Abstract
Main conclusion
MpBHY codes for a carotene β-ring 3(,3′)-hydroxylase responsible for both zeaxanthin and lutein biosynthesis in liverwort. MpCYP97C functions as an ε-ring hydroxylase (zeinoxanthin 3′-hydroxylase) to produce lutein in liverwort.
Abstract
Xanthophylls are oxygenated or hydroxylated carotenes that are most abundant in the light-harvesting complexes of plants. The plant-type xanthophylls consist of α-xanthophyll (lutein) and β-xanthophylls (zeaxanthin, antheraxanthin, violaxanthin and neoxanthin). The α-xanthophyll and β-xanthophylls are derived from α-carotene and β-carotene by carotene hydroxylase activities, respectively. β-Ring 3,3′-hydroxylase that mediates the route of zeaxanthin from β-carotene via β-cryptoxanthin is present in higher plants and is encoded by the BHY (BCH) gene. On the other hand, CYP97A (or BHY) and CYP97C genes are responsible for β-ring 3-hydroxylation and ε-ring 3′-hydroxylation, respectively, in routes from α-carotene to lutein. To elucidate the evolution of the biosynthetic routes of such hydroxylated carotenoids from carotenes in land plants, we identified and functionally analyzed carotenoid hydroxylase genes of liverwort Marchantia polymorpha L. Three genes homologous to higher plants, BHY, CYP97A, and CYP97C, were isolated and named MpBHY, MpCYP97A, and MpCYP97C, respectively. MpBHY was found to code for β-ring hydroxylase, which is responsible for both routes starting from β-carotene and α-carotene. MpCYP97C functioned as an ε-ring hydroxylase not for α-carotene but for zeinoxanthin, while MpCYP97A showed no hydroxylation activity for β-carotene or α-carotene. These findings suggest the original functions of the hydroxylation enzymes of carotenes in land plants, which are thought to diversify in higher plants. In addition, we generated recombinant Escherichia coli cells, which produced rare and novel carotenoids such as α-echinenone and 4-ketozeinoxanthin, through pathway engineering using bacterial carotenogenic genes that include crtW, in addition to the liverwort MpLCYb, MpLCYe and MpBHY genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are the most widespread group of pigments found in bacteria, fungi, plants, and animals. Carotenoids are synthesized in all photosynthetic organisms and in some bacteria and fungi. On the other hand, since animals are unable to synthesize carotenoids de novo, they must obtain them by dietary means. β-Carotene, α-carotene and β-cryptoxanthin serve as dietary precursors of vitamin A (Castenmiller and West 1998). Numerous reports have shown that several carotenoids, which include β-carotene, β-cryptoxanthin, lycopene, lutein, zeaxanthin, and astaxanthin, exert various beneficial effects on human health, e.g., potential anti-cancer therapeutic properties and protective roles against cardiovascular and eye diseases as well as bone loss (Mayne 1996; Nishino et al. 2000; Iwamoto et al. 2000; Krinsky et al. 2003; Talegawkar et al. 2008; Sugiura et al. 2012). Thus, these carotenoids are used as functional food supplements or food additives and in cosmetics.
Xanthophylls are oxygenated or hydroxylated carotenes that are most abundant in the light-harvesting complexes (Niyogi et al. 2001; Tian et al. 2003; Dall’Osto et al. 2007). The xanthophyll composition of higher plants is conserved and consists of α-xanthophyll (lutein) and β-xanthophylls (zeaxanthin, antheraxanthin, violaxanthin and neoxanthin). α-Xanthophyll and β-xanthophylls are derived from α-carotene and β-carotene, respectively. Lutein is unexceptionally present in land plants, including bryophytes, but only in limited divisions of algae (Rhodophyta, Cryptophyta, Euglenophyta, Chlorarachinophyta and Chlorophyta) (Takaichi 2011), whereas it has never found in prokaryotes, including cyanobacteria. On the other hand, zeaxanthin is distributed not only in land plants and algae but also in some species of bacteria that include cyanobacteria.
Plant-type α-xanthophyll and β-xanthophylls are converted from α-carotene [(6′R)-β,ε-carotene] and β-carotene (β,β-carotene) by carotene hydroxylase activities, respectively. Lutein [(3R,3′R,6′R)-β,ε-carotene-3,3′-diol] is biosynthesized from α-carotene by the action of both β-ring and ε-ring hydroxylases, while zeaxanthin [(3R,3′R)-β,β-carotene-3,3′-diol] is synthesized from β-carotene by only β-ring hydroxylase. Two different types of enzymes that catalyze these hydroxylation reactions have been found in higher plants, i.e., cytochromes P450 that belong to the CYP97 family, which catalyze the hydroxylations of β- and ε-rings, and the non-heme di-iron enzyme BHY (also called BCH, HYD, or CrtR-b) as an ortholog of bacterial CrtZ, which catalyzes the hydroxylation of β-rings (Kim et al. 2009). BHY enzymes function mainly as 3,3′-hydroxylase for the β-rings in the route of β-carotene to zeaxanthin via β-cryptoxanthin. In many plant species, the redundancy of the BHY (BCH) genes has been reported (Kim et al. 2009; Vallabhaneni et al. 2009; Li et al. 2010; Qin et al. 2012; Kim et al. 2001). Phylogenetic analyses suggest that the duplication of BHY occurred in higher plants after the split of monocot and dicot (Kim et al. 2009). The BHY genes of higher plants show gene-specific expression patterns, which provide different carotenoid levels in a tissue-specific manner. On the other hand, BHY in green algae investigated thus far is not redundant (Cui et al. 2013). In contrast to BHY type enzymes, P450s belonging to the CYP97 family can hydroxylate both β- and α-carotenes, while their preferred substrate is α-carotene. Arabidopsis CYP97A3 acts predominantly on the β-ring of α-carotene, whereas Arabidopsis CYP97C1 can efficiently hydroxylate both the β- and ε-rings of α-carotene (Kim et al. 2009). Moreover, Arabidopsis CYP97B3 is reported to be able to hydroxylate the β-ring of α-carotene (Kim et al. 2010). The CYP97 gene subfamilies (CYP97A, B and C) are thought to have originated before the separation of higher plants and green algae lineage (Bak et al. 2011; Cui et al. 2013). However, the molecular evolution of these genes is yet understood.
As described above, the physiology of several carotenoids, including β-carotene, β-cryptoxanthin, lycopene, lutein, zeaxanthin, and astaxanthin, has been well studied; however, others have not been investigated thus far because of their scarceness in nature. To produce these carotenoids in abundance, the pathway engineering (metabolic engineering) approach with Escherichia coli is one of the most powerful tools (Lee and Schmidt-Danner 2002; Das et al. 2007; Ye and Bhatia 2012; Misawa 2011). Since more than two decades ago, various carotenoids such as lycopene, β-carotene, zeaxanthin, and astaxanthin have been synthesized in E. coli (Cunningham et al. 1993; Misawa et al. 1990; Ruther et al. 1997). However, there are a number of rare and novel carotenoids that have not been synthesized in E. coli. In particular, there are few reports on the production of carotenoids that belong to α-xanthophylls in E. coli (Quinlan et al. 2007). Numerous kinds of carotenoids are synthesized among plants, algae, bacteria, and fungi (Britton et al. 2004). Therefore, the combination of carotenoid biosynthesis genes from different organisms makes it feasible to produce rare or novel carotenoids (Albrecht et al. 1997; Yokoyama et al. 1998; Shindo et al. 2008).
In this study, to gain insight into the evolution of carotenoid hydroxylases in plants, we isolated and functionally analyzed genes for carotenoid hydroxylases from liverwort Marchantia polymorpha L., which is thought to be one of the first land plants. We also report that we were able to effectively produce lutein in E. coli using the liverwort MpBHY and MpCYP97C genes. In addition, we successfully produced novel and rare carotenoids belonging to α-xanthophylls through pathway engineering approach of E. coli.
Materials and methods
Cloning of the carotenoid hydroxylase genes from M. polymorpha
Homology search to the M. polymorpha genome, cDNA and EST sequences of the liverwort (Kohchi et al. personal communication) was performed to find the homologous sequences to the known BHY (BCH), CYP97A and CYP97C genes. Based on the homologous sequences obtained, the primers were designed and the coding regions without the expected transit peptides of each gene were amplified by PCR of the liverwort cDNAs. The following primers were used:
-
MpBHYF: 5′-CATATGACAGAAATATTCGGAACA-3′.
-
MpBHYR: 5′-GGATCCTACTTGGAGGATGCAGAG-3′.
-
MpCYP97AF: 5′-CATATGCGAACTACAGTGGCAGTAA-3′.
-
MpCYP97AR: 5′-GGATCCCTAAGATTGCTCGAGTGTG-3′.
-
MpCYP97CF: 5′-CATATGTCGGATATGGAGAAAGAG-3′.
-
MpCYP97CR: 5′-GGATCCTTATATGCTTGCGAGCTC-3′.
(The underlined sequences were added for the cloning.)
Then, PCR products were cloned into the plasmid vector and sequenced.
Sequence analysis
DNA sequences of the genes were analyzed using DNASIS DNA analysis software (Hitachi Solutions, Tokyo, Japan). Homology search was performed by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences were aligned by CLUSTAL W (http://www.clustal.org/), and a neighbor-joining tree was constructed with a 500 bootstrap replication support using MEGA6 software (Tamura et al. 2013). Transit peptides of the gene products were predicted by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/). Transmembrane helical regions were predicted by TMHMM2.0 (http://www.cbs.dtu.dk/services/TMHMM).
Expression of the liverwort carotenoid biosynthesis genes in E. coli
The coding regions of the liverwort BHY (MpBHY), CYP97A (MpCYP97A) and CYP97C (MpCYP97C) were amplified with the M. polymorpha cDNA as the template by PCR and cloned into the pET21a or pRSFDuet (Merck Millipore, Darmstadt, Germany) independently. These plasmids were named pET21-MpBHY, pET21-MpCYP97A and pET21-MpCYP97C, or pRSFDuet-MpBHY, pRSFDuet-MpCYP97A and pRSFDuet-MpCYP97C. We also simultaneously inserted two of the genes MpBHY and MpCYP97C, and MpCYP97A and MpCYP97C into the pRSFDuet (Merck Millipore) and called the constructed vectors pRSFDuet-MpBHY/MpCYP97C and pRSFDuet-MpCYP97A/MpCYP97C, respectively. The crtW gene from Brevundimonas sp. strain SD212 (Nishida et al. 2005; accession no. AB181388) was also introduced with MpBHY into pRSFDuet. This plasmid was named pRSFDuet-MpBHY/crtW. They were transformed into the E. coli (BL21 (DE3)) that produced lycopene (with pACCRT-EIB) (Misawa et al. 1995), β-carotene (with pACHP-Beta), or α-carotene (with pACCRT-EIB and pETDuet-MpLCYb/MpLCYe) (Takemura et al. 2014). To construct pACHP-Beta, the Haematococcus pluvialis idi gene was isolated from pHP11 (Kajiwara et al. 1997; accession no. AB019034) with XhoI and NotI, and inserted into the AvaI-SalI site of pACCAR16ΔcrtX (Misawa et al. 1995) with an amplified NotI-SalI fragment including the Pantoea ananatis crtE gene. The transformed E. coli was grown in 2YT medium at 37 °C until an optical density of 0.8–1.0, induced with 0.05 mM of IPTG, and further cultured at 21 °C for 2 days.
An N-terminally truncated form of the Arabidopsis NADPH-P450 reductase 2 gene (Hull and Celenza 2000; Urban et al. 1997) was amplified by PCR and cloned into the CDF vector (Merck Millipore). This plasmid was named CDF-AtATR2 and co-transformed with the plasmids described above into E. coli.
Extraction and analysis of carotenoids from E. coli cells
Extraction of carotenoids from E. coli was performed by the method as previously described (Fraser et al. 2000). E. coli cultures were centrifuged and cell pellets were extracted in methanol using mixer for 5 min. Tris–HCl (50 mM, pH 7.5) (containing 1 M NaCl) was added and mixed. Then, chloroform was added to the mixture and incubated for 5 min. After centrifugation, the chloroform phase was removed and dried by centrifugal evaporation. Dried residues were re-suspended with ethyl acetate and applied to the HPLC–PDA.
Chromatography was carried out on a Waters Alliance 2695–2996 system (Waters, Milford, MA, USA) with a column, TSKgel ODS-80Ts (4.6 × 150 mm, 5 μm; Tosoh, Tokyo, Japan), according to the method described previously (Yokoyama and Miki 1995). Briefly, the extract was eluted at a flow rate of 1.0 ml/min at 25 °C with solvent A (water–methanol, 5:95, v/v) for 5 min, followed by a linear gradient from solvent A to solvent B (tetrahydrofuran–methanol, 3:7, v/v) for 5 min, solvent B alone for 8 min, and then back to solvent A. Carotenoids were identified by comparing both their retention times and absorption spectra monitored using PDA relative to those of the authentic standards.
Spectroscopic data of lutein, zeaxanthin, β-cryptoxanthin, rubixanthin, astaxanthin, canthaxanthin, echinenone, and the other ketocarotenoids in addition to β-carotene, α-carotene, δ-carotene and lycopene were described (Britton et al. 2004). α-Echinenone, zeinoxanthin, and 4-ketozeinoxanthin were identified by NMR, HR-MS and CD. Since 4-ketozeinoxanthin [(3R,6′R)-3-hydroxy-β,ε-caroten-4-one] was a novel compound, its detailed data are reported (Maoka et al. 2014).
Spectroscopic data
α-Echinenone [(6′R)-β,ε-caroten-4-one]: UV–vis λ max (Ether) 445–470 nm; HR-ESI MS; m / z 550.4195 (M+, calcd for C40H54O, 550.4175); 1H NMR (CDCl3, 500 MHz) δ 0.83 (H3-17′, s), 0.91 (H3-16′, s), 1.20 (H3-16, 17, s), 1.18 (H-2′β, m), 1.43 (H-2′α, m), 1.59 (H3-18′, s), 1.85 (H2-2, t, 7) 1.88 (H3-18, s), 1.91 (H3-19′, s), 1.95 (H3-20′, s), 1.98 (H3-20, s), 2.00 (H3-19, s), 2.00 (H2-3′ m), 2.51 (H2-4, t, 7), 5.42 (H-4′, br s), 5.53 (H-7′, dd, 15, 9.5), 6.11 (H-8′, d, 15), 6.12 (H-10′, d, 11), 6.24 (H-7, d, 16), 6.24 (H-14′, d, 11), 6.28 (H-10, d, 11), 6.30 (H-14, d, 11), 6.34 (H-12′, d, 15), 6.37 (H-8, d, 16), 6.45 (H-12, d, 15), 6.61 (H-11′, dd, 15, 11), 6.62 (H-15′, dd, 15, 11) and 6.67 (H-15, dd, 15, 11,) 6.65 (H-11, dd, 15,11); CD (Ether) λ (Δε) 210 (0), 245 (+4.3), 270 (0), 285 (-0.7), 298 (0), 344 (+3.7), 380 (0).
Zeinoxanthin [(3R,6′R)-β,ε-caroten-3-ol]: UV–vis λ max (Ether) 421, 444, and 471 nm; HR-ESI MS; m/z 552.4319 (M+, calcd for C40H56O2, 552.4331); 1H NMR (CDCl3, 500 MHz) δ 0.83 (H3-17′, s), 0.91 (H3-16′, s), 1.07 (H3-16, 17, s), 1.18 (H-2′β, m), 1.43 (H-2′α, m), 1.48 (H-2, β, dd, 12, 12), 1.59 (H3-18′, s), 1.74 (H3-18, s), 1.78 (H-2α, ddd, 12, 5, 1.5), 1.91 (H3-19′, s), 1.97 (H3-19, 20, 20′, s), 2.00 (H2-3′ m), 2.04 (H-4β dd, 16, 10), 2.39 (H-4α, ddd 16, 6, 1.5), 2.18 (H-6′, d, 9.5), 4.00 (H-3, m), 5.42 (H-4′, br s), 5.53 (H-7′, dd, 15, 9.5), 6.10 (H-7, d, 16), 6.11 (H-8′, d, 15), 6.12 (H-10′, d, 11), 6.15 (H-8, d, 16), 6.15 (H-10, d, 11), 6.24 (H-14, d, 11), 6.24 (H-14′, d, 11), 6.34 (H-12′, d, 15), 6.35 (H-12, d, 15), 6.61 (H-11′, dd, 15, 11), 6.62 (H-15 and H-15′, m), 6.64 (H-11, dd, 15,11); 13C NMR (CDCl3, 125 MHz) δ 12.8 (C-19, 20, 20′), 13.1 (C-19′), 21.6 (C-18), 23.1 (C-18′), 27.0 (C-3′, 16′), 27.7 (C-17′), 28.7 (C-16), 30.2 (C-17), 31.7 (C-2′), 32.5 (C-1′), 37.1 (C-1), 42.5 (C-4), 48.4 (C-2), 54.9 (C-6′), 65.1 (C-3), 120.8 (C-4′), 125.0 (C-11, 11′), 125.5 (C-7), 126.1 (C-5), 129.9 (C-15′), 130.1 (C-15, 10′), 131.1 (C-7′), 131.3 (C-7′), 131.3 (C-10), 132.3 (C-14′), 132.6 (C-14), 134.5 (C-5′), 135.6 (C-9, 9′), 136.2 (C-13, 8′, 13′), 137.1 (C-12′), 137.6 (C-6, 12), 138.5 (c-8), CD (Ether) λ (Δε) 245 (+3.4), 280 (0), 290 (−1.5), 340 (0), 360 (+1.0).
Expression analysis of the MpBHY, MpCYP97A, MpCYP97B, and MpCYP97C genes
Expression analysis of the four genes was performed by real-time PCR. Total RNAs were extracted from liverwort thalli using an RNeasy Plant mini kit (Qiagen, Hilden, Germany) and treated with DNaseI. One μg each of total RNA was reverse-transcribed with oligo-dT primer using PrimeScript RT Master Mix (Takara, Shiga, Japan). Amplification of real-time PCR was performed using SYBR Premix DimerEraser (Takara) and data analysis was carried out using the ABI 7300 Real-Time PCR System (Life Technologies).
For real-time PCR, the following primers were used:
-
MpBHYRT1F: 5′-CCGTCTCTCTGATGCTCTACGG-3′,
-
MpBHYRT1R: 5′-CATCGTGAACGAACATGTAG-3′,
-
MpCYP97ART1F: 5-GCATCTGGAGAGTTCACAGT-3′,
-
MpCYP97ART1R: 5′-CAACCTTCTCTCGTAATTGG-3′,
-
MpCYP97BRT1F: 5′-AAGGTTAGAAGGCGAGCTAT-3′,
-
MpCYP97BRT1R: 5′-GCTCTATGCAGACGAGTTTC-3′,
-
MpCYP97CRT1F: 5′- AGTATCTGGCGACTATGGTG-3′,
-
MpCYP97CRT1R: 5′-GGTCAGCTGAGAGAATCTTG-3′,
-
MpACTF: 5′-TGGCCGACTCTGAGGATGTT-3′,
-
MpACTR: 5′-TTCCAGATCCATTGTCGCAG-3′.
The expression levels of the various genes were normalized by MpACT as reference genes for internal control. The value relative to the expression level for female thalli was calculated.
Results
Isolation of the carotenoid hydroxylase genes from M. polymorpha
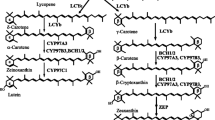
Figure 1 shows the xanthophyll biosynthetic pathway in higher plants, whose biosynthesis genes have been identified. Recently, we carried out detailed analysis of carotenoids in liverwort, M. polymorpha (Takemura et al. 2014). Consequently, no significant differences were found in the carotenoid content and composition between its male and female. In the male thalli, lutein (56.3 %) and β-carotene (30.9 %) were major carotenoids, and α-carotene (1.6 %), zeaxanthin (1.0 %), antheraxanthin (2.3 %), violaxanthin (1.6 %), and 9′-cis-neoxanthin (2.7 %) were also found. These results suggest that the liverwort has the same carotenoid biosynthetic pathway to that of higher plants and the same set of carotenoid biosynthesis genes. To find homologous genes with Arabidopsis BHY (BCH), CYP97A3, and CYP97C1 genes, homology searches for the EST and genome sequences were performed (Kohchi et al. personal communication). As a result, each gene was found and named MpBHY, MpCYP97A, and MpCYP97C. From in silico analysis of current genome sequencing and transcriptome data from different tissues and conditions, all were expected to be single copy genes (Fig. 2).
Xanthophyll biosynthesis pathways in higher plants. GGPP geranylgeranyl pyrophosphate, PSY phytoene synthase, PDS phytoene desaturase, Z-ISO 15-cis-ζ-carotene isomerase, ZDS ζ-carotene desaturase, CRTISO carotenoid isomerase, LCYb lycopene β-cyclase, LCYe lycopene ε-cyclase, BHY β-carotene hydroxylase, CYP97A cytochrome P450 97A, CYP97C cytochrome P450 97C, ZEP zeaxanthin epoxidase
Phylogenetic relationship between carotene hydroxylases. A rooted neighbor-joining tree was constructed. Numbers adjacent to branches are bootstrap values supporting the presented final tree. a Phylogenetic tree of β-carotene hydroxylases. Accession numbers are: Maize BCH1 (ZmBCH1), GQ131287; Maize HYD3 (ZmHYD3), AY844958; Maize HYD4 (ZmHYD4), AY844956; Arabidopsis BCH1 (AtBCH1), AY113923; Arabidopsis BCH2 (AtBCH2), AY117225; Tomato CrtR-b1 (SlCrtR-b1), Y14809; Tomato CrtR-b2 (SlCrtR-b2), Y14810; Liverwort BHY (MpBHY), AB981062; Chlamydomonas CHYB (CrCHYB), AY860819; Pantoea CrtZ (crtZ), D90087. b Phylogenetic tree of CYP97A, CYP97B and CYP97C proteins. Accession numbers are: Arabidopsis CYP97A3 (AtCYP97A3), NM_102914; Tomato CYP97A29 (SlCYP97A29), EU849605; Rice CYP97A4 (OsCYP97A4), AK068163; Chlamydomonas CYP97A5 (CrCYP97A5), EF587911; Liverwort CYP97A (MpCYP97A), AB981063; Physcomitrella CYP97A (PpCYP97A), A9TZA6; Arabidopsis CYP97B3 (AtCYP97B3), NM_117600; Rice CYP97B4 (OsCYP97B4), TC299269; Liverwort CYP97B (MpCYP97B), AB981064; Physcomitrella CYP97B (PpCYP97B), A9ST64; Arabidopsis CYP97C1 (AtCYP97C1), AY424805; Tomato CYP97C11 (SlCYP97C11), EU849604; Rice CYP97C2 (OsCYP97C2), AK065689; Chlamydomonas CYP97C3 (CrCYP97C3), EF587910; Liverwort CYP97C (MpCYP97C), AB981065 Physcomitrella CYP97C (PpCYP97C), A9TSP5
Four transmembrane domains were predicted in MpBHY peptides as in other plant BCHs (BHYs; Supplementary Fig. S1). MpBHY also had the Motif1 (Sun et al. 1996). MpBHY showed approximately 50 and 21 % amino acid identities to AtBCH and the P. ananatis CrtZ, respectively. The 137 amino acids of N-terminus were predicted to be a transit peptide to chloroplast.
Phylogenetic analysis of CYP97 proteins indicated that three gene subfamily members existed in the liverwort as in higher plants (Fig. 2b) (Kim et al. 2009). The CYP97A and CYP97C have been reported to be involved in carotenoid biosynthesis, while the function of CYP97B has not well been elucidated. The liverwort CYP97 s included consensus sequences for both oxygen-binding pocket and heme-binding domain (Chapple 1998) (Supplementary Fig. S2). The 60 and 79 amino acids of N-terminus of MpCYP97A and MpCYP97C, respectively, were predicted to be signal sequences to chloroplast.
MpBHY acts as the β-carotene hydroxylase
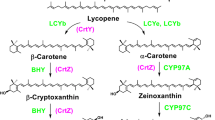
To identify the β-carotene hydroxylase of the liverwort, we used E. coli expression system (Misawa et al. 1995). The E. coli having plasmid pACHP-Beta that carries the Pantoea ananatis crtE, crtB, crtI, crtY genes in addition to the Haematococcus pluvialis idi [isopentenyl diphosphate (IPP) isomerase] gene produces β-carotene (β,β-carotene) (Figs. 3, 4a). We introduced MpBHY, MpCYP97A and MpCYP97C genes independently into this E. coli. Although 138 amino acids of the MpBHY N-terminal were predicted to be the transit signal to chloroplast, this region contained the first predicted TM-helix (Supplementary Fig. S1). Therefore, we constructed the pET-MpBHY without the first 120 amino acids. On the other hand, MpCYP97A and MpCYP97C, whose individual predicted transit peptides were eliminated, were fused with pET vector. When the empty vector pET21a was expressed in this E. coli, β-carotene was detected as expected (Fig. 4a). When pET-MpBHY was introduced, zeaxanthin [(3R, 3′R)-β,β-carotene-3,3′-diol] was detected (Fig. 4b). A small amount of the monohydroxylated carotenoid β-cryptoxanthin [(3R)-β,β-caroten-3-ol] and rubixanthin [(3R)-β,ψ-caroten-3-ol] were also detected. Rubixanthin is a 3-hydroxylated product of γ-carotene that is the monocyclic intermediate of β-carotene synthesis from lycopene. These results demonstrated that MpBHY exerts β-ring hydroxylation activity for β-carotene and γ-carotene to produce zeaxanthin (via β-cryptoxanthin) and rubixanthin, respectively. In contrast to MpBHY, neither MpCYP97A nor MpCYP97C influenced the composition of carotenoids when introduced into the β-carotene-producing E. coli (Fig. 4c, d). These results suggested that neither MpCYP97A nor MpCYP97C exerted β-carotene hydroxylase activity.
HPLC analysis of products formed from β-carotene in transformed E. coli. a–d HPLC chromatograms of the extracts from E. coli that carried a pACHP-Beta plus pET21a vector; b pACHP-Beta plus pET-MpBHY; c pACHP-Beta plus pET-MpCYP97A; d pACHP-Beta plus pET-MpCYP97C. e Absorption spectra of individual peaks. 1 β-carotene; 2 zeaxanthin; 3 rubixanthin; 4 β-cryptoxanthin
Most of the plant P450s involved in the secondary metabolism require a FAD and FMN-containing NADPH-P450 reductase to receive the electrons from NADPH and reduce P450 itself (Hannemann et al. 2007). For the functional expression of such plant P450 genes in E. coli, a gene for plant NADPH-P450 reductase, is necessary to be co-expressed (Harada et al. 2011; Schückel et al. 2012), while a redox partner flavoprotein endogenous in E. coli is sometimes likely to complementarily function with foreign P450s such as higher plant CYP97A and CYP97C (Tian et al. 2003, Quinlan et al. 2012, Christopher and Waterman 1998). However, we do not know whether the M. polymorpha CYP97s need a foreign redox partner such as a plant NADPH-P450 reductase. For this reason, we tested the requirement of a higher plant NADPH-P450 reductase for MpCYP97A or MpCYP97C by co-expressing the Arabidopsis NADPH-P450 reductase 2 (AtATR2) gene (Hull and Celeza 2000; Urban et al. 1997). As a result, no differences were found in the products between the presence and absence of AtATR2 (Supplementary Fig. S3).
MpBHY and MpCYP97C act to produce lutein
To further investigate the enzyme activities of these three genes, we used the E. coli producing α-carotene [(6′R)-β,ε-carotene] due to the presence of the plasmids pACCRT-EIB and pETDuet-MpLCYb/MpLCYe (Fig. 5a; Takemura et al. 2014). When either the MpCYP97A or MpCYP97C gene was introduced into this recombinant E. coli, α-carotene was not converted to any other carotenoids (Fig. 5b, c). In addition, the AtATR2 gene did not affect their enzyme activities (Supplementary Fig. S3). These results indicated that neither MpCYP97A nor MpCYP97C possesses ring hydroxylase activity for α-carotene. On the other hand, when the MpBHY gene was introduced into the α-carotene-producing E. coli, the monohydroxylated intermediate, zeinoxanthin, was detected, indicating that MpBHY has an activity to hydroxylate β-ring of α-carotene (Fig. 5d).
HPLC analysis of products formed from α-carotene in transformed E. coli. a–f HPLC chromatograms of the extracts from α-carotene-producing E. coli (pACCRT-EIB and pETDuet-MpLCYb/MpLCYe) transformed by a pRSFDuet vector; b pRSFDuet -MpCYP97A; c pRSFDuet -MpCYP97C; d pRSFDuet -MpBHY; e pRSFDuet-MpCYP97A/MpCYP97C; f pRSFDuet-MpBHY/MpCYP97C. g Absorption spectra of individual peaks. 1 α-carotene; 2 δ-carotene; 3 lycopene; 4 zeinoxanthin; 5 zeaxanthin; 6 lutein
We further investigated the combination of MpCYP97A/MpCYP97C and MpBHY/MpCYP97C. This is because MpBHY and MpCYP97A were expected as β-ring hydroxylase, while MpCYP97C was expected as ε-ring hydroxylase. When pRSF-MpCYP97A/MpCYP97C was introduced into the E. coli producing α-carotene, no significant modification of the carotenoid profile was found (Fig. 5e). In contrast, the introduction of pRSF-MpBHY/MpCYP97C resulted in the dominant production of lutein [(3R,3′R,6′R)-β,ε-carotene-3,3′-diol; Fig. 5f]. These results indicated that MpBHY and MpCYP97C function as β-ring and ε-ring hydroxylase of α-carotene, respectively, to produce lutein.
Expression of the MpBHY, MpCYP97A, MpCYP97B and MpCYP97C genes
To examine the expression of MpBHY, MpCYP97A, MpCYP97B and MpCYP97C, we performed a real-time PCR. The expression level of the MpCYP97C gene was similar in the female and male thalli (Fig. 6). On the other hand, the expressions of MpBHY, MpCYP97A and MpCYP97B in the male thalli were slightly higher than those in the female thalli.
Expression analysis of MpBHY, MpCYP97A, MpCYP97B and MpCYP97C. Total RNA was extracted from male and female thalli and subjected to real-time PCR analysis. Expression levels were normalized by MpACT as internal control and values relative to that of female thalli. Data are average ±SD of three independent measurements
Production of rare or novel carotenoids
Previous reports have indicated that astaxanthin (4,4′-ketozeaxanthin) is produced from zeaxanthin by the β-carotene ketolase (CrtW) in genetically engineered E. coli (Misawa et al. 1995; Fraser et al. 1997). Recently, a rare carotenoid, α-echinenone (4-keto-α-carotene) was reported as an unexpected by-product in transgenic rice callus expressing the Brevundimonas crtW (Breitenbach et al. 2014). However, it is unclear whether it is possible to produce ketocarotenoids from α-carotene and/or α-xanthophyll in transgenic E. coli. In this study, we introduced the crtW gene into the α-xanthophylls-producing E. coli. This recombinant E. coli was orange (Supplementary Fig. S4). Pigment analysis showed that α-echinenone and 4-ketozeinoxanthin were produced from α-carotene and zeinoxanthin, respectively (Table 1). This result showed that CrtW catalyzed the ketolation reaction of β-ring of α-carotene and zeinoxanthin in E. coli. α-Echinenone is a rare carotenoid reported to be present in sea urchin. 4-Ketozeinoxanthin is a novel carotenoid, which has never been isolated from any organism.
Discussion
In this study, we isolated and functionally analyzed carotenoid hydroxylase genes, named MpBHY, MpCYP97A and MpCYP97C, from a liverwort for the first time. Sequence analysis suggested that MpBHY and MpCYP97A coded for carotenoid β-ring hydroxylase and that MpCYP97C coded for carotenoid ε-ring hydroxylase. It has been reported that higher plants have more than two BHY (BCH) genes. For example, six different BHY paralogs were recently identified in maize (Vallabhaneni et al. 2009). Two of these clearly encoded β-carotene hydroxylase, two were pseudogenes, and the remaining two were functionally unclear (Vallabhaneni et al. 2009). Arabidopsis (Sun et al. 1996; Tian and DellaPenna 2001), citrus (Kim et al. 2001), and tomato (Galpaz et al. 2006) were also reported to have two BHY genes. In contrast, only one BHY gene was found in the liverwort M. polymorpha genome. In flowering plants, the carotenoid compositions among flowers, fruits, and leaves are significantly different, because the genes of carotenoid biosynthesis are regulated in a tissue- and/or development-specific manner (Galpaz et al. 2006; Li et al. 2010). It is likely that duplication and subsequent functional divergence of BHY (BCH) genes occurred after the split of the bryophytes and higher plants (Moore and Purugganan 2005; Kim et al. 2009). The BHY is presumed to be located in the thylakoid membranes of chloroplasts in plants. MpBHY contains an N-terminal extension of 170 amino acids more than the corresponding bacterial enzyme CrtZ. Sun et al. (1996) reported that truncation of the first 69 amino acids of the Arabidopsis BCH1 (AtBCH1), which was predicted as the signal peptide to chloroplast, did not impair enzyme activity in E. coli. They also found that removing the first 129 amino acids, including the first transmembrane helix, resulted in a high ratio of β-cryptoxanthin to zeaxanthin. Therefore, they speculated that amino acid residues 70–129 of AtBCH1 may play a role in formation of a functional homodimer (Sun et al. 1996). In this study, we produced a polypeptide lacking the first 170 amino acids, which was the extended region compared to CrtZ, and contained the first transmembrane region. The activity of this truncated enzyme was not impaired (Supplementary Fig. S5), suggesting that this extension was not required for the activity of MpBHY. Considering that the first transmembrane domain of AtBCH1 is important for this activity, this domain may have acquired new roles during plant evolution.
MpBHY catalyzed the reaction of β-ring hydroxylation of both β- and α-carotenes in the same manner as higher plant BCH enzymes. On the other hand, MpCYP97A did not show any carotenoid hydroxylation activity, which was different from the corresponding P450s of higher plants encoded by the CYP97A genes such as Arabidopsis LUT5 (CYP97A3) (Kim and DellaPenna 2006), rice CYP97A4 (Lv et al. 2012) and tomato CYP97A29 (Stigliani et al. 2011). MpCYP97C hydroxylated the ε-ring of α-carotene, which is the same as in the higher plant CYP97C. However, it is emphasized that this P450 functions as ε-ring hydroxylase not for α-carotene but for zeinoxanthin (Fig. 3). We detected zeinoxanthin but no α-cryptoxanthin in the lutein-producing E. coli cells, which included the MpBHY and MpCYP97C genes. Moreover, no monohydroxylated carotene (α-cryptoxanthin) was produced from α-carotene only with MpCYP97C. These results demonstrated that β-ring hydroxylation precedes ε-ring hydroxylation during lutein synthesis from α-carotene. This finding is supported by the analysis of Arabidopsis lut1 mutant, which produced zeinoxanthin but not α-cryptoxanthin (Tian et al. 2003, 2004). Quinlan et al. (2012) have proposed the protein–protein interaction between CYP97A and CYP97C in rice. Analysis of the interaction between MpBHY and MpCYP97C proteins is also required.
We also showed that MpBHY and MpCYP97C function together to produce lutein in the liverwort, whereas in higher plants CYP97C functions mainly along with CYP97A, not with BHY (Kim and DellaPenna 2006; Tian et al. 2003; Quinlan et al. 2012). In the liverwort, CYP97A might function in the different pathway. It is possible that MpCYP97A needs some factor which is absent in E. coli to exert its activity. Since we showed that AtATR2 conferred no effect on MpCYP97A activity, liverwort MpCYP97A is unlikely to be involved in carotenoid biosynthesis. It needs to be determined what function MpCYP97A has in the liverwort.
Among the CYP97 family, CYP97A and CYP97C are involved in carotenoid biosynthesis in higher plants, whereas the function of CYP97B has not yet been well clarified except for two reports. One report has shown that Arabidopsis CYP97B3 has the activity of β-carotenoid hydroxylase (Kim et al. 2010). Other has shown that Porphyra PuCHY1 has not only a β-carotenoid hydroxylation activity but also a possible ε-carotenoid hydroxylation activity (Yang et al. 2014). We also investigated whether liverwort CYP97B (MpCYP97B) was able to hydroxylate β-carotene or α-carotene, and it was found to have no activity as carotene hydroxylase (Supplementary Fig. S6).
In this study, we determined the carotenoid hydroxylase activities of each gene product using E. coli cells. Since it is important to know the physiological functions of them in the liverwort, we plan on analyzing the functions by constructing a transgenic liverwort.
From an evolutionary point of view, in the early plants, BHY (BCH) genes were mainly involved in β-ring hydroxylation of both β- and α-carotenes to synthesize zeaxanthin and zeinoxanthin, respectively. These plants also possessed CYP97C genes, which hydroxylate the ε-ring of zeinoxanthin to produce lutein. On the other hand, there were CYP97A genes in the early land plants; however, they did not function for carotenoid biosynthesis. It is interesting to know when CYP97A acquired activity as a β-carotene hydroxylase, and was replaced partially with BHY. The functional analysis of carotenoid hydroxylase genes from divergent plants, including algae, bryophyte, spermatophytes and so on, should reply to this question.
By the combination of carotenoid biosynthesis genes derived from different organisms, the production of novel and rare carotenoids has been successful, as noted in the Introduction. Transplastomic tobacco and lettuce plants, in which Brevundimonas sp. SD212 crtZ and crtW genes were introduced into their plastids, have been constructed (Hasunuma et al. 2008; Harada et al. 2014). In these plants, astaxanthin and fritschiellaxanthin (4-keto-lutein) were produced from zeaxanthin and lutein, respectively. However, α-echinenone and 4-ketozeinoxanthin that should be derived from α-carotene and zeinoxanthin, respectively, were not found in the transplastomic plants. This may be because α-carotene and zeinoxanthin were converted rapidly to lutein in the plastids, and/or because the activity of CrtZ (and BHY) is stronger than that of CrtW. In contrast, α-echinenone (4-keto-α-carotene) was produced in transgenic rice callus expressing the Zmpsy, crtI (PacrtI), and crtW (BrcrtW) (Breitenbach et al. 2014). In this case, CrtW may show a higher activity than the native BHY, so that the majority of α-carotene could be converted to α-echinenone but not to lutein. We generated recombinant E. coli cells, which produced rare and novel carotenoids such as α-echinenone and 4-ketozeinoxanthin, through pathway engineering using bacterial carotenogenic genes that include Brevundimonas sp. SD212 crtW, in addition to the liverwort MpLCYb, MpLCYe and MpBHY genes (Fig. 7).
Our results indicate the feasibility of producing novel or rare carotenoids, which have not yet or hardly been produced in any hosts, in E. coli by appropriate combinations of carotenogenic genes derived from various organisms.
Abbreviations
- BCH:
-
β-Carotene 3,3′-hydroxylase
- BHY:
-
β-Carotene 3,3′-hydroxylase
- CD:
-
Circular dichroism
- CYP:
-
Cytochrome P450
- LCYb:
-
Lycopene β-cyclase
- LCYe:
-
Lycopene ε-cyclase
- NMR:
-
Nuclear magnetic resonance
References
Albrecht M, Takaichi S, Misawa N, Schnurr G, Böger P, Sandmann G (1997) Synthesis of atypical cyclic and acyclic hydroxy carotenoids in Escherichia coli transformants. J Biotechnol 58:177–185
Bak S, Beisson F, Bishop G, Hamberger B, Höfer R, Paquette S, Werck-Reichhart D (2011) Cytochromes P450. Arabidopsis Book. doi:10.1199/tab.0144
Breitenbach J, Bai C, Rivera SM, Canela R, Capell T, Christou P, Zhu C, Sandmann G (2014) A novel carotenoid, 4-keto-α-carotene, as an unexpected by-product during genetic engineering of carotenogenesis in rice callus. Phytochemistry 98:85–91
Britton G, Liaaen-Jensen S, Pfander H (2004) Carotenoids handbook. Birkhäuser Verlag, Basel
Castenmiller JJ, West CE (1998) Bioavailability and bioconversion of carotenoids. Annu Rev Nutr 18:19–38
Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49:311–343
Christopher MJ, Waterman MR (1998) NADPH-flavodoxin reductase and flavodoxin from Escherichia coli: characteristics as a soluble microsomal P450 reductase. Biochemistry 37:6106–6113
Cui H, Yu X, Wang Y, Cui Y, Li X, Liu Z, Qin S (2013) Evolutionary origins, molecular cloning and expression of carotenoid hydroxylases in eukaryotic photosynthetic algae. BMC Genom 14:457–476
Cunningham FX Jr, Chamovitz D, Misawa N, Gantt E, Hirschberg J (1993) Cloning and functional expression in Escherichia coli of a cyanobacterial gene for lycopene cyclase, the enzyme that catalyzes the biosynthesis of β-carotene. FEBS Lett 328:130–138
Dall’Osto L, Fiore A, Cazzaniga S, Giuliano G, Bassi R (2007) Different roles of α- and β-branch xanthophylls in photosystem assembly and photoprotection. J Biol Chem 282:35056–35068
Das A, Yoon S-H, Lee S-H, Kim J-Y, Oh D-K, Kim S-W (2007) An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl Microbiol Biotechnol 77:505–512
Fraser PD, Miura Y, Misawa N (1997) In vitro characterization of astaxanthin biosynthetic enzymes. J Biol Chem 272:6128–6135
Fraser PD, Elisabete M, Pinto S, Holloway DE, Bramley PM (2000) Application of high-performance liquid chromatography with photodiode array detection to the metabolic profiling of plant isoprenoids. Plant J 24:551–558
Galpaz N, Ronen G, Khalfa Z, Zamir D, Hirschberg J (2006) A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18:1947–1960
Hannemann F, Bichet A, Ewen KM, Bernhardt R (2007) Cytochrome P450 systems-biological variations of electron transport chains. Biochim Biophys Acta 1770:330–344
Harada H, Shindo K, Iki K, Teraoka A, Okamoto S, Yu F, Hattan J, Utsumi R, Misawa N (2011) Efficient functional analysis system for cyanobacterial or plant cytochromes P450 involved in sesquiterpene biosynthesis. Appl Microbiol Biotechnol 90:467–476
Harada H, Maoka T, Osawa A, Hattan J, Kanamoto H, Shindo K, Otomatsu T, Misawa N (2014) Construction of transplastomic lettuce (Lactuca sativa) dominantly producing astaxanthin fatty acid esters and detailed chemical analysis of generated carotenoids. Transgenic Res 23:303–315
Hasunuma T, Miyazawa S, Yoshimura S, Shinzaki Y, Tomizawa K, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55:857–868
Hull AK, Celenza JL (2000) Bacterial expression and purification of the Arabidopsis NADPH-cytochrome P450 reductase ATR2. Protein Expr Purif 18:310–315
Iwamoto T, Hosoda K, Hirano R, Kurata H, Matsumoto A, Miki W, Kamiyama M, Itakura H, Yamamoto S, Kondo K (2000) Inhibition of low-density lipoprotein oxidation by astaxanthin. J Atheoscler Thomb 7:216–222
Kajiwara S, Fraser PD, Kondo K, Misawa N (1997) Expression of an exogenous isopentenyl diphosphate isomerase gene enhances isoprenoid biosynthesis in Escherichia coli. Biochem J 324:421–426
Kim J, DellaPenna D (2006) Defining the primary route for lutein synthesis in plants: the role of Arabidopsis carotenoid β-ring hydroxylase CYP97A3. Proc Natl Acad Sci 103:3474–3479
Kim I-J, Ko K-C, Kim C-S, Chung W-I (2001) Isolation and characterization of cDNAs encoding β–carotene hydroxylase in Citrus. Plant Sci 161:1005–1010
Kim J, Smith JJ, Tian L, DellaPenna D (2009) The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol 50:463–479
Kim J-E, Cheng KM, Craft NE, Hamberger B, Douglas CJ (2010) Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a β-carotene ketolase provides insight into in vivo function. Phytochemistry 71:168–178
Krinsky NI, Landrum JT, Bone RA (2003) Biological mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr 23:171–201
Lee PC, Schmidt-Danner C (2002) Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl Microbiol Biotechnol 60:1–11
Li Q, Farre G, Naqvi S, Breitenbach J, Sanahuja G, Bai C, Sandmann G, Capell T, Christou P, Zhu C (2010) Cloning and functional characterization of the maize carotenoid isomerase and β–carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res 19:1053–1068
Lv M-Z, Chao D-Y, Shan J-X, Zhu M-Z, Shi M, Gao J-P, Lin H-X (2012) Rice carotenoid β-ring hydroxylase CYP97A4 is involved in lutein biosynthesis. Plant Cell Physiol 53:987–1002
Maoka T, Takemura M, Tokuda H, Suzuki N, Misawa N (2014) 4-Ketozeinoxanthin, a novel carotenoid produced in Escherichia coli through metabolic engineering using carotenogenic genes of bacterium and liverwort. Tetrahedron Lett 55:6708–6710
Mayne ST (1996) Beta-carotene, carotenoids, and disease prevention in humans. FASEB J 10:690–701
Misawa N (2011) Pathway engineering for functional isoprenoids. Curr Opin Biotechnol 22:627–633
Misawa N, Nakagawa M, Kobayashi K, Yamano S, Izawa Y, Nakamura K, Harashima K (1990) Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J Bacteriol 172:6704–6712
Misawa N, Satomi Y, Kondo K, Yokoyama A, Kajiwara S, Saito T, Ohtani T, Miki W (1995) Structure and functional analysis of a marine bacterial carotenoid biosynthesis gene cluster and astaxanthin biosynthetic pathway proposed at the gene level. J Bacteriol 177:6575–6584
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, Sawabe A, Komemushi S, Miki W, Misawa N (2005) Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol 71:4286–4296
Nishino H, Tokuda H, Murakoshi M, Satomi Y, Masuda M, Onozuka M, Yamaguchi S, Takayasu J, Tsuruta J, Okuda M, Khachik F, Narisawa T, Takasuka N, Yano M (2000) Cancer prevention by natural carotenoids. BioFactors 13:89–94
Niyogi KK, Shih C, Soon Chow W, Pogson BJ, DellaPenna D, Bjökman O (2001) Photoprotection in a zeaxanthin- and lutein- deficient double mutant of Arabidopsis. Photosynth Res 67:139–145
Qin X, Zhang W, Dubcovsky J, Tian L (2012) Cloning and comparative analysis of carotenoid β-hydroxylase genes provides new insights into carotenoid metabolism in tetraploid (Triticum turgidum ssp. durum) and hexaploid (Triticum aestivum) wheat grains. Plant Mol Biol 80:631–646
Quinlan RF, Jaradat TT, Wurtzel ET (2007) Escherichia coli as a platform for functional expression of plant P450 carotene hyrdoxylases. Arch Biochem Biophys 458:146–157
Quinlan RF, Shumskaya M, Bradbury LMT, Beltrán J, Ma C, Kennelly EJ, Wurtzel ET (2012) Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol 160:204–214
Ruther A, Misawa N, Böger P, Sandman G (1997) Production of zeaxanthin in Escherichia coli transformed with different carotenogenic plasmids. Appl Microbiol Biotechnol 48:162–167
Schückel J, Rylott EL, Grogan G, Bruce NC (2012) A gene-fusion approach to enabling plant cytochromes p450 for biocatalysis. Chem BioChem 13:2758–2763
Shindo K, Hasunuma T, Asagi E, Sano A, Hotta E, Minemura N, Miyake C, Maoka T, Misawa N (2008) 4-Ketoantheraxanthin, a novel carotenoid produced by the combination of the bacterial enzyme β-carotene ketolase CrtW and endogenous carotenoid biosynthetic enzymes in higher plants. Tetrahedron Lett 49:3294–3296
Stigliani AL, Giorio G, D’Ambrosio C (2011) Characterization of P450 carotenoid β- and ε-hydroxylases of tomato and transcriptional regulation of xanthophyll biosynthesis in root, leaf, petal and fruit. Plant Cell Physiol 52:851–865
Sugiura M, Nakamura M, Ogawa K, Ikoma M, Yano N (2012) High serum carotenoids associated with lower risk for bone loss and osteoporosis in post-menopausal Japanese female subjects: prospective cohort study. PLoS One 7:e52643
Sun Z, Gantt E, Cunningham FX Jr (1996) Cloning and functional analysis of the β-carotene hydroxylase of Arabidopsis thaliana. J Biol Chem 271:24349–24352
Takaichi S (2011) Carotenoids in algae: distributions, biosyntheses and functions. Mar Drugs 9:1101–1118
Takemura M, Maoka T, Misawa N (2014) Carotenoid analysis of a liverwort Marchantia polymorpha and functional identification of its lycopene β- and ε-cyclase genes. Plant Cell Physiol 55:194–200
Talegawkar SA, Johnson EJ, Carithers TC, Taylor HA Jr, Bogle ML, Tucker KL (2008) Carotenoid intakes, assessed by food-frequency questionnaires (FFQs), are associated with serum carotenoid concentrations in the Jackson Heart Study: validation of the Jackson Heart Study Delta NIRI Adult FFQs. Public Health Nutr 11:989–997
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tian L, DellaPenna D (2001) Characterization of a second carotenoid β-hydroxylase gene from Arabidopsis and its relationship to the LUT1 locus. Plant Mol Biol 47:379–388
Tian L, Magallanes-Lundback M, Musetti V, DellaPenna D (2003) Functional analysis of β- and ε-ring carotenoid hydroxylases in Arabidopsis. Plant Cell 15:1320–1332
Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc Natl Acad Sci 101:402–407
Urban P, Mignotte C, Kazmaier M, Delorme F, Pompon D (1997) Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADPH-cytochrome P450 reductases with P450 CYP73A5. J Biol Chem 272:19176–19186
Vallabhaneni R, Gallagher CE, Licciardello N, Cuttriss AJ, Quinlan RF, Wurtzel ET (2009) Metabolite sorting of a germplasm collection reveals the Hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol 151:1635–1645
Yang L-E, Huang X-Q, Hang Y, Deng Y-Y, Lu Q-Q, Lu S (2014) The P450-type carotene hydroxylase PuCHY1 from Porphyra suggests the evolution of carotenoid metabolism in red algae. J Integr Plant Biol 56:902–915
Ye VM, Bhatia SK (2012) Pathway engineering strategies for production of beneficial carotenoids in microbial hosts. Biotechnol Lett 34:1405–1414
Yokoyama A, Miki W (1995) Composition and presumed biosynthetic pathway of carotenoids in the astaxanthin-producing bacterium Agrobacterium aurantiacum. FEMS Microbiol Lett 128:139–144
Yokoyama A, Shizuri Y, Misawa N (1998) Production of new carotenoids, astaxanthin glucosides, by Escherichia coli transformants carrying carotenoid biosynthetic genes. Tetrahedron Lett 39:3709–3712
Acknowledgments
The authors thank Dr. Hiroshi Shimada, Hiroshima University, for the construction of plasmid pACHP-Beta. We also thank Dr. Takayuki Kohchi, Kyoto University; Dr. Katsuyuki T. Yamato, Kinki University and Dr. Kimitsune Ishizaki, Kobe University for their assistance in searching liverwort sequences.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gene accession numbers
The nucleotide sequence reported in this paper has been submitted to DDBJ under accession numbers, AB981062 (MpBHY), AB981063 (MpCYP97A), AB981064 (MpCYP97B) and AB981065 (MpCYP97C).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Takemura, M., Maoka, T. & Misawa, N. Biosynthetic routes of hydroxylated carotenoids (xanthophylls) in Marchantia polymorpha, and production of novel and rare xanthophylls through pathway engineering in Escherichia coli . Planta 241, 699–710 (2015). https://doi.org/10.1007/s00425-014-2213-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2213-0