic process.
One way of estimating the angiogenetic activity is to measure the microvessel density (MVD) in the tumour tissue. The MVD method [1], counting of microvessels in hot spots, has been the preferred method. Alternative techniques such as the Chalkley method, the vascular grading system and interactive stereologic counting procedures using computer techniques have also been applied [2]. Visualization of endothelial cells is essential in order to count microvessels, and previously, antibodies targeting the cell markers CD31, CD34, and factor VIII were the most commonly used for this purpose. Recently, antibodies against CD105 have been introduced, and evidence suggests that this is a more specific tumour endothelial cell marker [3–6]. CD105 is a co-receptor for transforming growth factor β1 and β3 [7] and associated with proliferation of tumour endothelial cells [6]. CD105 is predominantly expressed on endothelial cells [6, 8], but a recent report suggests that it may not be specific for blood vascular endothelium only [9].
The MVD estimate seems to be higher at the invasive front compared to the central parts of epithelial tumours [6, 10]. Evidence also suggests that MVD assessed at the invasive front may have clinical relevance [6, 10, 11]. Furthermore, a differentiation between mature and immature microvessels may provide additional information [12].
The literature seems to support the use of MVD as a prognostic marker in patients with colorectal cancer (CRC) [13–15], but the prognostic value might depend on the choice of endothelial cell marker [16]. Furthermore, several studies have found a positive association between MVD and VEGF-A [10, 11, 17–19], but less is known about a possible association between MVD and the receptors in the VEGF system. These previous studies have been based on quantitative or semi-quantitative immunohistochemical (IHC) analyses, and the question on a similar association between MVD and quantitative measures of the VEGF system remains to be answered.
Genetic variations in genomic DNA, which are easily accessible through blood samples, are thought to be rather stable molecular markers compared to similar, less stable, variations in the tumour DNA. Variations in genomic DNA therefore constitute attractive candidates in relation to clinical cancer research. Also, such genetic variations in the VEGF system might change the expression of the relevant genes, but they have not previously been addressed in relation to MVD. If such associations were to be demonstrated, single nucleotide polymorphism (SNP) analysis could represent a future alternative to protein analysis and should be considered among other prognostic markers. A recent study demonstrated an association between SNPs in the VEGF-A gene and prognosis [20], but the possible associations between SNPs in the VEGF receptors and prognosis remain to be elucidated.
The aims of the present descriptive study were primarily to compare MVD with SNPs in the genes of VEGFR-1 and VEGFR-2 and to correlate MVD with quantitative measures of VEGFR-1 and VEGFR-2 in CRC tissue. Finally, we assessed the prognostic value of the MVD and the SNPs.
Materials and methods
Patients
This retrospective study consecutively included 110 patients with CRC entering the study in the period from February 2004 to July 2005, all undergoing surgical resection for histologically verified adenocarcinomas of the colon or rectum at the Department of Surgery, Vejle Hospital, Denmark. Patients who had received preoperative chemoradiation of rectal cancer were excluded. Pre-treatment examinations included a chest X-ray and ultrasound or CT scan of the abdomen. Postoperatively, the tumours were histologically classified and staged according to the tumor–lymph nodes–metastases (TNM) system. The study was approved by the Regional Scientific Ethical Committee for southern Denmark according to Danish law, J.nr. S-VF-20040047, and informed consent was obtained from all patients enrolled in the study.
Tissue sampling
Immediately after surgery, the removed bowel segment was brought to the Department of Clinical Pathology and a pathologist collected samples from the tumour. Samples for protein analysis were fresh frozen and stored at −80°. All samples were stored less than 30 min after the surgical removal. Based on pathological examination, it was semi-quantitatively estimated from frozen sections that the tumour tissue used for protein analyses consisted of at least 80% tumour cells.
Analysis of single nucleotide polymorphism
Genomic DNA was isolated from whole blood by the NucleoSpin® Tissue method according to the user manual (Machery-Nagel, Germany, May 2005/Rev.04; www.bioke.com/blobs/Macherey-Nagel/GenomicDNABlood_R04.pdf). The PCR analysis was performed using the ABI PRISM 7900 HT fast real-time PCR system (Applied Biosystem, USA). Commercial assays from the same company were used for the analysis of the VEGFR-2 SNPs. Assay number and the approximate length of the amplification products (estimated by gel electrophoresis) are shown in Table 1. Primer and probes for the VEGFR-1 SNP analysis were designed according to the study by Schneider et al. [21]. Tests were optimized and a probe concentration of 250 nM and a primer concentration of 600 nM were chosen.
Extraction of proteins
Tissue samples, 10–50 mg, were homogenised by the Ultra-Turrax system (Ika, Germany), at 4°C, with 10 vol.% (w/v) buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.4, 1 mM EDTA, 10% glycerol, protease inhibitor cocktail; cat. no. P8340, Sigma-Aldrich, USA). Triton X-100 was added after homogenisation to a final sample concentration of 1% (v/v) in order to stabilize membrane bound proteins, but only under the protein extraction process, and hence did not affect the binding capacities of neither standards nor controls during the following ELISA analysis. Lysates were mixed and incubated for 30 min at 4°C followed by centrifugation at 16,000×g for 10 min. After supernatant recovery, protein concentration was determined using the Bicinchoninic Acid Protein Assay (Thermo Scientific, Pierce Protein Research Products, Rockford, IL, USA).
VEGFR protein analysis
The enzyme-linked immunosorbent assays, Quantikine (R&D Systems, USA; Catalog Number DVR100B and DVR200), were used to quantify VEGFR-1 and VEGFR-2 in CRC tissue. Controls were supplied from R&D Systems as well. The assay employs the quantitative sandwich enzyme immunoassay technique. Analysis was performed on a 96-well microtiter plate, pre-coated with a mouse monoclonal antibody specific for the two receptors. Samples were diluted by a factor 5 for VEGFR-1 and by a factor 10 for VEGFR-2 in sample diluents provided with the kit.
To each well was added 100 μl of Assay Diluents (RD1W, a buffered protein base) and 100 μl of protein extract, standard or control, followed by incubation for 2 h at room temperature. After four washes with wash buffer (the washing was performed on the Wellwash 4 MK2 from Thermo Electron Corporation; Thermo Fisher Scientific Inc., Waltham, MA, USA), 200 μl of VEGFR conjugate (polyclonal antibody against the two receptors conjugated to horseradish peroxidase) was added to each well and then again incubated for 2 h at room temperature. Wells were washed again and 200 μl of substrate solution (consisting of hydrogen peroxide and chromogen) was added. Then 30 min later, 50 μl of stop solution (sulphuric acid) was added, and colour change appeared. The optical density was determined using a Novo Biolabs kinetic microplate reader set to 450 nm (Molecular Devices, Sunnyvale, CA, USA). Data were analysed using Multicalc 2000 software. Wavelength correction for optical imperfections in the plates was executed automatically.
Results are presented in picograms per milligram of total protein. Tissue samples, standards and controls were assayed in duplicate and the mean was recorded. The total coefficients of variation on three levels were VEGFR1 low 12.4%, medium 12.8% and high 11.3% and VEGFR2 low 11.3%, medium 10.1% and high 8.3%.
CD34 immunostaining
The CRC specimens were fixed for 24 h in 4% buffered neutral formalin. Subsequent fixation standardized sampling was carried out followed by paraffin-embedding according to routine procedures.
Sections of 4-μm were cut and mounted on coated slides, dried for 30 min at 60°, then overnight at 37°. The tissue was deparaffinised in Estisol for 10 min at room temperature and rehydrated in graded alcohol solutions (99–70%). Hydrogen peroxide 3% was added for 5 min to block endogenous peroxidase. Washing with water was performed before and after this blocking procedure. Heat-induced epitope retrieval was performed in order to demascade antigenes. Sections to be stained with the anti-CD34 antibody (mouse, monoclonal, Dako, code M7165, Clone QBend-10, class II) were boiled in a TEG buffer (TRIS 10 mM, EGTA 0.5 mM, Titriplex®—VI) at pH 9 for 15 min. After cooling for 15 min at room temperature, Tris-buffered saline (TBS)/Tween was added for 5 min. The anti-CD34 antibody diluted 1:100 in 1% bovine serum albumin/TBS buffer was added, and tissue sections were incubated for 30 min. After washing in TBS/Tween, final visualisation was performed using EnVision + DAB (Dako, Denmark). A solution of 0.5% copper sulphate in TBS was added for 10 min to intensify the staining pattern. The cell nuclei were stained by the addition of a Mayer haematoxylin solution. Parts of the staining procedure were performed on the Tissue-Tek Prisma, Sakura.
CD105 and caldesmon immunostaining
We also measured MVD using tissue sections stained by antibodies against CD105 and caldesmon aiming at differentiation between immature and more mature microvessels. We used a kit from Dako (code K5361, EnVision™ G│2 Doublestain System, Rabbit/Mouse, DAB+/Permanent Red) following the manufacturer’s instructions with the following exceptions: antibodies, polymer/HRP and polymer/AP incubated for 30 min; incubation in DAB + working solution and permanent red for 10 min and counterstaining with hematoxylin for 1 min; no copper sulphate added. The manual procedures were used. The anti-CD105 antibody was provided from Novo Castra (endoglin NCL-CD105, Clone 4G11) and used in a 1:25 dilution. The anti-caldesmon antibody was provided from Dako (code M3557, Clone h-CD) and used in a 1:50 dilution. Heat-induced epitope retrieval in TEG buffer was performed in order to break the formalin bindings.
Microvessel counting
Initially, we compared the MVD method as described by Weidner et al. [1] and the Chalkley method as described by Fox et al. [22]. Microvessels were counted by two investigators in an independent sample of 21 tissue sections from CRC, which were not included in the study. Linear correlation analysis showed acceptable correlation between the results, MVD (r = 0.80, p < 10−4) and Chalkley (r = 0.60, p < 10−4). A correlation on the same level was demonstrated when the interobserver variability was analysed using difference plots. The MVD method was chosen for the later analysis because of a marginally better correlation between the two investigators. It was easier and faster to use and most of the published studies in this field also use the MVD method.
Microvessels were counted at the invasive tumour front without knowledge of patient outcome. Tumour sections were scanned for hot spots at ×40 and ×100, and counting was performed at ×200. Microvessels were counted in three hot spots, and the mean MVD was used for the later analysis. Any stained endothelial cell or endothelial cell clusters clearly separated from adjacent microvessels by tumour cells and/or stroma elements were considered a single countable microvessel. Vessel lumen was not necessary for a structure to be counted as a microvessel. Hot spots near tumour necrosis or ulcerations were not chosen, as microvessels in such areas may be associated with an inflammatory response rather than with the tumour. Also, hot spots near the section edges were avoided because of a more intense and non-specific staining in these areas. Consensus resolved any discrepancy between the observers. Regarding the CD105 (brown) and caldesmon (red) staining, only microvessels without associated red staining were counted in order to estimate the MVD of the immature vessels.
Statistical analysis
Median values were compared using the Wilcoxon’s signed rank test. Chi-square statistics were used to test for Hardy–Weinberg equilibrium. Linear regression analysis was used to describe the relationship between MVD by CD34 and CD105. Prognostic associations were tested according to the Kaplan–Meier method, and the log-rank test was used to test for differences between the survival curves. Six patients with missing data were excluded from the progression-free survival (PFS) analysis. One patient had no blood sample available and hence no SNP status was analyzed, and five patients were missing survival data. All statistical calculations were carried out using the NCSS statistical software (NCSS Statistical Software, version 2007, Kaysville, UT 84037, USA). p values <0.05 were considered significant, and all tests were two-sided.
Results
Patient characteristics
Clinicopathological characteristics of the 110 included patients are listed in Table 2. The median age was 72 years (range 41–91 years). The median follow-up was 42 months (range 0.1–56.2 months), and during that period, 45 patients had either progressed or died. Five were out of reach for follow-up. PFS was defined as the time from operation until progression or death.
Blood samples were available for 109 patients. Table 1 shows the distribution of genotypes for all four SNPs. They all followed the Hardy–Weinberg equilibrium.
No significant associations between the clinicopathological characteristics (age, gender, TNM category, stage and tumour localization) and MVD or SNPs were detected.
CD34/CD105 expression and MVD
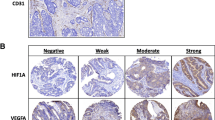
Figure 1a, b shows sections of CRC with representative staining of CD34 and CD105 positive microvessels. In general, the anti-CD34 antibody gave a more intense staining pattern than the anti-CD105 antibody, but they were both easy to quantify. More microvessels were positive for CD34 compared to CD105: median CD34 MVD 11.67 (95% confidence interval (CI) 11.00–12.33) and median CD105 MVD 6.00 (95% CI 5.33–6.00; p < 10−6). A significant correlation was detected between the two estimates (r = 0.62 and p < 10−4; Fig. 2).
a, b Immunohistochemical expression of CD34 and CD105 plus caldesmon in tumour vasculature. The two colorectal cancer sections are from different patients. a CD34-positive microvessels in a hot spot at the invasive tumour front. The unbiased counting frame is also illustrated. Endothelial cell-derived structures inside the frame or in contact with the non-solid line were counted. Structures in contact with the solid line were not counted. In this case, eight microvessels were counted. b CD105-positive microvessels in another hot spot (circle) at the invasive tumour front. Note the caldesmon-positive vessel indicated with the arrow (original magnification ×200)
MVD and SNP status
Figure 3 shows the significant association between the VEGFR-2 1192 C/T (p = 0.002) and −604 T/C (p = 0.009) SNPs and MVD as estimated by CD105 (n = 109). A possible association, although non-significant, was found for the CD34 MVD (p = 0.051 and p = 0.052, respectively). MVD showed no association with the VEGFR-1 519 C/T and the VEGFR-2 1719 SNPs (data not shown).
a Median CD105 MVD scores and corresponding confidence intervals according to the genotypes for the 1192 C/T and −604 T/C VEGFR-2 SNPs. Vertical CD105 MVD. Horizontal genotypes. The differences between the median MVD score according to the genotype groups were significant, p = 0.002 for the 1192 SNP and p = 0.009 for the −604 SNP (n = 109). b Median CD34 MVD scores and corresponding confidence intervals according to the genotypes for the 1192 C/T and −604 T/C VEGFR-2 SNPs. Vertical CD34 MVD. Horizontal genotypes. The differences between the median MVD score according to the genotype groups were marginally significant, p=0.051 for the 1192 SNP and p=0.052 for the −604 SNP (n=109)
MVD and VEGFR protein level
CRC samples from 104 patients were available for protein analysis. Table 3 shows the associations between MVD and VEGFR-1 and VEGFR-2 protein concentrations. No significant differences were found.
SNP status and VEGFR protein level
The VEGFR-2 1192 C/T SNP was significantly associated with VEGFR-2 protein concentrations in normal colorectal tissue. The median protein concentration for the CC genotype was 68 pg/mg (95% CI 59–83 pg/mg) compared to 99 pg/mg (95% CI 79–133 pg/mg) for the CT + TT genotypes (p = 0.01; n = 104). No significant associations was demonstrated in CRC tissue (p = 0.67). The three remaining SNPs were not associated with the protein concentrations of the relevant receptors in either normal colorectal or in CRC tissue (data not shown).
Prognosis
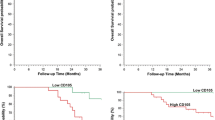
The prognostic importance of MVD, all SNPs and VEGFR-1 and VEGFR-2 protein concentrations were assessed (n = 104). Patients were classified into two groups using median MVD as a cutoff point. In the Kaplan–Meier survival analysis, no significant effect on PFS was demonstrated using either CD34 or CD105 as illustrated in Fig. 4, which shows the survival curves in relation to the CD105 MVD (p = 0.73). Similar curves were seen for the CD34 MVD (p = 0.73; data not shown).
The VEGFR-2 −604 T/C SNP showed a significant correlation with PFS (p = 0.03), and a marginal significant correlation was demonstrated for the VEGFR-2 1192 C/T SNP (p = 0.05; Fig. 5). No significant correlations were demonstrated for the remaining two SNPs. No associations between PFS and VEGFR-1 and VEGFR-2 protein levels were demonstrated (data not shown).
a, b Kaplan–Meier survival curves according to the VEGFR-2 SNPs. a The survival curves according to the −604 T/C SNP. The solid line represents the TT + TC genotypes and the hatched line represents the CC genotype, p = 0.03. b The survival curves according to the 1192 C/T SNP. The solid line represents the CC genotype and the hatched line represents the CT + TT genotypes, p = 0.05 (n = 104)
Discussion
Several studies on the association between MVD and VEGF-A have been published recently [11, 17, 19, 23]. No publications on the possible correlations between the VEGF receptors and MVD exist, and there is only sparse literature on the VEGF receptors in general. These receptors may be of crucial importance to the biological function of the system. This motivated us to investigate this further in the present pilot study.
CD34 and CD105 staining was consistently present in all the cases. Microvessels were seen at a higher concentration in or around the tumour, which is in agreement with previous studies [5, 6]. Median MVD by CD105 was significantly lower than median MVD by CD34. Furthermore, CD105 seems to be preferentially expressed on small microvessels with a lack of smooth muscle cells in their walls. This may suggest a higher specificity of the CD105 marker for the newly formed vessels, which has also been proposed in previous studies [5–7, 24, 25]. Our results suggest a correlation between the mature and more immature microvessels as expected. When the density of immature microvessels increases, one would expect a similar increase in maturing vessels.
We tested whether genetic differences in genomic DNA were associated with differences in MVD. Genomic DNA was chosen because it is easily accessible in blood samples, and it represents a stable molecular marker compared to the genetic variations in the tumour DNA. The prognostic and predictive values of these germ line polymorphisms have recently been documented [20, 26]. In the present study, two of the SNPs, the VEGFR-2 1192 C/T and −604 T/C SNPs, had a significant relationship with MVD as estimated by CD105. A possible association, although non-significant, was found for MVD estimated by CD34. Sample size may be an explanation for the lack of significance regarding CD34. A subgroup analysis based on tumour location revealed similar associations for the patients with colon cancer only. Sample size and distribution of genotypes do not allow for meaningful statistical analyses of the patients with rectal cancer. To the best of our knowledge, this is the first report on SNPs in the VEGF system, which may have an influence on MVD in tumour tissue. These findings are interesting for several reasons. First of all, it demonstrates an association between results from a PCR-based technique and results from a technique based on visualisation of the tumour architecture and metabolism. Secondly, it gives another dimension to the term ‘functional importance’, which in relation to SNPs is often used to describe an influence on gene expression and protein levels. Finally, of course, it contributes to the rather limited knowledge regarding SNPs in the VEGFR genes and their possible role in CRC. One could speculate whether a SNP with an influence on the density of immature microvessels, known to depend on VEGF-A as a survival factor, could harbour predictive information regarding the response to the anti-VEGF-A antibody bevacizumab.
We found no association between MVD and VEGFR-1 and VEGFR-2 protein concentrations in CRC tissue. A possible explanation could be tumour heterogeneity. Although tumour cells constituted more than 80% of the cells in the tumour tissue, different amounts of non-tumour cells in the remaining fraction could still blur a possible association. Protein levels are dynamic and influenced not only by the biology of the system, and this could explain the lack of association. Searching the literature we found no reports on studies comparing MVD with quantitative protein measures of VEGFR-1 and VEGFR-2 in CRC. Further studies analysing the association between MVD and VEGF receptors are needed. A better understanding of these associations might be useful in the development of future targeted therapies.
A significant association between the VEGFR-2 1192 C/T SNP and protein concentrations of VEGFR-2 was demonstrated in normal colorectal tissue, but not in CRC. This association between SNP status in genomic DNA and protein concentrations in normal tissue is not surprising, and the lack of association with protein concentrations in the neoplastic tissue may be explained by auto-regulation of the tumour cells due to mutations in the tumour DNA. The possible clinical importance of these findings is uncertain and validation is necessary.
Approximately one out of five patients had metastatic disease at time of operation. Sub-group analysis revealed no significant associations between M stage and MVD, SNPs or protein concentrations of VEGFR-1 and VEGFR-2.
In 2006, Des et al. published a meta-analysis on MVD and its prognostic importance analysing 22 studies. They concluded that using CD31 or CD34, MVD was inversely related to survival, whereas no prognostic value was demonstrated using factor VIII [14]. Romani et al. studied the prognostic role of MVD by CD105 in 125 CRC stages I–IV patients and found an increased MVD to be associated with a decreased rate of overall survival [15]. We found no association between MVD and survival, although traditional prognostic markers like the N and M stage both demonstrated a highly significant association with PFS (p < 0.001; data not shown), indicating that our cohort of patients with CRC are representative for the disease. Other studies have also failed to show an association between MVD and survival [27, 28]. In these studies, CD31 was used as an endothelial cell marker. Based on the literature, the evidence seems to support MVD as a useful prognostic marker, although several differences in methodology may affect the results, e.g. choice of antibody, type of fixative used, use of the periphery or centre of the tumour for assessment, definition of cutoff values, observer variability in choosing hot spots etc. The lack of association between MVD and survival in our study may likewise be explained by differences in methodology or the heterogeneity in our patient material, and large scaled studies seem necessary.
We demonstrated an association between survival and the VEGFR-2 1192 C/T and −604 C/T SNPs. To our knowledge, no reports have been published regarding the prognostic value of these SNPs in patients with CRC. In a study from 2008 by Schneider et al., the association between several SNPs in the VEGF system, including the 1192 C/T and 1719 T/A SNPs and result of paclitaxel plus bevacizumab, was evaluated in 363 breast cancer patients [26]. An association between the treatment outcome and two of the VEGF-A SNPs was demonstrated, but no association with the VEGFR-2 SNPs was documented. It is worth noticing that in our study, the genotypes associated with low MVD, the 1192 CC genotype and the −604 TT + TC genotypes, where the same genotypes associated with superior survival. This association needs to be validated in another cohort of patients, but if an association indeed exists between these polymorphisms and prognosis, then it would be another argument for testing their possible predictive role in relation to anti-VEGF-A therapy in a randomised prospective trial.
A few patients in this study died within a week after their operation. Running the survival analysis with or without these patients did not change any of the conclusions. Dividing the patient material in patients with colon cancer and rectum cancer did not change the results either.
Regarding the future potential of MVD as a predictive marker, it is of some concern that MVD is not sufficient to reveal the angiogenic status of tumour neovasculature and that it offers no indication as to which patients might best respond to anti-angiogenic therapy. Assessing the maturation state of the microvessels, however, might give a better characterization of the vasculature and could prove useful in guiding anti-angiogenic treatment [29, 30]. Results from prospective randomized trials will hopefully clarify the possible predictive role of MVD in patients with CRC.
This study has the standard limitations related to retrospective studies and studies dealing with IHC. The results should be considered preliminary, and validation in another cohort of patients is of course necessary for further evidence.
In conclusion, this study showed that the VEGFR-2 1192 C/T and −604 T/C SNPs, assessed in germ line DNA, may have a functional influence on neoangiogenesis in CRC, especially indicating an impact on immature microvessels. Furthermore, an association between the same SNPs and survival was demonstrated. The clinical implications of these findings, especially the potential role of the VEGFR-2 SNPs in relation to angiogenesis in general, need further investigation.
References
Weidner N, Semple JP, Welch WR et al (1991) Tumor angiogenesis and metastasis—correlation in invasive breast carcinoma. N Engl J Med 324:1–8
Hansen S, Grabau DA, Rose C et al (1998) Angiogenesis in breast cancer: a comparative study of the observer variability of methods for determining microvessel density. Lab Invest 78:1563–1573
Saad RS, Liu YL, Nathan G et al (2004) Endoglin (CD105) and vascular endothelial growth factor as prognostic markers in colorectal cancer. Mod Path 17:197–203
Akagi K, Ikeda Y, Sumiyoshi Y et al (2002) Estimation of angiogenesis with anti-CD105 immunostaining in the process of colorectal cancer development. Surgery 131:S109–S113
Minhajat R, Mori D, Yamasaki F et al (2006) Endoglin (CD105) expression in angiogenesis of colon cancer: analysis using tissue microarrays and comparison with other endothelial markers. Virchows Arch 448:127–134
Miller DW, Graulich W, Karges B et al (1999) Elevated expression of endoglin, a component of the TGF-beta-receptor complex, correlates with proliferation of tumor endothelial cells. Int J Cancer 81:568–572
Fonsatti E, Del VL, Altomonte M et al (2001) Endoglin: an accessory component of the TGF-beta-binding receptor-complex with diagnostic, prognostic, and bioimmunotherapeutic potential in human malignancies. J Cell Physiol 188:1–7
Fonsatti E, Sigalotti L, Arslan P et al (2003) Emerging role of endoglin (CD105) as a marker of angiogenesis with clinical potential in human malignancies. Curr Cancer Drug Targets 3:427–432
Royston D, Jackson DG (2009) Mechanisms of lymphatic metastasis in human colorectal adenocarcinoma. J Pathol 217:608–619
Takahashi Y, Kitadai Y, Bucana CD et al (1995) Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res 55:3964–3968
Kaio E, Tanaka S, Kitadai Y et al (2003) Clinical significance of angiogenic factor expression at the deepest invasive site of advanced colorectal carcinoma. Oncology 64:61–73
Yonenaga Y, Mori A, Onodera H et al (2003) Absence of smooth muscle actin-positive pericyte coverage of tumor vessels correlates with hematogenous metastasis and prognosis of colorectal cancer patients. Oncology 69:159–166
Rajaganeshan R, Prasad R, Guillou PJ et al (2007) The influence of invasive growth pattern and microvessel density on prognosis in colorectal cancer and colorectal liver metastases. Br J Cancer 96:1112–1117
Des GG, Uzzan B, Nicolas P et al (2006) Microvessel density and VEGF expression are prognostic factors in colorectal cancer. Meta-analysis of the literature. Br J Cancer 94:1823–1832
Romani AA, Borghetti AF, Del RP et al (2006) The risk of developing metastatic disease in colorectal cancer is related to CD105-positive vessel count. J Surg Oncol 93:446–455
Li C, Gardy R, Seon BK et al (2003) Both high intratumoral microvessel density determined using CD105 antibody and elevated plasma levels of CD105 in colorectal cancer patients correlate with poor prognosis. Br J Cancer 88:1424–1431
Xiong B, Sun TJ, Yuan HY et al (2003) Cyclooxygenase-2 expression and angiogenesis in colorectal cancer. World J Gastroenterol 9:1237–1240
Cheng J, Slavin RE, Gallagher JA et al (2004) Expression of vascular endothelial growth factor and receptor flk-1 in colon cancer liver metastases. J Hepatobiliary Pancreat Surg 11:164–170
Nakasaki T, Wada H, Shigemori C et al (2002) Expression of tissue factor and vascular endothelial growth factor is associated with angiogenesis in colorectal cancer. Am J Hematol 69:247–254
Kim JG, Chae YS, Sohn SK et al (2008) Vascular endothelial growth factor gene polymorphisms associated with prognosis for patients with colorectal cancer. Clin Cancer Res 14:62–66
Schneider BP, Radovich M, Sledge GW et al (2008) Association of polymorphisms of angiogenesis genes with breast cancer. Breast Cancer Res Treat 111:157–163
Fox SB, Leek RD, Smith K et al (1994) Tumor angiogenesis in node-negative breast carcinomas—relationship with epidermal growth factor receptor, estrogen receptor, and survival. Breast Cancer Res Treat 29:109–116
Perrone G, Vincenzi B, Santini D et al (2004) Correlation of p53 and bcl-2 expression with vascular endothelial growth factor (VEGF), microvessel density (MVD) and clinico-pathological features in colon cancer. Cancer Lett 208:227–234
Yu JX, Cui L, Zhang QY et al (2006) Expression of NOS and HIF-1alpha in human colorectal carcinoma and implication in tumor angiogenesis. World J Gastroenterol 12:4660–4664
Yu JX, Zhang XT, Liao YQ et al (2003) Relationship between expression of CD105 and growth factors in malignant tumors of gastrointestinal tract and its significance. World J Gastroenterol 9:2866–2869
Schneider BP, Wang M, Radovich M et al (2008) Association of vascular endothelial growth factor and vascular endothelial growth factor receptor-2 genetic polymorphisms with outcome in a trial of paclitaxel compared with paclitaxel plus bevacizumab in advanced breast cancer: ECOG 2100. J Clin Oncol 26:4672–4678
Bossi P, Viale G, Lee AK et al (1995) Angiogenesis in colorectal tumors: microvessel quantitation in adenomas and carcinomas with clinicopathological correlations. Cancer Res 55:5049–5053
Gonzalez FJ, Vicioso L, Alvarez M et al (2007) Association between VEGF expression in tumour-associated macrophages and elevated serum VEGF levels in primary colorectal cancer patients. Cancer Biomark 3:325–333
Hlatky L, Hahnfeldt P, Folkman J (2002) Clinical application of antiangiogenic therapy: microvessel density, what it does and doesn’t tell us. J Natl Cancer Inst 94:883–893
Vermeulen PB, Gasparini G, Fox SB et al (2002) Second international consensus on the methodology and criteria of evaluation of angiogenesis quantification in solid human tumours. Eur J Cancer 38:1564–1579
Acknowledgements
We are very thankful for the technical assistance provided by Lone Frischknecht, Lone Hartmann Hansen, Sara Egsgaard, Birgit Roed Sørensen and Karin Larsen. The study was supported by The Cancer Foundation.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
Support: The Cancer Foundation
Rights and permissions
About this article
Cite this article
Hansen, T.F., Sørensen, F.B., Spindler, KL.G. et al. Microvessel density and the association with single nucleotide polymorphisms of the vascular endothelial growth factor receptor 2 in patients with colorectal cancer. Virchows Arch 456, 251–260 (2010). https://doi.org/10.1007/s00428-009-0878-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-009-0878-8