Abstract
The plastid genome of lettuce (Lactuca sativa L.) cv. Berkeley was site-specifically modified with the addition of three transgenes, which encoded β,β-carotenoid 3,3′-hydroxylase (CrtZ) and β,β-carotenoid 4,4′-ketolase (4,4′-oxygenase; CrtW) from a marine bacterium Brevundimonas sp. strain SD212, and isopentenyl diphosphate isomerase from a marine bacterium Paracoccus sp. strain N81106. Constructed transplastomic lettuce plants were able to grow on soil at a growth rate similar to that of non-transformed lettuce cv. Berkeley and generate flowers and seeds. The germination ratio of the lettuce transformants (T0) (98.8 %) was higher than that of non-transformed lettuce (93.1 %). The transplastomic lettuce (T1) leaves produced the astaxanthin fatty acid (myristate or palmitate) diester (49.2 % of total carotenoids), astaxanthin monoester (18.2 %), and the free forms of astaxanthin (10.0 %) and the other ketocarotenoids (17.5 %), which indicated that artificial ketocarotenoids corresponded to 94.9 % of total carotenoids (230 μg/g fresh weight). Native carotenoids were there lactucaxanthin (3.8 %) and lutein (1.3 %) only. This is the first report to structurally identify the astaxanthin esters biosynthesized in transgenic or transplastomic plants producing astaxanthin. The singlet oxygen-quenching activity of the total carotenoids extracted from the transplastomic leaves was similar to that of astaxanthin (mostly esterified) from the green algae Haematococcus pluvialis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids are C40 isoprenoid pigments (tetraterpenes) with long conjugated double bonds that possess color ranging from yellow through to orange and red. Pigments are biosynthesized in all photosynthetic prokaryotes including cyanobacteria, in algae and higher plants, and also in some non-photosynthetic bacteria and fungi. Carotenoids fulfil two essential functions during photosynthesis in photosynthetic organisms: light harvesting and the protection of the photosynthetic apparatus from photooxidative damage (Demmig-Adams and Adams 1996). More than 750 different carotenoids have been isolated from natural sources (Britton et al. 2004). However, only a few carotenoid species can be obtained in sufficient amounts, including lycopene, β-carotene, β-cryptoxanthin, zeaxanthin, lutein, fucoxanthin, and astaxanthin. All these carotenoids except for astaxanthin are present in edible plant sources, and their beneficial health effects such as prevention against cancer, cardiovascular disease, eye disease, and bone loss have been extensively examined (e.g., Krinsky et al. 2003; Maeda et al. 2005; Nishino et al. 2002; Sugiura et al. 2012; Talegawkar et al. 2008). Astaxanthin [IUPAC-IUB semisystematic name: (3S,3′S)-3,3′-dihydroxy-β,β-carotene-4,4′-dione] commercialized as functional food and cosmetics is produced by a unicellular green alga belonging to the Chlorophyceae class, Haematococcus pluvialis. This carotenoid comprises a fatty acid diester (28 %), monoester (70 %), and free form (2 %) (Okada et al. 2009). This green-algal red pigment has recently attracted a lot of attention because of its diverse clinical benefits against age-related functional decline, and muscle or eye fatigue (Guerin et al. 2003; Kidd 2011), e.g., it was shown to improve fine lines/wrinkles and elasticity in the skin of middle-aged women (Yamashita 2006). Stability during storage or during food extrusion at elevated temperatures was shown to be higher with the ester form than with the free form (Gloor and Simon 2007). Ingested astaxanthin was found to be present in the cell membrane or mitochondrion membrane, in which it exerts strong antioxidant activity (Miki 1991; Tatsuzawa et al. 2000; Wolf et al. 2010), i.e., it neutralizes free radicals or other oxidants by either accepting or donating electrons, without being destroyed or becoming a pro-oxidant in the process (Kidd 2011). The global market of Haematococcus astaxanthin for human uses is growing fast, and has been estimated at US$ 35–60 million, according to a Frost and Sullivan 2008 report.
Among higher plants, only the petal of inedible Adonis plants such as Adonis aestivalis, which belong to the Ranunculaceae family, can produce astaxanthin (fatty acid diester, 82.6 %; monoester, 15.8 %; free form, 1.6 %; Maoka et al. 2011), while they biosynthesize benzylisoquinoline alkaloids that can be toxic to humans, which function to defend plants against herbivores and pathogens (Liscombe et al. 2005). Thus, pathway engineering (metabolic engineering) approaches have been widely performed to confer astaxanthin biosynthesis ability on various crop plants (Farré et al. 2010; Misawa 2009). Consequently, it is feasible that astaxanthin could be produced in the edible organs of several crops, e.g., maize (Zea mays; Zhu et al. 2008), potato (Solanum phureja and Solanum tuberosum; Morris et al. 2006; Gerjets and Sandmann 2006), tomato (Solanum lycopersicum; Fraser et al. 2009; Huang et al. 2013), carrot (Daucus carota; Jayaraj et al. 2008), and rapeseed (Brassica napus; Fujisawa et al. 2009). Only the free form of astaxanthin was observed to accumulate in these crop plants, except for the tomato fruit (Huang et al. 2013). In the present study, we constructed transplastomic lettuce plants dominantly producing astaxanthin fatty acid esters and identified the structures of the carotenoids generated in the leaves.
Materials and methods
General molecular cloning experiments
Escherichia coli DH5α (ECOS Competent E. coli DH5α; Nippon Gene, Tokyo, Japan) was utilized as the host for DNA manipulations. PCR amplifications were carried out using a thermal cycler (Applied Biosystems, Foster City, CA). Restriction enzymes and DNA-modifying enzymes were purchased from New England BioLabs (Beverly, CA) or Takara Bio. A Ligation-Convenience Kit (Nippon Gene) was also used. Plasmid DNA was prepared with a QIAprep Miniprep Kit (Qiagen, Hilden, Germany). Nucleotide sequences were confirmed with Bigdye terminator cycle sequencing ready reaction kit version 3.1 (Applied Biosystems) and a model 3730 DNA analyzer (Applied Biosystems). General molecular cloning experiments were carried out according to the suppliers’ manuals or Sambrook and Russell (2001).
Construction of chloroplast transformation plasmids
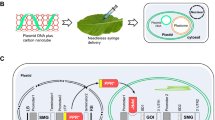
The pRL200 vector, which contains the rbcL and accD genes from the chloroplast (plastid) genome of lettuce (Lactuca sativa L.) cv. Cisco, was constructed for the transformation of lettuce chloroplasts by Kanamoto et al. (2006). Hasunuma et al. (2008) described the plasmid pLD7–rrnP–crtZ–crtW, which included the crtZ and crtW genes encoding the CrtZ and CrtW proteins from Brevundimonas sp. strain SD212 (GenBank/DDBJ accession no. AB377272 and AB377271, respectively) in pLD7–rrnP–MCS (accession no. AB375764), which contained the aadA (aminoglycoside 3′-adenyltransferase; streptomycin/spectinomycin resistance) gene expression cassette [the rrn promoter (P rrn )-aadA-the psbA terminator (T psbA )], P rrn , multiple cloning sites (MCS), and the rps16 terminator (T rps16 ). The aadA–crtZ–crtW fragment was cut out with NotI and SalI from pLD7–rrnP–crtZ–crtW, and inserted into the corresponding site of pRL200, yielding the final construct pRL–crtZW (Supplementary Fig. 1). An idi PCR fragment was amplified from the chemically synthesized idi [isopentenyl diphosphate (IPP) isomerase] gene encoding the Idi protein from Paracoccus sp. strain N81106 (GenBank/DDBJ accession no. AB453829), using a primer set Fidi (5′-AATCAATTGAAAGAGGAGAAATTACATATGACTGAAATCTCTCGTAGAAAG-3′; italic and underlined letters indicate the MfeI and Shine–Dalgarno sequence, respectively.) and Ridi (5′-GGAATTCTTAGAGTCCAGCATCACCAG-3′; italics indicate the EcoRI sequence). This PCR fragment was cut out with MfeI and EcoRI, and inserted into the EcoRI site of pLD7–rrnP–crtZ–crtW, yielding pLD7–rrnP–crtZ–crtW–idi. The aadA–crtZ–crtW–idi fragment was cut out with NotI and SalI from this plasmid, and inserted into the corresponding site of pRL200, yielding the other final construct pRL–crtZWidi (Supplementary Fig. 1).
Chloroplast transformation of lettuce plants
Two cultivars of lettuce, Lactuca sativa L. cv. Berkeley and L. sativa L. cv. Cisco, were purchased from Takii Co. (Kyoto, Japan). The seeds of each cultivar were put on MS medium (Murashige and Skoog 1962) containing 3 % (wt/vol) sucrose and 0.8 % (wt/vol) agar (for Plant Culture Medium; Wako Pure Chemical Industries, Osaka, Japan) after sterilization, and were cultivated for 4 weeks under long-day conditions (16 h/8 h light/dark) at 20 °C. Several young leaves were placed adaxial side up on the lettuce regeneration medium [MS medium supplemented with 3 % sucrose, 0.5 mg/l 6-benzylaminopurine (BA), 0.1 mg/l α-naphthaleneacetic acid, and 0.8 % agar, pH 5.8]. After 1 day of incubation, the leaves of cv. Berkeley and cv. Cisco were bombarded using 0.6 μm gold particles coated with DNA, under the conditions of 900 psi and a 6-cm distance, using a PDS-1000/He Biolistic Particle Delivery System (Bio-Rad, Hercules, USA). Five bombardments were carried out for each plasmid. Two days after the bombardment, the leaves were cut into pieces (4 mm × 4 mm), placed adaxial side down on the lettuce selection medium [lettuce regeneration medium containing 50 mg/l spectinomycin dihydrochloride (Spe; Nacalai Tesque, Kyoto, Japan) and 500 mg/l polyvinylpyrrolidone (PVP)], and were then cultivated for 3–8 weeks under long-day conditions at 20 °C. Regenerated shoots with a red color were transferred onto the lettuce selection medium or the lettuce rooting medium [phytohormone-free 1/2 MS medium supplemented with 1.5 % sucrose, and 0.8 % agar, pH5.8] containing 50 mg/l Spe, and were cultivated continuously. Leaves with a more reddish color were cut into pieces (4 mm × 4 mm), placed adaxial side down on the lettuce regeneration medium, and were cultivated for 3–8 weeks under long-day conditions at 20 °C (reselection). Regenerated shoots were similarly grown to obtain leaves with a more reddish color (reselection). Such a reselection process was repeated three to four times to obtain transplastomic lettuce plants, in which the recombinant genome was almost replaced with the native genome in the chloroplasts. In order to obtain seeds, transplastomic lettuce plants were transferred to soil in pots, and were cultivated at 20–25 °C in a green house for recombinant plants.
Southern blot and PCR analysis
Total DNA was isolated from the leaf of a non-transformed or chloroplast-transformed lettuce, using Wizard Genomic DNA Purification Kit (Promega, Madison, WI). After the DNA samples were digested with NotI and SalI, 1 μg of each was subjected to agarose gel electrophoresis, and was transferred onto a nylon membrane (Amersham, Uppsala, Sweden). A 0.6 kb crtW fragment was amplified from plasmid pLD7–rrnP–crtZ–crtW, using a prime set crtWf (5′-GCTGAGCCTAGAATCGTTCC-3′) and crtWr (5′-CAGAAGATCTAGCGTGGTGAG-3′), and was labeled with PCR DIG Probe Synthesis Kit (Roche Diagnostics, Tokyo, Japan). The blot was hybridized with this probe, and was washed under the conditions of 2× SSC with 0.1 % (w/v) SDS for 15 min at room temperature (two times) and 0.5× SSC with 0.1 % (w/v) SDS for 15 min at 68 °C (two times). The resultant blot was detected with DIG Luminescent Detection Kit for Nucleic Acids (Roche), using LAS-3000 (Fujifilm, Tokyo, Japan).
PCR was performed with the total leaf DNA being isolated from non-transformed or chloroplast-transformed lettuce, using Kaneka High-Speed DNA Polymerase (Kaneka, Osaka, Japan) under the following conditions: 94 °C for 20 s, 55 °C for 10 s, 72 °C for 2 min 30 s; 35 cycles. The primer pair that possessed the sequences in the rbcL and accD genes flanking the transgene integration site in the lettuce chloroplast genome was pRL200-F1844 (5′-GATACTTTGGATCAATAACTTTCGTTCTCTTAAT-3′) and pRL200-R2323 (5′-TTCCTATCAGAATAAGCATATCAATACAATCACTA-3′), respectively.
Extraction and analysis of pigments
Transplastomic lettuce cv. Berkeley (T1) and non-transformed lettuce cv. Berkeley (wild-type; WT) plants were germinated, and grown on soil for 4–8 weeks under long-day conditions at 20 °C. Several leaves [T1, 5.11 g (fresh weight, FW); WT, 6. 86 g (FW)] were cut into narrow pieces, submerged in 20 ml of acetone, and incubated for 5 h at room temperature. Lettuce leaves were moved to mortar and ground with pestle while 50 ml of chloroform (CHCl3)-methanol (MeOH) (1:1) was added stepwise, and incubated for 6 h at room temperature. This extract was merged with the above acetone extract, partitioned with 200 ml of hexane-water [1:1 (v/v)], and the upper solvent layer (colored) was collected. After each upper layer was dehydrated with sodium sulfate anhydrate, total carotenoids were determined quantitatively by spectrophotometry (Schiedt and Liaaen-Jensen 1995). The extinction coefficients (absorbance of 1 % concentration in a 1 cm path-length cuvette), 2,100 at 470 nm and 2,400 at 447 nm, were respectively adopted for the overall quantification of ketocarotenoids including astaxanthin (ester and free forms) and the other carotenoids.
This was evaporated to dryness and re-extracted with acetone/hexane (4:6). Each carotenoid extract was then subjected to HPLC on silica gel (Cosmosil 5SL-II, 4.6 × 250 mm, Nacalai Tesque Inc., Kyoto, Japan), developed with acetone/hexane (2:8) for 20 min, then with acetone/hexane (4:6) at a flow rate of 1.0 ml/min, and was monitored at 450 nm. Carotenoid compositions were estimated from the peak area on HPLC. Individual carotenoids were fractionated during HPLC, and analyzed by UV–VIS, FAB MS, 1H NMR (500 MHz), and CD, as described (Maoka et al. 2011).
Spectroscopic data
(3S,3′S)-Astaxanthin diester:
UV–VIS 470 nm (diethylether; Et2O). FAB-MS m/z 1,072 (Astaxanthin dimyristate), m/z 1,044 (Astaxanthin myristyl palmitate), m/z 1,016 (Astaxanthin dipalmitate). Astaxanthin dimyristate: Astaxanthin myristyl palmitate: Astaxanthin dipalmitate (19:47:34; according to the ratio of their ionic strengths; See Supplementary Fig. 2a). 1H NMR δ (CDCl3) 0.88 (t, J = 7.5 Hz, CH3 in the fatty acid moiety), 1.23 (6H, s, CH3-17, 17′), 1.26 (s, CH2 in the fatty acid moiety), 1.34 (6H, s CH3-16, 16′), 1.70 (t, J = 7.5 Hz, CH2 in the fatty acid moiety), 1.91 (6H, s, CH3-18, 18′), 1.99 (6H, s, CH3-19, 19′), 2.00 (6H, s, CH3-20, 20′), 2.01 (2H, overlapped, H-2, 2′ eq), 2.07 (2H, dd J = 12, 6 Hz, H-2, 2′ ax), 2.08 (t, J = 7.5 Hz, CH2 in the fatty acid moiety), 2.43 (m, CH2 in fatty acid moiety), 5.54 (2H, dd, J = 13.5, 5.5 Hz, H-3, 3′), 6.20 (2H, d, J = 16 Hz, H-7, 7′), 6.30 (2H, d, J = 11.5 Hz, H-10, 10′), 6.32 (2H, m, H-14, 14′), 6.40 (2H, d, J = 16 Hz, H-8, 8′), 6.45 (2H, d, J = 15.5 Hz, H-12, 12′), 6.65 (2H, dd, J = 15.5, 11.5 Hz, H-11, 11′), 6.67 (2H, m, H-15, 15′). CD (Et2O) λ nm (Δε) 240 (−20.0), 252 (0), 270 (+20.0), 284 (0), 314 (−34.0), 355 (0), 372 (+3.0).
(3S,3′S)-Astaxanthin monoester:
UV–VIS 470 nm (Et2O). FAB-MS m/z 834 (Astaxanthin myristate), m/z 806 (Astaxanthin palmitate). Astaxanthin myristate: Astaxanthin dipalmitate (45:55; according to the ratio of their ionic strengths; See Supplementary Fig. 2b). 1H-NMR δ (CDCl3) 0.88 (t, J = 7.5 Hz, CH3 in the fatty acid moiety), 1.21 (3H, s, CH3-17′), 1.23 (3H, s, CH3-17), 1.26 (s, CH2 in the fatty acid moiety), 1.32 (3H, s, CH3-16′), 1.34 (3H, s, CH3-16), 1.70 (t, J = 7.5 Hz, CH2 in the fatty acid moiety), 1.82 (1H, dd, J = 13, 13 Hz, H-2′ax), 1.91 (3H, s, CH3-18), 1.95 (3H, s, CH3-18′), 1.99 (6H, s, CH3-19, 19′), 2.00 (6H, s, CH3-20, 20′), 2.01 (1H, overlapped, H-2 eq), 2.07 (1H, dd, J = 12, 6 Hz, H-2 ax), 2.08 (t, J = 7.5 Hz, CH2 in the fatty acid moiety), 2.16 (1H, dd, J = 12, 5.5 Hz, H-2′ eq), 2.43 (m, CH2 in the fatty acid moiety), 3.69 (1H, d, J = 1.5 Hz, OH-3′), 4.33 (1H, ddd, J = 13.5, 5.5, 1.5 Hz, H-3′), 5.54 (1H, dd, J = 13.5, 5.5 Hz, H-3), 6.20 (1H, d, J = 16 Hz, H-7), 6.21 (2H, d, J = 16 Hz, H-7′), 6.30 (2H, d, J = 11.5 Hz, H-10, 10′), 6.31 (2H, m, H-14, 14′), 6.40 (2H, d, J = 16 Hz, H-8), 6.43 (2H, d, J = 16 Hz, H-8′), 6.45 (2H, d, J = 15.5 Hz, H-12, 12′), 6.66 (2H, dd, J = 15.5, 11.5 Hz, H-11, 11′), 6.67 (2H, m, H-15, 15′). CD (Et2O) λ nm (Δε) 240 (−20.0), 252 (0), 270 (+20.0), 284 (0), 314 (−34.0), 355 (0), 372 (+3.0).
Canthaxanthin:
UV–VIS 470 nm (Et2O). FAB-MS m/z 564 [M]+.
(3S)-Adonirubin:
UV–VIS 470 nm (Et2O). FAB-MS m/z 580 [M]+. CD (Et2O) λ nm (Δε) 220 (+12.0), 230 (0), 234 (−12.0), 252 (0), 275 (+10.0), 283 (0), 310 (−17.0), 340 (0), 370 (+4.4).
(3′S)-Asteroidenone:
UV–VIS 455 nm (Et2O). FAB-MS m/z 566 [M]+.
(3S,3′S)-Astaxanthin:
UV–VIS 470 nm (Et2O). FAB-MS m/z 596 [M]+. CD (Et2O) λ nm (Δε) 240 (−20.0), 252 (0), 270 (+20.0), 284 (0), 314 (−34.0), 355 (0), 372 (+3.0).
Fritschiellaxanthin:
UV–VIS 452 nm (Et2O). FAB-MS m/z 582 [M]+.
4-Ketoantheraxanthin:
UV–VIS 452 nm (Et2O). FAB-MS m/z 598 [M]+ (Shindo et al. 2008).
cis-4-Ketoantheraxanthin:
UV–VIS 448 nm (Et2O). FAB-MS m/z 598 [M]+.
Lactucaxanthin:
UV–VIS 416, 439, 468 nm (Et2O). FAB-MS m/z 568 [M]+.
Lutein:
UV–VIS 422, 445, 474 nm (Et2O). FAB-MS m/z 568 [M]+.
Singlet oxygen suppression experiment
The carotenoid extract from the leaves of the transplastomic lettuce (T1) plants or WT was adsorbed to the Toyopearl-DEAE 650 M column (OH− form, 20 × 200 mm), eluted with acetone/hexane (1/1), and red or yellow fractions were collected and merged. Chlorophylls were removed during this process. The merged fraction (40 ml) was added to 40 ml H2O, and partitioned in the separating funnel. The acetone/hexane layer (upper layer) containing carotenoids was evaporated to dryness. The singlet oxygen (1O2)-quenching activity of total carotenoids from the transplastomic lettuce (T1) or non-transformed lettuce leaves was examined by measuring the methylene blue-sensitized photooxidation of linoleic acid (Kobayashi and Sakamoto 1999). 40 μl of 0.05 mM methylene blue and 10 μl of 2.4 M linoleic acid with or without 40 μl of carotenoid solution [final concentration 1–100 μM, each was dissolved into ethanol (EtOH)] were added to micro glass vials (5.0 ml). The vials were tightly closed with a screw cap and septum, and the mixtures were illuminated at 7,000 lux at 22 °C for 3 h in corrugated cardboard. 50 μl of the reaction mixture was then removed, diluted to 1.5 ml with EtOH, and the absorbance at 235 nm was measured to estimate the formation of conjugate dienes (Hirayama et al. 1994). The value in the absence of carotenoids was determined, and 1O2-quenching activity was calculated relative to this reference value. Activity was indicated as the IC50 value representing the concentration, at which 50 % inhibition was observed.
Results
Construction of transplastomic plants
From the results of the bombardment experiments with the plasmids pRL–crtZWidi and pRL–crtZW (Supplementary Fig. 1) for the two cultivars of lettuce, cv. Berkeley and cv. Cisco, we individually isolated 3–4 seedlings that showed Spe resistance from each cultivar. During reselection processes on the lettuce selection medium, we found that the growth rate of Berkeley was significantly higher than that of Cisco as hosts, and that transformants with pRL–crtZWidi had a higher growth rate than that of transformants with pRL–crtZW. Consequently, we obtained three lines (named ZWidi1, ZWidi2, and ZWidi3) as red seedlings from Berkeley transformed with pRL–crtZWidi, after reselection processes were repeated three to four times. The chloroplast-transformed lettuce lines ZWidi1 and ZWidi3 were able to grow on soil with a growth rate similar to that of non-transformed Berkeley (wild-type; WT) plants and generated flowers and seeds. Figure 1a shows a typical example of such flowers. The size of the transplastomic flowers appeared to be slightly smaller than WT ones. Thus, we measured the length of seeds generated from ZWidi1 or ZWidi3, with the result being shown in Supplementary Fig. 3. There were no statistical differences between the size of the transplastomic and WT seeds. We then examined the germination percentage of the transplastomic lettuce (ZWidi1) seeds. Twenty seeds each from ZWidi1-1, ZWidi1-2, and ZWidi1-3 were found to germinate with a frequency of 98.8 %. On the other hand, 67 WT seeds germinated from 72 WT seeds (at a frequency of 93.1 %). In further experiments, we used transplastomic lettuce plants (T1: second generation) that were raised from the seeds harvested first from several pot plants belonging to the ZWidi1 line. Figure 1b shows the typical outlook of the T1 transplastomic lettuce young plants. A pot plant of T1 in the green house was also shown in Fig. 2. Supplementary Fig. 4 shows the typical outlook of (T2: third generation) transplastomic lettuce seedlings, which appeared to stably inherit the red phenotype.
Confirmation of site-specific integration of the transgenes
Supplementary Fig. 5a shows results of Southern blot analysis of a transplastomic lettuce (T1). The plasmid pRL–crtZWidi contained the 4,061-bp NotI–SalI transgene fragment. Only a band of 4.1 kb appeared in the transplastomic lettuce, which indicated that the full-length transgene fragment was introduced into the genome of this chloroplast-transformed lettuce. We then investigated whether this transgene fragment was site-specifically inserted between the neighboring plastid genes rbcL and accD. PCR was performed with total genome DNA from the leaves of a WT plant and a transplastomic lettuce (T1), using the primer pair that possessed sequences in the rbcL and accD genes flanking the transgene integration site (Supplementary Fig. 5b). The 0.5 and 4.5 kb fragments were expected to appear in the native plastid genome and recombinant plastid genome containing the transgenes, respectively. The results showed that the NotI–SalI transgene fragment was site-specifically inserted between the neighboring plastid genes. However, a 0.5-kb band was also observed in the transplastomic lettuce. This indicates that the native genome was not completely replaced with the recombinant genome in the chloroplasts, even though the reselection processes had been repeated three to four times in these chloroplast-transformed plants.
Carotenoid analysis of transplastomic lettuce
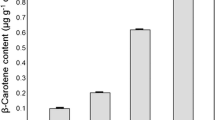
Figure 3a and b show the results of HPLC analysis in the leaves of a WT plant and a transplastomic lettuce (T1), respectively. The carotenoid profile of WT leaves was the same as that of leaves from other higher plants, except for the existence of lactucaxanthin (Britton et al. 2004). The carotenoid content in the WT and transplastomic lettuce (T1) leaves is shown in Table 1. Individual carotenoids produced in the transplastomic leaves were identified by spectroscopic analyses, as described in the Materials and Method section. Carotenoid compositions in the WT and transplastomic leaves are also shown in Table 1. Lactucaxanthin is a carotenoid specific in lettuce plants (Britton et al. 2004), which is also included in this transplastomic lettuce, but in a small amount. Native carotenoids detected in the transplastomic leaves were lutein and lactucaxanthin only. In this chloroplast-transformed lettuce, astaxanthin was the predominant carotenoid, which was present as its fatty acid diester (49.2 %), monoester (18.2 %), and free form (10.0 %), and ketocarotenoids reached 94.9 % of total carotenoids by 4- and/or 4′-ketolation (oxygenation) reactions of β-ring-containing carotenoids. The proposed biosynthetic routes of these carotenoids are show in Fig. 4.
Normal phase HPLC profiles of carotenoids extracted from the leaves of lettuce cv. Berkeley (WT) (a) and from the leaves of transplastomic lettuce cv. Berkeley [ZWidi1-4 (T1)] (b). a 1 β-carotene; 2 β-cryptoxanthin; 3 lactucaxanthin; 4 lutein; 5 zeaxanthin; 6 lutein-5,6-epoxide; 7 antheraxanthin; 8 chlorophyll degradation products; 9 9′-cis-neoxanthin. 9′-cis-Neoxanthin was thought to be generated from neoxanthin with an unknown isomerase (see Fig. 4). b 1 (3S,3′S)-astaxanthin diesters; 2 canthaxanthin; 3 (3S,3′S)-astaxanthin monoesters; 4 (3S)-adonirubin (phoenicoxanthin); 5 (3′S)-asteroidenone (3′-hydroxyechinenone); 6 (3S,3′S)-astaxanthin; 7 fritschiellaxanthin; 8 lactucaxanthin; 9 lutein; 10 4-ketoantheraxanthin; 11 cis-4-ketoantheraxanthin; 12 chlorophyll degradation products
Carotenoid biosynthetic pathway in lettuce leaves and proposed carotenoid biosynthetic pathway in the transplastomic lettuce cv. Berkeley (T1) leaves. The names of carotenoids detected in the transplastomic leaves were shown with pink letters. R–C=O in the astaxanthin diester or monoester refers to the acyl chain of palmitate (C16) or myristate (C14). (Color figure online)
Singlet oxygen suppression of carotenoids from transplastomic lettuce
Total carotenoids were extracted from the leaves of a WT plant and transplastomic lettuce line [ZWidi1-4 (T1)]. The result of 1O2 suppression experiment showed that their IC50 values were 42.1 and 8.1 μM, respectively. Astaxanthin was collected as a reference from a capsule of DHC (Tokyo, Japan) commercially available as health food astaxanthin, and its IC50 value was found to be 7.6 μM.
Discussion
Plastid genetic engineering offers several attractive features and advantages over nuclear transformation, e.g., they include very high transgene expression levels because of plastid genome polyploidy (plastome; up to 10,000 copies/cell) and/or the high stability of foreign proteins, and an absence of the pollen transmission of transgenes because of transgene containment via the maternal inheritance of plastids in most higher plants (Day and Goldschmidt-Clermont 2011; Rogalski and Carrer 2011). The latter feature appears to be important specifically for the modern biotechnology of crops such as lettuce, which belongs to the Asteraceae family, since many weeds belong to the same family in temperate zone countries including Japan. Hasunuma et al. (2008) generated transplastomic tobacco (Nicotiana tabacum) plants, which contained two genes that coded for β,β-carotenoid (or β-carotene) 3,3′-hydroxylase (CrtZ) and β,β-carotenoid (or β-carotene) 4,4′-ketolase (4,4′-oxygenase; CrtW) from a marine bacterium Brevundimonas sp. strain SD212 belonging to the Alphaproteobacteria class (Nishida et al. 2005). These two genes (crtZ and crtW) were found to be the best combination for the efficient production of astaxanthin from β-carotene using the E. coli complementation system (Choi et al. 2006, 2005). Leaves of the generated transplastomic tobacco plant biosynthesized 5.13 or 5.44 mg/g dry weight (513 or 544 μg/g FW if calculated as 90 % of water content) of astaxanthin (free form), which corresponded to 71 or 74 % of total carotenoids (Hasunuma et al. 2008). In the present study, we utilized the idi gene encoding IPP isomerase (type 2) from the marine bacterium Paracoccus sp. strain N81106 belonging to the Alphaproteobacteria class (Maruyama et al. 2007) in addition to the same crtZ and crtW genes. The idi gene was found to be effective in obtaining transplastomic lettuce plants of interest. Surprisingly, the transplastomic lettuce (T1), which contained the crtZ, crtW and idi genes as transgenes, produced astaxanthin fatty acid esters predominantly (67 % of total carotenoids) in the leaves of the photosynthesis apparatus. The leaves of the transplastomic tobacco (T0) as well as various organs of many transgenic plants biosynthesized only the free form of astaxanthin (Hasunuma et al. 2008; and see the Introduction section), while the Adonis flowers and transgenic tomato fruits predominantly synthesized astaxanthin esters (Maoka et al. 2011; Huang et al. 2013). Therefore, we concluded that astaxanthin is occasionally esterified in higher plants, although the reason for this is unclear. We identified fatty acid moieties included in the astaxanthin esters of the transplastomic lettuce as myristate and palmitate. This is the first report to structurally identify the astaxanthin esters biosynthesized in transgenic or transplastomic plants producing astaxanthin.
Astaxanthin, which we ingest as food, is included in red fishes such as salmon, trout, and sea bream, and in crustaceans such as crab, krill, shrimp, prawn, and lobster. Astaxanthin in the integument of crustaceans or red fishes is esterified, while the muscle (flesh) and egg of salmon or trout contain the free form. Astaxanthin contents in the integument of crayfish, the skin of salmon, and the muscle or egg of salmon were shown to be 1–3, 1, and 20–38 μg/g FW, respectively (Ando et al. 1989; Matsuno et al. 1980; Schiedt et al. 1995). On the other hand, the transplastomic lettuce leaves (Table 1) contained 178 μg/g FW of astaxanthin, if calculated as the content of the free form. Thus, the transplastomic lettuce is promising as a functional (health) food supplying astaxanthin. AstaReal Co., (Tokyo, Japan) recommends the intake of 6 mg/day of astaxanthin as a health food, which corresponds to 34 g FW of the transplastomic lettuce leaves. An in vitro antioxidative experiment demonstrated that the carotenoid extract from the transplastomic lettuce had a similar 1O2-quenching activity to that of astaxanthin from H. pluvialis. Whereas, the carotenoid extract from non-transformed lettuce exhibited a markedly lower antioxidative effect than that from the transplastomic lettuce.
Interestingly, β-carotene, zeaxanthin, antheraxanthin, violaxanthin, and (9′-cis-)neoxanthin were not detected in the transplastomic lettuce leaves, while small amounts of lactucaxanthin and lutein were present as native carotenoids (Table 1). Such transgenic plants, i.e., higher plants without β-carotene, zeaxanthin, antheraxanthin, and violaxanthin, have never been constructed through nuclear transformation by pathway engineering for astaxanthin production, although many transgenic plants of interest have been generated (see the Introduction section). This finding strongly suggests that the expression of carotenoid biosynthesis genes or the stable accumulation of gene products in the plastids was more efficient with genetic engineering of the plastid genome than with that of the nucleus genome. The absence of these native carotenoids in the transplastomic lettuce indicates that its chloroplasts never possessed the xanthophyll cycle mediated by the conversion from violaxanthin to zeaxanthin via antheraxanthin, which safely dissipates excess light energy (Demmig-Adams and Adams 1996). This absence also means that light-harvesting complexes (LHCs) cannot possess lutein, violaxanthin, and 9′-cis-neoxanthin in their appropriate proportions in the transplastomic lettuce. The artificial ketocarotenoids that exist in the transplastomic leaves may fulfil compensatory functions of the xanthophyll cycle or the carotenoids in LHCs; however, relevant physiological experiments using the transplastomic lettuce are needed. β-Carotene, which is one of the main carotenoids that protect plant leaves from damage caused by 1O2, was also missing in the transplastomic lettuce leaves. Ketocarotenoids, such as astaxanthin and canthaxanthin, which possess stronger 1O2-quenching activity (Miki 1991), may mimic or cover this function of β-carotene. Plant hormone abscisic acid is considered to be generated from 9-cis-epoxycarotenoids including 9′-cis-neoxanthin by 9-cis-epoxycarotenoid dioxygenase (Milborrow 2001). 9′-cis-Neoxanthin was the only 9-cis-epoxycarotenoid that existed in non-transformed lettuce, and was missing in the transplastomic lettuce (Table 1). Instead, this transplastomic plant included a small amount of cis-4-ketoantheraxanthin, which is considered to be 9′-cis-ketoantheraxanthin, although its structural confirmation was not feasible because of its small amount. 9′-cis-Ketoantheraxanthin is likely to be metabolized to abscisic acid, because this carotenoid possesses the same 9-cis-epoxycarotenoid moiety to 9′-cis-neoxanthin, and disorders caused by the total loss of abscisic acid, including early germination before seed harvest, were not observed in the transplastomic lettuce. Omics analyses containing transcriptomes, proteomes, and metabolomes (metabolomics) are currently being conducted using the lettuce transformants, and the results of these analyses will be published in the near future. Finally, we want to emphasize that lettuce is one of the most suitable edible plants for their production in indoor environments. For example, when 49 lettuce cv. Berkeley (WT) plants were examined for their growth within a 20-cm distance in a hydroponics factory for vegetables, only 40 days were needed for a harvest of 93 ± 19 (SD) g FW of Berkeley after seeding (Smile Leaf Spica Co., kindly carried out this trial).
In conclusion, we constructed reproducible chloroplast-transformed lettuce that produced astaxanthin fatty acid esters (67.4 % of total carotenoids), and the free forms of astaxanthin (10.0 %) and the other ketocarotenoids (17.5 %), which indicated that artificial ketocarotenoids corresponded to 94.5 % of total carotenoids. Native carotenoids were there lactucaxanthin (3.8 %) and lutein (1.3 %) only. This transplastomic lettuce may not only be a new astaxanthin- and other ketocarotenoids-supplying vegetable as a candidate of functional food (health food) or functional feed, but may also be a model plant for physiological studies on the roles of carotenoids in light harvesting and the protection of plant leaves from photooxidative damage.
References
Ando S, Osada K, Hatano M, Saneyoshi M (1989) Comparison of carotenoids in muscle and ovary from four genera of salmonoid fishes. Comp Biochem Physiol 93B:503–508
Britton G, Liaaen-Jensen S, Pfander H (2004) Carotenoids handbook. Birkhäuser Verlag, Basel
Choi SK, Nishida Y, Matsuda S, Adachi K, Kasai H, Peng X, Komemushi S, Miki W, Misawa N (2005) Characterization of β-carotene ketolases, CrtW, from marine bacteria by complementation analysis in Escherichia coli. Mar Biotechnol 7:515–522
Choi SK, Matsuda S, Hoshino T, Peng X, Misawa N (2006) Characterization of bacterial β-carotene 3,3′-hydroxylases, CrtZ, and P450 in astaxanthin biosynthetic pathway and adonirubin production by gene combination in Escherichia coli. Appl Microbiol Biotechnol 72:1238–1246
Day A, Goldschmidt-Clermont M (2011) The chloroplast transformation toolbox: selectable markers and marker removal. Plant Biotechnol J 9:540–553
Demmig-Adams B, Adams WW III (1996) The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci 1:21–26
Farré G, Sanahuja G, Naqvi S, Bai C, Capell T, Zhu C, Christou P (2010) Travel advice on the road to carotenoids in plants. Plant Sci 179:28–48
Fraser PD, Enfissi EMA, Bramley PM (2009) Genetic engineering of carotenoid formation in tomato fruit and the potential application of systems and synthetic biology approaches. Arch Biochem Biophys 483:196–204
Fujisawa M, Takita E, Harada H, Sakurai N, Suzuki H, Ohyama K, Shibata D, Misawa N (2009) Pathway engineering of Brassica napus seeds using multiple key-enzyme genes involved in ketocarotenoid formation. J Exp Bot 60:1319–1332
Gerjets T, Sandmann G (2006) Ketocarotenoid formation in transgenic potato. J Exp Bot 57:3639–3645
Gloor A, Simon W (2007) Astaxanthin esters. United States Patent US 7,253,297 B2
Guerin M, Huntley ME, Olaizola M (2003) Haematococcus astaxanthin: applications for human health and nutrition. Trends Biotechnol 21:210–216
Hasunuma T, Miyazawa SI, Yoshimura S, Shinzaki Y, Tomizawa KI, Shindo K, Choi SK, Misawa N, Miyake C (2008) Biosynthesis of astaxanthin in tobacco leaves by transplastomic engineering. Plant J 55:857–868
Hirayama O, Nakamura K, Hamada S, Kobayashi Y (1994) Singlet oxygen quenching ability of naturally occurring carotenoids. Lipids 29:149–150
Huang JC, Zhong YJ, Liu J, Sandmann G, Chen F (2013) Metabolic engineering of tomato for high-yield production of astaxanthin. Metab Eng 17:59–67
Jayaraj J, Devlin R, Punja Z (2008) Metabolic engineering of novel ketocarotenoid production in carrot plants. Transgenic Res 17:489–501
Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa KI (2006) Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res 15:205–217
Kidd PM (2011) Astaxanthin, cell membrane nutrient with diverse clinical benefits and anti-aging potential. Altern Med Rev 16:355–364
Kobayashi M, Sakamoto Y (1999) Singlet oxygen quenching ability of astaxanthin esters from the green alga Haematococcus pluvialis. Biotechnol Lett 21:265–269
Krinsky NI, Landrum JT, Bone RA (2003) Biological mechanisms of the protective role of lutein and zeaxanthin in the eye. Annu Rev Nutr 23:171–201
Liscombe DK, MacLeod BP, Loukanina N, Nandi OI, Facchini PJ (2005) Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry 66:1374–1393
Maeda H, Hosokawa M, Sashima T, Funayama K, Miyashita K (2005) Fucoxanthin from edible seaweed, Undaria pinnatifida, shows antiobesity effect through UCP1 expression in white adipose tissues. Biochem Biophys Res Commun 332:392–397
Maoka T, Etoh T, Kishimoto S, Sakata S (2011) Carotenoids and their fatty acid esters in the petals of Adonis aestivalis. J Oleo Sci 60:47–52
Maruyama T, Kasai H, Choi SK, Ramasamy AK, Inomata Y, Misawa N (2007) Structure of a complete carotenoid biosynthesis gene cluster of marine bacterium Paracoccus sp. strain N81106. Carotenoid Sci 11:50–55
Matsuno T, Katsuyama M, Nagata S (1980) Comparative biochemical studies of carotenoids in fishes-XIX Carotenoids of chum salmon, coho salmon, biwa trout, red-spotted masu salmon, masu salmon, kokanee. Bull Jpn Soc Sci Fish 46:879–884
Miki W (1991) Biological functions and activities of animal carotenoids. Pure Appl Chem 63:141–146
Milborrow BV (2001) The pathway of biosynthesis of abscisic acid in vascular plants: a review of the present state of knowledge of ABA biosynthesis. J Exp Bot 52:1145–1164
Misawa N (2009) Pathway engineering of plants toward astaxanthin production. Plant Biotechnol 26:93–99
Morris WL, Ducreux LJ, Fraser PD, Millam S, Taylor MA (2006) Engineering ketocarotenoid biosynthesis in potato tubers. Metab Eng 8:253–263
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol Plant 15:473–497
Nishida Y, Adachi K, Kasai H, Shizuri Y, Shindo K, Sawabe A, Komemushi S, Miki W, Misawa N (2005) Elucidation of a carotenoid biosynthesis gene cluster encoding a novel enzyme, 2,2′-β-hydroxylase, from Brevundimonas sp. strain SD212 and combinatorial biosynthesis of new or rare xanthophylls. Appl Environ Microbiol 71:4286–4296
Nishino H, Murakoshi M, Ii T, Takemura M, Kuchide M, Kanazawa M, Mou XY, Wada S, Masuda M, Ohsaka Y, Yogosawa S, Satomi Y, Jinno K (2002) Carotenoids in cancer chemoprevention. Cancer Metastasis Rev 21:257–264
Okada Y, Ishikura M, Maoka T (2009) Bioavailability of astaxanthin in Haematococcus algal extract: the effects of timing of diet and smoking habits. Biosci Biotechnol Biochem 73:1928–1932
Rogalski M, Carrer H (2011) Engineering plastid fatty acid biosynthesis to improve food quality and biofuel production in higher plants. Plant Biotechnol J 9:554–564
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Schiedt K, Liaaen-Jensen S (1995) Isolation and analysis. In: Britton G, Liaaen-Jensen S, Pfander H (eds) Carotenoids, vol 1A. Birkhäuser, Basel, pp 81–108
Schiedt K, Bischof S, Glinz E (1995) Example 5: fish isolation of astaxanthin and its metabolites from skin of Atlantic salmon (Salmo salar). In: Britton G, Liaaen-Jensen S, Pfander H (eds) Carotenoids, vol 1A. Birkhäuser, Basel, pp 243–252
Shindo K, Hasunuma T, Asagi E, Sano A, Hotta E, Minemura N, Miyake C, Maoka T, Misawa N (2008) 4-Ketoantheraxanthin, a novel carotenoid produced by the combination of the bacterial enzyme β-carotene ketolase CrtW and endogeneous carotenoid biosynthetic enzymes in higher plants. Tetrahedron Lett 49:3294–3296
Sugiura M, Nakamura M, Ogawa K, Ikoma Y, Yano M (2012) High serum carotenoids associated with lower risk for bone loss and osteoporosis in post-menopausal Japanese female subjects: prospective cohort study. PLoS ONE 7:e52643
Talegawkar SA, Johnson EJ, Carithers TC, Taylor HA Jr, Bogle ML, Tucker KL (2008) Carotenoid intakes, assessed by food-frequency questionnaires (FFQs), are associated with serum carotenoid concentrations in the Jackson Heart Study: validation of the Jackson Heart Study Delta NIRI Adult FFQs. Public Health Nutr 11:989–997
Tatsuzawa H, Maruyama T, Misawa N, Fujimori K, Nakano M (2000) Quenching of singlet oxygen by carotenoids produced in Escherichia coli-attenuation of singlet oxygen-mediated bacterial killing by carotenoids. FEBS Lett 484:280–284
Wolf AM, Asoh S, Hiranuma H, Ohsawa I, Iio K, Satou A, Ishikura M, Ohta S (2010) Astaxanthin protects mitochondrial redox state and functional integrity against oxidative stress. J Nutr Biochem 21:381–389
Yamashita E (2006) The effects of a dietary supplement containing astaxanthin on skin condition. Carotenoid Sci 10:91–95
Zhu C, Naqvi S, Breitenbach J, Sandamnn G, Christou P, Capell T (2008) Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc Natl Acad Sci USA 105:18232–18237
Acknowledgments
The authors are grateful to Central Laboratories for Frontier Technology, Kirin Holdings Co., Ltd., since this work was initially performed there under support from the New Energy and Industrial Technology Development Organization (NEDO). We thank Dr. Tomohisa Hasunuma for the gift of the pLD7–rrnP–crtZ–crtW plasmid. The authors also thank Mss Miyuki Murakami, Megumi Hashida, and Kazuko Arai for their assistance in experiments on plant transformation and cultivation. We also thank Mss Kumiko Ito and Nami Fukuo, Nihon Women’s University, for their in vitro antioxidative experiments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Harada, H., Maoka, T., Osawa, A. et al. Construction of transplastomic lettuce (Lactuca sativa) dominantly producing astaxanthin fatty acid esters and detailed chemical analysis of generated carotenoids. Transgenic Res 23, 303–315 (2014). https://doi.org/10.1007/s11248-013-9750-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-013-9750-3