Abstract
Hedychium coronarium, a perennial herb belonging to the family Zingiberaceae, is cultivated as a garden plant or cut flower as well as for medicine and aromatic oil. Its flowers emit a fresh and inviting scent, which is mainly because of monoterpenes present in the profile of the floral volatiles. However, fragrance produced as a result of monoterpenes has not been well studied. In the present study, two novel terpene synthase (TPS) genes (HcTPS7 and HcTPS8) were isolated to study the biosynthesis of monoterpenes in H. coronarium. In vitro characterization showed that the recombinant HcTPS7 was capable of generating sabinene as its main product, in addition to nine sub-products from geranyl diphosphate (GPP). Recombinant HcTPS8 almost specifically catalyzed the formation of linalool from GPP, while it converted farnesyl diphosphate (FPP) to α-bergamotene, cis-α-bisabolene, β-farnesene and other ten sesquiterpenes. Subcellular localization experiments revealed that HcTPS7 and HcTPS8 were located in plastids. Real-time PCR analyses showed that HcTPS7 and HcTPS8 genes were highly expressed in petals and sepals, but were almost undetectable in vegetative organs. The changes of their expression levels in petals were positively correlated with the emission patterns of sabinene and linalool, respectively, during flower development. The results indicated that HcTPS7 and HcTPS8 were involved in the biosynthesis of sabinene and linalool in H. coronarium flowers. Results on these two TPSs first characterized from H. coronarium provide new insights into molecular mechanisms of terpene biosynthesis in this species and also lay the basis for biotechnological modification of floral scent profile in Hedychium.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Floral scent provides an important means of communication between flowering plants and pollinators during their long evolution, especially in long-distance communication (Dudareva and Pichersky 2000). Potential pollinators can easily locate and identify the scented flowers promoting a connection between the plants and their pollinators through special compounds or unique ratios of general compounds (Raguso 2008). The efficient “attraction and pollination model”, which increases fertilization rates and reduces energy consumption by the plants and their pollinators, improves the plants adaptability to its environment (Pichersky and Dudareva 2007). The anti-microbial or anti-herbivore activity of volatiles emitted from flowers protects the vulnerable reproductive organs of plants from their enemies (Dudareva et al. 2004). Like floral color, longevity and form, floral fragrance is a significant ornamental characteristic that enhances the esthetic values of the ornamental plants (Pichersky and Dudareva 2007).
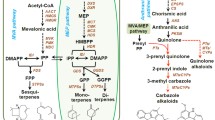
Floral scent is determined by a complex mixture of low-molecular weight volatile substances belonging to three major groups: terpenoids, phenylpropanoids/benzenoids, and fatty acid derivatives (Vainstein et al. 2001). The largest class of volatiles is the terpenoids, which is a very large and structurally diverse group of secondary metabolites. All terpenoids are derived from the five-carbon compound isopentenyl pyrophosphate (IPP) and its allylic isomer dimethylallyl pyrophosphate (DMAPP) via two alternative pathways: the cytosolic mevalonate (MVA) pathway and the plastidial methylerythritol phosphate (MEP) pathway (Pichersky et al. 2006). The successive head-to-tail condensation of IPP and DMAPP by the action of prenyltransferases generates the direct precursors of terpenes, GPP and geranylgeranyl diphosphate (GGPP) in plastids, as well as FPP in cytosol. Thereafter, TPSs convert GPP, FPP and GGPP into structurally diverse monoterpenes, sesquiterpenes and diterpenes, respectively (Yu and Utsumi 2009). TPSs are the primary enzymes in the terpenoid biosynthetic pathways (Tholl 2006). It is generally believed that the biosynthesis of monoterpenes and sesquiterpenes is compartmentalized wherein the monoterpene is produced in the plastids where GPP is synthesized, and the sesquiterpene is formed in the cytosol where FPP is generated. However, metabolic engineering in Arabidopsis, tobacco and tomato uncovered the existence of a small FPP pool in plastids and a GPP pool in the cytosol (Aharoni et al. 2003; Davidovich-Rikanati et al. 2008; Gutensohn et al. 2013; Wu et al. 2006).
So far, some TPSs responsible for the formation of floral volatile terpenes have been characterized in several ornamental plants including Clarkia breweri (Dudareva et al. 1996; Pichersky et al. 1995), rose (Guterman et al. 2002), snapdragon (Dudareva et al. 2003; Nagegowda et al. 2008) and Alstroemeria (Aros et al. 2012) and in the model plant Arabidopsis (Chen et al. 2003; Tholl et al. 2005). Volatile terpenes are often released from specific floral tissues at particular times or developmental stages to attract pollinators (Tholl 2006). However, terpene production is always correlated with the transcriptional activity of the corresponding TPS genes. For example, in Arabidopsis, two flower-specific TPS genes (TPS11 and TPS21) account for the formation of almost all sesquiterpenes emitted from flowers (Tholl et al. 2005). In snapdragon, two nearly identical TPS genes are responsible for linalool and nerolidol biosynthesis in flowers. The expression pattern of both TPS genes is positively correlated to the emission of linalool and nerolidol (Nagegowda et al. 2008).
Hedychium coronarium (commonly named as white ginger lily or butterfly ginger) is a perennial herb that belongs to the family Zingiberaceae. It is commonly found in tropical and subtropical regions, such as Southern China, India, Brazil, and Southeast Asian countries. This herb is often cultivated as a garden plant or for cut flowers because of its distinct and highly fragrant flower, and also grown for medicine and aromatic oil (Wu and Raven 2001). The blooming of H. coronarium flowers results in the emission of a large number of volatile compounds, which mainly consist of the monoterpene linalool, (E)/(Z)-β-ocimene, 1,8-cineole, sabinene, α-thujene, myrcene, α/β-pinene, and sesquiterpene α-farnesene, β-caryophyllene, as well as some benzenoids (Baez et al. 2011; Fan et al. 2003, 2007). The volatile compounds from this herb are more fragrant than other studied species, such as snapdragon and rose. Therefore, it is an ideal material to study the formation and emission of floral scents. However, there has been very little research on the formation of this fragrance in H. coronarium. Until now, one TPS gene with unsure function (Li and Fan 2011) and one FPP synthase gene possibly related to sesquiterpene synthesis (Lan et al. 2013) have been reported in H. coronarium. The genes that are responsible for the biosynthesis of floral volatile components are largely unknown. In addition, many species in Hedychium are of high ornamental values because of their showy flowers with myriad colors and shapes, or their fragrances. However, some species with fine ornamental traits in Hedychium lack the fragrance, such as H. coccineum (Wu and Raven 2001). Therefore, researches on the formation of floral scent will facilitate the breeding of scent-related traits in Hedychium. The present study describes the cloning, biochemical characterization, subcellular localization and expression pattern of two monoterpene synthases involved in the formation and emission of sabinene and linalool in H. coronarium flowers.
Materials and methods
Plant materials
Hedychium coronarium, H. forrestii, H. coccineum and H. gardnerianum were grown in the horticulture chamber in South China Agricultural University under natural light from April to November, and the materials used in the experiments were collected in September with ~12-h day length. Hedychium coronarium flowers for gene expression and volatiles analysis of different flower developmental stages bought from a local H. coronarium cut flowers farm. The cut flowers were immediately cultured in MS liquid medium after they were brought back to laboratory. They had the similar flower developmental process with natural flowers. Arabidopsis thaliana ecotype Col-0 for subcellular localization experiments was grown in a growth room at 22 °C with 12-h light and 12-h dark cycles.
Reagents and radiochemicals
Unlabeled GPP, FPP and terpenoid standards were purchased from Sigma-Aldrich. [1-3H]-GPP (20 Ci/mmol) and [1-3H]-FPP (20 Ci/mmol) were purchased from American Radiolabeled Chemicals. All other reagents or solvents were obtained from Sangon Biotech, Sigma-Aldrich unless otherwise stated. The enzyme preparations were purchased from TaKaRa unless otherwise stated.
Cloning of HcTPS7 and HcTPS8 genes
Two putative partial ginger TPS sequences (DY354445 and DY357133) were found in the ginger-expressed sequence tag database (http://www.agcol.arizona.edu/cgi-bin/pave/GT/index.cgi) using a previous cloned H. coronarium TPS gene as the inquiry sequence in BLAST searches. Total RNA from H. coronarium flowers for genes cloning was isolated with RNAiso Plus reagent (TaKaRa) following the manufacturer’s protocol. To obtain 3′ and 5′ ends of the two clones, first-strand cDNA synthesis and rapid amplification of cDNA ends (RACE) were performed using Full RACE Kit (TaKaRa) according to manufacturer’s introduction. For 3′ RACE, Gene-specific forward primers were 5′-GGAGCTTTGAAACCTGGTTTGATC-3′ for HcTPS7 and 5′-CTGCACGTCTCTTTAGGCTTCTC-3′ for HcTPS8. Nested gene-specific primers for second round were 5′-TGAACTGGAGATTCCAGCGATTGCAC-3′ for HcTPS7 and 5′-GGAATGCTGAGTTTGTATGAGGC-3′ for HcTPS8. Based on the sequence information from 3′ end amplicons, Gene-specific reverse primers for 5′ RACE were 5′-TCTTTGTGTCCATCTCCCTAAAAGC-3′ for HcTPS7 and 5′-AAAGCAGTTTCCTTTTGTTTGTGCG-3′ for HcTPS8. Nested gene-specific reverse primers were 5′-CGTTGAAGTCCAACTTGGCCAACAG-3′ for HcTPS7 and 5′-CAAGAAGAAGAGGGTTGTCGATGAC-3′. The full-length cDNAs were amplified using high-fidelity DNA polymerase KOD-Plus (TOYOBO) following the manufacturer’s recommendation. Forward primers were 5′-GGCATTAAGTTAAGATGTCTGTCTC-3′ for HcTPS7 and 5′-TCGGATCATCAGGAAATCCAAGT-3′ for HcTPS8. Reverse primers were 5′-TTATTAACGGTTACACGAGTTCG-3′ for HcTPS7 and 5′-GCTGATGGGCGAGGCTTTATAT-3′ for HcTPS8. PCR conditions were as follows: 94 °C for 2 min, followed by 35 cycles of 98 °C for 15 s, 55 °C for 30 s, 68 °C for 2 min. The product was ligated into pMD-19 vector after the A base was added on 3′ end by means of Taq DNA polymerase. The resulting plasmid DNA was sequenced.
Genomic DNA was isolated from the flowers using the Plant Genomic DNA Kit (TIANGEN). Genomic sequences of HcTPS7 and HcTPS8 were amplified with primers for full-length cDNA amplification. The alignment was performed by ClustalX and shaded with GeneDoc. For phylogenetic analysis, the tree was constructed with MEGA4 after the amino acid sequences were aligned using ClustalX.
Heterologous expression of HcTPSs in Escherichia coli
Most monoterpene synthases have an N-terminal transit peptide that could cause the formation of inclusion body in bacterial hosts (Williams et al. 1998). So, the open reading frames (ORFs) excluding the N-terminal transit peptides (Fig. 1) were amplified by PCR using high-fidelity DNA polymerase KOD-Plus (TOYOBO) and then cloned into pET30a vector (Novagen). The truncated HcTPSs were fused to six C-terminus histidine residues to facilitate protein purification. The resulting constructs were transformed into E. coli Rosetta (DE3) competent cells (Invitrogen). Single positive colonies verified by sequencing were incubated overnight at 37 °C with shaking at 180 rpm in Luria–Bertani (LB) liquid medium supplemented with kanamycin (100 μg/ml) and chloramphenicol (34 μg/ml). The next day the grown cultures were diluted 1:50 with LB medium under the same conditions as above until OD600 of 0.4–0.6 was achieved. Cell cultures were inducted with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and then incubated overnight at 16 °C and 180 rpm. The cells were separated from the medium by centrifugation at 5,000g and 4 °C for 5 min. The pellets were resuspended in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole) containing 0.5 mg/ml lysozyme and 1 mM phenylmethanesulfonyl fluoride (PMSF). The reconstituted pellets were incubated on ice for 20 min. The cells were then sonicated thirty times (work 3 s with 3 s intervals for each time) on ice and the clarified lysate was collected by centrifugation at 12,000g and 4 °C for 10 min. The lysate was loaded onto a Ni–NTA His·Bind Resins (Novagen) to purify His-tagged recombinant protein following manufacture’s introduction. Partially purified proteins were dialyzed twice against a buffer containing 30 mM HEPES (pH 7.5) and 5 mM dithiothreitol (DTT). Protein concentrations were determined by the Bradford method (Bradford 1976) using bovine serum albumin as the standard. Bacterial lysates and purified enzyme samples were examined on SDS-PAGE stained with Coomassie brilliant blue. The specific amount of TPS protein was determined by quantitative comparison of the intensity of the Coomassie brilliant blue-stained TPS protein band with those of bovine serum albumin standards.
Alignment of deduced amino acid sequences of HcTPS7 and HcTPS8 with S. pomifera sabinene synthase (Sp-SabS1, ABH07678), L. angustifolia linalool synthase (LaLINS, ABB73045) and M. citrata linalool synthase (McLINS, AAL99381). Amino acid residues shaded in black, gray and light gray represent 100, 80 and 60 % conserved identity, respectively. Dashes indicate gaps inserted for optimal alignment. The conserved RRX8W and DDXXD motifs of monoterpene synthases are underlined. Triangle indicates truncation sites of HcTPS7 and HcTPS8 for heterologous expression assay in E. coli
Enzyme assay and production identification
For enzyme characterization, standard assays were performed in a total volume of 100 μl containing buffer (30 mM HEPES, pH 7.5, 5 mM DTT, 20 mM MgCl2), 20 μM [1-3H]-GPP (50 mCi/mmol) and 1–10 μg partially purified protein. The mixture was immediately overlaid with 500 μl hexane and incubated at 30 °C for 20 min. The reaction was stopped by vigorous mixing and centrifugation to separate the phase. A volume of 400 μl hexane containing radioactive reaction products was counted by liquid scintillation counter (PerkinElmer Tri-carb 2910TR, efficiency for 3H = 62.51 %). Five reaction time points (5, 10, 15, 20 and 30 min), seven temperature levels (22.5, 25, 27.5, 30, 32.5, 35 and 37.5 °C) and HEPES buffer with eight pH levels (5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5 and 9.0) were selected to evaluate enzyme characterization. For the determination of the metal ion requirement, assays were performed in buffer with varying concentrations of MgCl2 (0–100 mM) or MnCl2 (0–10 mM). For evaluation of the K m value, different concentrations of GPP were applied in standard assays in three replications. Calculations of the K m value and V max were obtained by hyperbolic regression of resulting data using SigmaPlot 10.0 (Systat Software). Controls in all assays were performed under the same conditions, but using the purified product of the empty vector or heat-denatured enzyme.
For the identification of reaction products, enzyme assays were carried out in 5-ml sealed glass vials in a total volume of 1 ml containing recombinant HcTPS proteins, unlabeled GPP/FPP (20 μM) and buffer as described above for standard assays. A polydimethylsiloxane (PDMS, with 50/30 μm divinylbenzene/Carboxen) fiber (Supelco) for solid-phase microextraction was inserted into the vial to collect volatiles. The mixture was incubated at 30 °C for 1 h and then at 45 °C for 15 min. After incubation, the PDMS fiber was injected into a gas chromatography–mass spectrometry (GC–MS) system for analysis. GC–MS analyses were performed on Agilent 7890A gas chromatography system coupled with Agilent 5975C mass spectrometry detector. The instrument was equipped with an Agilent DB-5MS capillary column (30 m × 0.25 mm) and helium as a carrier gas at a constant flow of 1 ml/min. The oven temperature was initially maintained at 40 °C for 2 min, followed by an increase to 250 °C at a rate of 5 °C/min, and held at 250 °C for 5 min. The products were identified by comparing the mass spectra and retention times with authentic standards or known volatile compounds of H. coronarium, or by comparing mass spectra with the NIST 08 mass spectra library.
Subcellular localization experiments
The various fragments used for localization experiments were fused upstream of, and in frame with, green fluorescent protein (GFP) in the cloning sites Spe I and Sma I (at the 5′ and 3′ end of both inserts, respectively) of the p35S-EGFP-1 vector. Sequencing proved that no errors had been introduced. Arabidopsis protoplasts were isolated and transformed as described by Sheen (Yoo et al. 2007). Protoplasts were visualized 16 h after transformation using Leica TCS SP2 AOBS Spectral Confocal Scanner mounted on a Leica DM RXA2 upright fluorescent microscope with 40 × 0.75 numerical aperture objective. GFP fluorescence was excited at 488 nm and chlorophyll autofluorescence at 543 nm using Ar ion laser and HeNe laser (LASOS Lasertechnik GmbH), respectively. Triple dichroic filter (TD) was used to collect fluorescence (500–535 nm for green and 555–700 nm for red). Images were processed from optical sections taken along the optical axis and projected into one image using the Leica LCS 2.61 software.
Validation of reference genes for quantitative real-time PCR
Total RNA for expression analysis of different tissues was isolated using an improved hexadecyl trimethyl ammonium bromide (CTAB) method (Jaakola et al. 2001; Wang and Zhou 2008). The genomic DNA was digested with DNase I at 37 °C for 10 min following the manufacturer’s instruction. Total RNA was then purified using RNA clean kit (TIANGEN) according to the manufacturer’s protocol. Total RNA from other samples was isolated as described above and quantified by spectrophotometry. One microgram of total RNA was reverse transcribed using the PrimeScript Reverse Transcriptase and oligo (dT)18 primers according to the manufacturer’s instructions. To the primers for real-time reverse transcription PCR (RT-PCR), the sequences of reference genes in H. coronarium were first cloned based on high conservation in plants. Then, the primers of reference genes were designed with Primer 5.0 software. Real-time RT-PCR was performed in a volume of 20 μl containing SYBR premix ExTaq, cDNA solution, forward and reverse primers following the manufacturer’s recommendations, with an iQ5 real-time PCR detection system (Bio-Rad). The PCR conditions were as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 30 s, 72 °C for 30 s. The specificity of each primer pair was verified by agarose gel electrophoresis analysis and melting curve analysis (60–95 °C). A standard curve using a dilution series of the cDNA derived from petals as the template was made to calculate the amplification efficiency and correlation coefficient (R 2) of each primer pair. The primer sequences and amplicon characteristics of reference genes are listed in Table 1. The stability of the potential reference genes was assessed using geNorm (Vandesompele et al. 2002) and NormFinder (Andersen et al. 2004) under different experimental conditions.
Real-time RT-PCR
The primer sequences designed in the 3′ untranslated region and their amplicon characteristics are listed in Table 1. The specificity of each primer pair was verified by agarose gel electrophoresis analysis, melting curve analysis (60–95 °C) and sequencing of cloned amplicons. Real-time RT-PCRs were performed as described above. Three independent amplifications were performed from each sample which comprised at least three biological replicates. The relative expression levels and standard deviation of target genes were calculated by iQ5 optical system software (Bio-Rad). ACT1 or RPS was used to normalize the relative quantity of target genes under different experimental conditions. The histograms were created using SigmaPlot 10.0. Analysis of variance was performed by SPSS software using Dunnett’s T3 test (P = 0.05).
RT-PCR
To compare the gene expressive abundance of HcTPS7 and HcTPS8, the RT-PCRs were performed using the primers for Real-time RT-PCR (Table 1) with the cDNA from the petals of 48 h. The reaction systems and PCR conditions were the same as described for Real-time RT-PCR except 24/28 cycles were conducted.
Headspace analysis of floral volatiles
The flower developmental process from squaring stage to senescence stage was divided into nine stages (Fig. 10a) at 9:00 (sample 0, 24, 48 h), 15:00 (8, 36, 56 h) and 23:00 (16, 40, 64 h), respectively. The whole flower of each stage was enclosed in a 500-ml glass bottle with the addition of 1.728 μg ethyl caprate as internal standard. After equilibrium of volatiles for 30 min, a PDMS fiber was inserted into the bottle to adsorb volatiles for 30 min. Then, the fiber was injected into a GC–MS system (Agilent) for analysis as described above.
The nucleotide sequences reported in this article have been deposited in the GenBank database under accession numbers KF765488 (HcTPS7), KF358246 (HcTPS8), KJ816318 (ACT1), KJ816319 (ACT2), KJ816320 (UBQ), KJ816321 (GAPDH) and KJ816322 (RPS).
Results
Cloning and sequence analysis of TPSs from H. coronarium
A ginger-expressed sequence tag database was blasted using a previous cloned TPS gene (GenBank accession no. JN695016) from H. coronarium to obtain the TPS genes that could account for the production of terpene compounds identified in H. coronarium flowers. Among several candidate TPS clones, two clones, DY354445 and DY357133 were expressed in the flowers of H. coronarium, but not in leaves and rhizomes (data not shown). Next, 3′ RACE and 5′ RACE were carried out to get the 3′ and 5′ ends of the two genes designated as HcTPS7 and HcTPS8. The full-length cDNAs of HcTPS7 and HcTPS8 contained putative ORFs of 1779 and 1773 bp encoding 593 and 591 amino acid (aa) residues with predicted molecular weights of 68.5 and 68.4 kDa, respectively. The motif DDXXD, which is highly conserved in almost all TPSs and is responsible for metal ion binding and substrate ionization, is present in the sequences of HcTPS7 and HcTPS8 (Christianson 2006; Degenhardt et al. 2009) (Fig. 1). Both sequences also have the N-terminal RRX8W motif essential for most monoterpene synthases (Degenhardt et al. 2009; Williams et al. 1998) (Fig. 1).
Alignment of the amino acid sequence revealed that HcTPS7 and HcTPS8 share 60 % identity with each other. However, they exhibit low identity with TPSs from other species (Fig. 1). For example, HcTPS7 shows 41 % sequence identity to Salvia pomifera sabinene synthase (Kampranis et al. 2007), while HcTPS8 shows 41 and 42 % identity with Mentha citrata linalool synthase (Crowell et al. 2002) and Lavandula angustifolia linalool synthase (Landmann et al. 2007), respectively (Fig. 1). Phylogenetic analysis showed that plant terpene synthases were classified into seven different subfamilies, designated TPS-a through TPS-g (Bohlmann et al. 1998; Dudareva et al. 2003; Martin et al. 2004). Both TPSs from H. coronarium belonged to TPS-b subfamily (Fig. 2a), which predominantly consists of angiosperm monoterpene synthases. However, the TPSs of monocots were distinctly clustered in another clade diverged from the clade of dicots (Fig. 2a). This divergence also appeared in the TPS-a clade (Fig. 2a) (Chen et al. 2011a) suggesting that the ancestral mono- and sesqui-TPSs appear to exist before the split of monocot and dicot lineages. To examine the genomic structure of HcTPS7 and HcTPS8, the PCR amplicons of genomic clones were sequenced and were compared with their corresponding cDNAs. The two genomic clones containing six introns and seven exons (Fig. 2b) belonged to the Class III TPS genes, which consist of angiosperm monoterpene, sesquiterpene, and diterpene synthases genes involved in secondary metabolism (Trapp and Croteau 2001). The nucleotide numbers of exons and phases of introns were also similar to the other Class III TPS genes (Trapp and Croteau 2001) (Fig. 2b).
a Phylogenetic tree of plant terpene synthases based on the neighbor-joining method. The copalyl diphosphate synthases were defined as an out-group when rooting the tree. All the terpene synthases are classified into seven subfamilies (TPS-a through TPS-g). In TPS-a and TPS-b subfamilies, the TPSs isolated from monocots are shadowed in gray and clustered to the disparate clade form dicots (shadeless). The numbers at each branch indicate bootstrap percentages from 1,000 replicates. The evolutionary distances were computed using the model of Poisson correction. Positions containing gaps and missing data were eliminated. The scale bar indicates 10 % sequence divergence. GenBank accession numbers are shown in Supplementary Table S1. Aa, Artemisia annua; Ap, Alstroemeria peruviana; Ag, Abies grandis; Am, Antirrhinum majus; At, Arabidopsis thaliana; Cb, Clarkia breweri; Cj, Citrus junos; Cl, Citrus limon; Cm, Cucurbita maxima; Cu, Citrus unshiu; Fa, Fragaria x ananassa; Fh, Freesia hybrid; Ga, Gossypium arboretum; La, Lavandula angustifolia; Le, Lycopersicon esculentum; Mc, Mentha citrata; Mp, Mentha x piperita; Nt, Nicotiana tabacum; Ob, Ocimum basilicum; Os, Oryza sativa; Pa, Picea abies; Pf, Perilla frutescens; Ps, Picea sitchensis; So, Salvia officinalis; Sp, Salvia pomifera; Vv, Vitis vinifera; Zm, Zea mays; Zo, Zingiber officinale; Zz, Zingiber zerumbet. b Genomic organization of HcTPS7 and HcTPS8. The frames and solid lines represent exons and introns, respectively. Dashed lines on two sides indicate the 5′-UTR (left) and 3′-UTR (right). The numerals in the frames and on the lines indicate the number of nucleotides. Black triangles represent the position of the DDXXD motif
Functional characterization of HcTPS7
The coding regions of HcTPSs excluding N-terminal transit peptides were expressed in E. coli, and their activity was analyzed in vitro. Recombinant HcTPS7 generated sabinene (74.1 %) as its major product in vitro when it was incubated with GPP (Fig. 3). Furthermore, nine by-products identified as α-thujene (5.0 %), α-pinene (3.2 %), β-pinene (8.2 %), myrcene (1.7 %), α-phellandrene (0.4 %), α-terpinene (1.8 %), β-phellandrene (2.2 %), γ-terpinene (2.7 %), and terpinolene (0.7 %) were synthesized from GPP (Fig. 3a and Supplementary Fig. S1). However, no sesquiterpene product was detected when FPP was used as a substrate (data not shown). The multiple reaction products derived from GPP were synthesized arising from a common reaction intermediate, α-terpinyl cation, following a series of hydride shifts, ring closures and deprotonations (Supplementary Fig. S2). Maximal activity was detected at pH 7.5 and similar content of each product was observed at pH 6.0 and 9.0. However, at pH 5.5, only trace level of sabinene was detected, indicating negligible enzyme activity of HcTPS7 under this acid–base condition (data not shown). HcTPS7 exhibited a divalent cation requirement (Mg2+ and Mn2+) for activity, but no activity only in presence of K+ (data not shown). For kinetic analysis, a range of GPP (0.5–100 μM) concentrations was assayed to yield the hyperbolic saturation curve (Fig. 6h). The K m and V max were calculated to be 26.49 ± 6.31 μM and 828.92 ± 76.57 pkat/mg, while K cat and catalytic efficiency K cat/K m were 0.057 s−1 and 2.17 s−1 mM−1, respectively.
Analyses of products generated by the recombinant HcTPS7 enzyme from GPP. a Total ion chromatogram of the monoterpene products formed by HcTPS7. b Mass spectra of the peak 3. c Mass spectra of sabinene in floral scent of H. coronarium. d Mass spectra of sabinene in the NIST08 library. Mass spectra of other peaks are provided in Supplementary Fig. S1. 1 α-thujene, 2 α-pinene, 3 sabinene, 4 β-pinene, 5 myrcene, 6 α-phellandrene, 7 α-terpinene, 8 β-phellandrene, 9 γ-terpinene, and 10 terpinolene
Functional characterization of HcTPS8
Recombinant HcTPS8 functioned as a bifunctional TPS that could utilize GPP and FPP as substrates. When incubated with GPP, HcTPS8 almost specifically catalyzed the formation of linalool (96.2 %), and trace levels of myrcene (1.6 %), limonene (1.4 %), (Z)-β-ocimene (0.8 %) were detected in the reaction products (Fig. 4). All of these products were not found in compounds formed by the extracts of E. coli expressing empty vector in the presence of GPP (Fig. 4a). The proposed reaction mechanism of linalool started with the ionization of GPP and was terminated through the water capture of geranyl cation (Supplementary Fig. S2). With FPP as a substrate, HcTPS8 synthesized α-bergamotene (36.6 %), β-farnesene (11.9 %), α-curcumene (5.0 %), (Z)-α-bisabolene (18.3 %), β-bisabolene (8.2 %), β-sesquiphellandrene (3.6 %) and seven other sesquiterpenes (16.4 %) that could not be identified due to lack of corresponding authentic standards, and low matching mass spectra to references in the NIST08 library (Fig. 5).
Analyses of products generated from GPP by the recombinant HcTPS8 enzyme. a Total ion chromatogram of the products formed by incubating extracts of empty vector (control) with GPP. b Total ion chromatogram of the monoterpene products formed by HcTPS8. c Total ion chromatogram of the linalool authentic standard. d Mass spectra of the peak 4. e Mass spectra of linalool in floral scent of H. coronarium. f Mass spectra of linalool authentic standard. Mass spectra of other peaks are provided in Supplementary Fig. S3a. 1 myrcene, 2 limonene, 3 (Z)-β-ocimene, and 4 linalool
Analyses of products generated from FPP by the recombinant HcTPS8 enzyme. a Total ion chromatogram of the sesquiterpene products formed by HcTPS8. b Mass spectra of the peak 1. c Mass spectra of α-bergamotene in the NIST08 library. Mass spectra of other peaks are provided in Supplementary Fig. S3b. d Chemical structures of α-bergamotene (peak 1), β-farnesene (peak 2), α-curcumene (peak 3), (Z)-α-bisabolene (peak 4), β-bisabolene (peak 5), and β-sesquiphellandrene (peak 6). Unlabeled peaks were unidentified sesquiterpene products
To further investigate H. coronarium TPS, the basic enzymatic features of HcTPS8 were characterized. Using GPP as a substrate, HcTPS8 showed a linear activity ranging from 5 to 30 min (Fig. 6a). The enzyme exhibited a catalytic optimum at 30 °C, with more than 70 % of its maximal activity between 22.5 and 37.5 °C (Fig. 6b). The optimal reaction pH for the enzyme was 7.5, and only very low catalytic activity was observed at pH 5.5 (Fig. 6c). Since, a divalent metal ion cofactor is required for the TPS activity (Schnee et al. 2002), the effect of different concentrations of Mg2+ (0–100 mM) and Mn2+ (0–10 mM) was tested (Fig. 6d, e). The maximum enzyme activity was achieved at 20–30 mM Mg2+, while higher concentration caused an obvious reduction in catalytic activity (Fig. 6d). The highest activity for Mn2+ was measured at 5 mM, further increase in Mn2+ concentration to 10 mM led to a slight reduction in catalytic activity (Fig. 6e). The enzyme activities showed a preference for Mg2+ and the maximum velocity was 1.49-fold higher in the presence of Mg2+ than Mn2+. The K m values were 673 and 35 μM for Mg2+ and Mn2+, respectively. No reaction product was detected by GC–MS in the presence of K+ instead of divalent metal ion (data not shown). Kinetic characterization of HcTPS8 for GPP was carried out under optimal conditions. The saturation curve was generated using the Michaelis–Menten equation by hyperbolic regression (Fig. 6f). The K m and V max were calculated to be 20.54 ± 4.52 μM and 1272.33 ± 101.06 pkat/mg, while K cat and catalytic efficiency K cat/K m were 0.088 s−1 and 4.31 s−1 mM−1, respectively. Under saturating substrate concentration, specific activity of HcTPS8 for GPP was 53 times higher than for FPP (Fig. 6g), suggesting HcTPS8’s preference for GPP.
Biochemical analysis of HcTPS7 and HcTPS8 proteins. Effect of reaction time (a), temperature (b), pH (c), Mg2+ concentration (d), and Mn2+ concentration (e) on HcTPS8 activity with GPP as the substrate. f Kinetic analysis for HcTPS8 under different GPP concentrations. The saturation curve was generated using Michaelis–Menten equation by hyperbolic regression (n = 3). g Specific activity of HcTPS8 with GPP and FPP analyzed by GC–MS. h Kinetic analysis for HcTPS7 under different GPP concentrations (n = 3)
Subcellular localization
To understand the active compartmentalization of HcTPS7 and HcTPS8, various prediction softwares (SignalP, http://www.cbs.dtu.dk/services/SignalP/; TargetP, http://www.cbs.dtu.dk/services/TargetP/; ChloroP, http://www.cbs.dtu.dk/services/ChloroP/; Predotar, http://urgi.versailles.inra.fr/predotar; WoLF PSORT, http://wolfpsort.seq.cbrc.jp/) were used to analyze their subcellular localization. The results of prediction suggested HcTPS7 was localized in the plastids with high probability in all software, while no consistent localization was attained for HcTPS8 (Supplementary Table S2). To experimentally confirm subcellular localization, the N-terminal 80-aa peptide of HcTPS7 and HcTPS8 as well as intact coding sequence of HcTPS8 was fused to a GFP reporter gene and transferred to Arabidopsis protoplasts, which were subsequently analyzed for transient GFP expression by confocal laser scanning microscopy (Fig. 7). The results showed that the green fluorescence of HcTPS7tp-GFP located in the plastids (Fig. 7e–h), which clearly indicated that the HcTPS7 protein is targeted to the plastid. Both fusions of the entire HcTPS8 protein and its N-terminal 80-aa peptide to GFP localized to the plastids (Fig. 7i–p), suggesting that N-terminal 80 amino acids were sufficient for plastid targeting to HcTPS8. Similar GFP expression patterns were observed for fusion of snapdragon AmNES/LIS-2 transit peptide (Nagegowda et al. 2008) used as positive control (Fig. 7q–t). Thus, HcTPS7 and HcTPS8 proteins were located in the plastids.
Confocal images of transiently expressed GFP fusions in Arabidopsis protoplasts. Images in the same row are from the same protoplast observed in different channels. The ‘Green’ column (a, e, i, m and q) shows GFP fluorescence, detected in the green channel. Chlorophyll autofluorescence detected in the red channel is shown in the ‘Red’ column (b, f, j, n and r). The ‘Merged’ column (c, g, k, o and s) shows merged green and red channel images. The images of bright field are shown in the ‘BF’ column (d, h, l, p and t). Schematic representation of the fusion constructs is as follows: 7tp and 8tp, N-terminal 80-amino acid peptide of HcTPS7 and HcTPS8, respectively; HcTPS8, full-length of HcTPS8; M, transit peptide of AmNES/LIS-2 in snapdragon as a plastidic marker. p35S, 35S cauliflower mosaic viral promoter, GFP enhanced green fluorescent protein as a cytosolic control, NOS NOS terminator, Bars 25 μm
Validation of reference genes for real-time RT-PCR
Real-time RT-PCR is an efficient technique for quantifying gene expression, but its accuracy is strongly influenced by the stability of the internal reference gene used for data normalization. However, no general reference gene is appropriate for each assay because of their varying expression model under different experimental conditions. Therefore, it is necessary to validate the stability of reference genes in each experimental condition prior to their use for real-time RT-PCR normalization (Chen et al. 2011b; Exposito-Rodriguez et al. 2008; Gutierrez et al. 2008). Five candidate reference genes were assessed by geNorm and NormFinder, and their amplicon lengths varied from 110 to 178 bp and amplification efficiency ranged between 93.2 and 105.7 % (Table 1). The results revealed that the most stable genes (lowest M value) were ACT1 and ACT2 in different tissues and GAPDH and RPS showed the highest stability at different flower developmental stages with geNorm algorithm (Fig. 8a, b). Similar results were obtained using NormFinder program with ACT1 being the most stable in different tissues and RPS being the most stable at different flower developmental stages (Fig. 8c). Therefore, ACT1 and RPS were used as endogenous controls for consequent expression analysis in different tissues and at different flower developmental stages, respectively.
Expression stability values of the candidate reference genes. a, b Average expression stability values of candidate reference genes in different H. coronarium tissues (a) and during different flower developmental stages (b) calculated by geNorm. c Stability values of the candidate reference genes calculated by NormFinder
Expression analysis of HcTPS7 and HcTPS8 gene
Real-time RT-PCR analysis showed that HcTPS7 and HcTPS8 were highly expressed in labella apex, labella base (except at lower level for HcTPS7), lateral petals and sepals (Fig. 9), which are the major parts for scent emission (Fan et al. 2003). HcTPS7 and HcTPS8 were also expressed at lower levels in other floral tissues (styles and stigmas (except at undetectable levels for HcTPS7), anthers, filaments and pedicels), but were almost undetectable in bracts, leaves, pseudostems, rhizomes and roots (Fig. 9). Meanwhile, sabinene and linalool were not or rarely detected in essential oils of leaves and rhizomes in H. coronarium (Dos Santos et al. 2010; Joy et al. 2007), indicating their correlation with gene expression.
Expression analysis of HcTPS7 and HcTPS8 in different tissues. a Photographs of floral organs at developmental stage of 40 h and vegetative organs of H. coronarium. Tissues used for the gene expression analysis are indicated. Bars 2 cm. b, c Relative expression analysis of HcTPS7 (b) and HcTPS8 (c) was performed by real-time RT-PCR. ACT1 was used as an endogenous control. Relative transcription level of tissue with highest expression quantity was set to 1 (100 %). Error bars indicate standard deviation of three replicates. Different lowercase letters labeled on bars indicate statistically significant differences at the level of P < 0.05. SS styles and stigmas, A anthers, F filaments, LA labella apex (the white part), LB labella base (the pale yellow part), LP lateral petals, Se sepals, Pe pedicels, B bracts, Le leaves, Ps pseudostems, Ri rhizomes, Ro roots
During flower development, there was almost no expression for HcTPS7 mRNA transcripts in petals before 40 h (Fig. 10c). The HcTPS7 expression levels increased sharply between 32 and 48 h, peaking at 48 h, and then decreased with flower senescence (Fig. 10c). Emission of sabinene was consistent with HcTPS7 gene expression between 40 and 64 h (Fig. 10e). However, emission of sabinene decreased gradually from 8 to 32 h when almost no HcTPS7 expression occurred (Fig. 10e). HcTPS8 mRNA levels in petals increased gradually between 0 and 40 h, peaking at 40 h with full bloom, and then decreased from 40 to 64 h (Fig. 10d). The change of HcTPS8 expression level showed a positive correlation with the emission pattern of linalool (Pearson correlation coefficient r = 0.632, significant correlation at the level of P < 0.1) (Fig. 10d, f). In sepals, the gene expression trends of HcTPS7 and HcTPS8 were similar to these in petals, but the expression peaks emerged 8 h ahead (Supplementary Fig. S4), indicating that the biosynthesis events of terpenes in sepals may precede that in petals.
HcTPS7 and HcTPS8 expression analyses, and the emission of sabinene and linalool in flowers of H. coronarium during flower development. a Representative photographs of flowers in different flower developmental stages. b RT-PCR analysis of HcTPS7 and HcTPS8 using cDNA of 48 h flowers to compare gene expressive abundance. c, d Expression analysis of HcTPS7 (c) and HcTPS8 (d) in petals in different flower developmental stages. RPS was used as an endogenous control. Relative transcription level of 32 h for HcTPS7 and 0 h for HcTPS8 was set to 1. Error bars indicate standard deviation of three replicates. e, f Emission of sabinene (e) and linalool (f) from flowers of H. coronarium. Error bars indicate standard deviation of two to five biological replicates. Different lowercase letters labeled on bars indicate statistically significant differences at the level of P < 0.05
Relative gene expressive abundance of HcTPS7 and HcTPS8 was estimated with RT-PCR using cDNA of petals at 48 h (Fig. 10b). The results showed that HcTPS7 amplicon could not be detected when 24 cycles were performed, while a weak band was observed at 28 cycles, which was similar in intensity with HcTPS8 at 24 cycles. Thus, the expressive abundance of HcTPS8 was approximately 16 times than HcTPS7.
The mRNA expression of HcTPSs was analyzed in blooming flowers of different species in Hedychium (Fig. 11). HcTPS7 gene was highly expressed in H. coronarium and at very low level in H. gardnerianum, H. coccineum and H. forrestii. The highest expression of HcTPS8 was noted in H. coronarium, and lower levels were observed in the scented H. gardnerianum and it was barely expressed in non-scented H. coccineum and H. forrestii. This result was consistent with previous northern blot analysis (data not shown).
Expression analysis of HcTPS7 and HcTPS8 in blooming flowers of four Hedychium species. RPS was used as an endogenous control. The transcription level of H. coronarium flowers was set to 1 (100 %). Error bars indicate standard deviation of three replicates. Different lowercase letters labeled on bars indicate statistically significant differences at the level of P < 0.05. 1 H. coronarium (scent), 2 H. gardnerianum (scent), 3 H. coccineum (scentless), 4 H. forrestii (scentless)
Discussion
Terpenoids are the most abundant and structurally diverse class of plant secondary metabolites. TPSs are responsible for the terpenoid diversity to a large extent (Yu and Utsumi 2009). In this research, two monoterpene synthases (HcTPS7 and HcTPS8) were isolated and functionally characterized. HcTPS7 designated as sabinene synthase was a multiproduct enzyme that synthesizes sabinene (74.5 %) as its main product and nine by-products (α-thujene, α-pinene, β-pinene, myrcene, α-phellandrene, α-terpinene, β-phellandrene, γ-terpinene, and terpinolene) (Fig. 3). Until now, sabinene synthases were characterized from S. officinalis, S. pomifera, and Citrus jambhiri. Sabinene synthase (SSS) from S. officinalis produces sabinene (63 %), γ-terpinene (21 %), terpinolene (7.0 %), limonene (6.5 %) and myrcene (2.5 %) (Wise et al. 1998). The K m value for SSS (7.4 μM) is lower than K m value for HcTPS7 (26.49 ± 6.31 μM). Sabinene synthase (Sp-SabS1) from S. pomifera synthesizes 98.4 % sabinene and 1.6 % myrcene (Kampranis et al. 2007). For Citrus jambhiri RlemTPS2, the dominant product is sabinene, and the minor products are α/β-pinene, and β-phellandrene (Kohzaki et al. 2009). The products of these TPSs also show that the multiproduct monoterpene synthases frequently produce limonene, myrcene, sabinene, and α/β-pinene in the product spectra (Faehnrich et al. 2011). The amino acid sequences of SSS and Sp-SabS1 share 95 % identity with each other; while they share only 40 and 41 % identity with HcTPS7, respectively. However, the identity between HcTPS7 and HcTPS8 at amino acid level is 60 %. The result shows that TPSs synthesizing different products in species with closer affinity are more similar to each other than TPSs producing the same products in distantly related species, suggesting that the divergent and convergent evolution of TPS gene function is common in higher plant (Chen et al. 2011a).
Linalool synthases are identified widely from both angiosperms and gymnosperms, and they generate linalool as a unique or predominant product from GPP. Among them, strawberry FaNES (Aharoni et al. 2004), tomato LeMTS1 (van Schie et al. 2007), snapdragon AmNES/LIS (Nagegowda et al. 2008), Populus trichocarpa PtTPS3/4 (Danner et al. 2011), and kiwifruit AcNES1 (Green et al. 2012) are bifunctional enzymes that produce acyclic terpene alcohols linalool from GPP and nerolidol from FPP. HcTPS8 was also a bifunctional enzyme, which synthesized linalool from GPP (Fig. 4). However, differently, the sesquiterpene products catalyzed by HcTPS8 were not nerolidol, but multiple cyclic sesquiterpenes (Fig. 5). Recently, one turmeric (Curcuma longa) monoterpene (MT17A2) shows similar catalytic function to HcTPS8 (Koo and Gang 2012). Both the size and the functional groups in the cavity of the active site are important for the substrate availability and outcome of the reaction (Green et al. 2011; Kampranis et al. 2007). We speculate that there may be something peculiar about the contour of the active site of HcTPS8, which serves as a template for the binding of GPP or FPP and reactive intermediates that follow the formation of the initial carbocation intermediate. Perhaps the active site is sufficiently large and unconstrained to permit water capture of the GPP upon ionization. With FPP, perhaps the larger substrate is more constrained in the active site; therefore, the reactive carbocationic intermediates can have many metabolic fates, instead of direct water capture, thereby resulting in the formation of multiple cyclic sesquiterpenes. The Km value of HcTPS8 for GPP was 20.54 ± 4.52 μM, which was lower than linalool synthases from L. angustifolia (42.7 ± 4.6 μM) (Landmann et al. 2007) and M. citrate (56 ± 5 μM) (Crowell et al. 2002), but was higher than most of the linalool synthases. The catalytic efficiency for GPP was similar to other bifunctional linalool synthases (Landmann et al. 2007).
It is generally believed that the biosynthesis of monoterpenes and sesquiterpenes is compartmentalized wherein the monoterpene is produced in the plastids where GPP is synthesized, and the sesquiterpene is formed in the cytosol where FPP is generated. Thus, the subcellular localization of TPS protein and substrate availability regulated the terpene biosynthesis in planta (Gutensohn et al. 2013; Nagegowda et al. 2008). Subcellular localization experiments confirmed the plastid targeting of HcTPS7 and HcTPS8 (Fig. 7). Thus, the enzymes are supposed to be plastidic sabinene synthase and linalool synthase, respectively. Furthermore, two plastid-targeted GPP synthases showing similar gene expression pattern with HcTPSs in H. coronarium were identified (unpublished data), indicating sufficient GPP supply for HcTPS7 and HcTPS8 enzymes. However, metabolic engineering of TPS genes revealed the presence of the small amount of plastidic FPP in Arabidopsis and tobacco (Aharoni et al. 2003; Wu et al. 2006). In the monocot, Phalaenopsis bellina, a PbGDPS enzyme exhibits dual prenyltransferase activity, producing both GPP and FPP, suggesting the presence of a possible FPP pool in plastids (Hsiao et al. 2008). Therefore, a bifunctional TPS enzyme characterized in vitro can possibly show bifunctional behavior in planta. HcTPS8 can accept GPP and FPP as substrates and produce monoterpene alcohol linalool and several sesquiterpenes in vitro (Figs. 4, 5). Therefore, the possible sesquiterpene activity of HcTPS8 in plastids does not exclude completely in planta. Nevertheless, HcTPS8 is likely to mainly act as a linalool synthase, because no sesquiterpene products catalyzed by HcTPS8 were detected in the floral volatile compound under recent experimental conditions.
In higher plants, emission of volatile terpenes is often regulated at the transcriptional level. In Arabidopsis, two TPSs that are responsible for the biosynthesis of sesquiterpenes emitted from flowers are only expressed in some parts of the Arabidopsis flower (Tholl et al. 2005). The emission of linalool from C. breweri flowers is positively correlated with the levels of linalool synthase mRNA (Dudareva et al. 1996). In snapdragon flowers, the biosynthesis and emission of the (E)-β-ocimene and myrcene correlate with the specific expression patterns of their corresponding TPS genes in the upper and lower petal lobes during flower development (Dudareva et al. 2003). Flowers of H. coronarium emit a large number of monoterpenes including linalool, (E)/(Z)-β-ocimene, 1,8-cineole, sabinene, α-thujene, myrcene and α/β-pinene when flower blooms (Baez et al. 2011; Fan et al. 2003, 2007). However, there are very little linalool and sabinene in the essential oil of leaves and rhizomes (Dos Santos et al. 2010). The HcTPS7 and HcTPS8 genes were also highly expressed in flower tissues and were very low in leaves and rhizomes (Fig. 9). During flower development, the gene expression patterns of HcTPS7 (40–64 h) and HcTPS8 in petals also coincide with the emission of sabinene and linalool (Fig. 10). However, emission of sabinene decreased gradually from 8 to 32 h when almost no HcTPS7 expression occurred (Fig. 10e). It seems that another TPS generated sabinene as product or by-product accounts for the biosynthesis of sabinene between 8 and 32 h.
In Hedychium, some species have many excellent ornamental traits, but lack fragrance, such as H. coccineum and H. forrestii (Wu and Raven 2001). Checking the volatile profile of H. coccineum flowers in previous papers, no sabinene and very little linalool were observed (Fan et al. 2007). That is mainly because TPS genes are barely expressed in the flower of these species, such as HcTPS7 and HcTPS8 (Fig. 11). Cseke et al. (1998) reported that scented C. breweri and scentless C. concinna showed vastly different expression characteristics of LIS gene, which was responsible for the formation of linalool in C. breweri. The reason resulting in this difference is likely to be the sequence variation in the region of LIS promoters. In Santalum, the genome sequence of santalene synthase in S. murrayanum with oil-deficient phenotype also contains a complete ORF that is highly similar with its orthologous genes in other three essential oil-containing species (Jones et al. 2011). It is likely that factors regulating the TPS gene expression, rather than the absence of, or mutations of the genes themselves, are responsible for the low- or no-oil phenotype (Jones et al. 2011). But, the reasons causing the TPS expression difference in Hedychium remain to be verified experimentally.
Previous phylogenetic analysis of the plant TPS super-family revealed seven subfamilies, designated TPS-a through TPS-g (Bohlmann et al. 1998; Dudareva et al. 2003; Martin et al. 2004). However, Chen et al. (2011a, b) recognized a new plant TPS subfamily (TPS-h) including eight TPSs possibly evolved from CPS/KS prototype TPS in lycopod Selaginella moellendorffii. Interestingly, in this nonseed vascular plant, another unusual type of TPSs, more closely related to microbial TPSs than other plant TPSs, were found (Li et al. 2012), indicating the evolutionary complexity of plant TPS gene family. Previous studies indicated that plant mono- and sesqui-TPSs appeared to derive from an ancestral diterpene synthase involved in primary metabolism by gene duplication, domain loss and functional divergence (Hillwig et al. 2011; Trapp and Croteau 2001). This evolution process seemed to occur independently in gymnosperms and angiosperms (Fig. 2a) (Martin et al. 2004; Chen et al. 2011a). Recent work based on the genome sequence of the cultivated tomato revealed that most of the TPS genes in the tomato genome are located in clusters (Falara et al. 2011). The cluster of TPS genes was also observed in the genomes of Arabidopsis and grape (Aubourg et al. 2002; Martin et al. 2010) as well as the monocot sorghum, rice and mays (Zhuang et al. 2012), representing relatively recent duplications and divergences. Analyses on the genomes of two monocots (rice and sorghum) revealed that very little TPS belonged to the TPS-b subfamily (Chen et al. 2011a). To data, none of the maize TPS belonging to this subfamily was reported. In this study, two H. coronarium mono-TPSs belonging to TPS-b subfamily were identified. Phylogenetic analysis indicates that two H. coronarium mono-TPSs, together with the other two functional characterized monocot mono-TPSs A. peruviana myrcene synthase (Aros et al. 2012) and Freesia hybrid linalool synthase (unpublished), form a distinct clade that diverges from dicot mono-TPSs in TPS-b subfamily (Fig. 2a). The evolution divergence between monocot and dicot also appears in the TPS-a subfamily containing mostly sesqui-TPSs in angiosperms (Fig. 2a), suggesting that the ancestral mono- and sesqui-TPSs evolved from di-TPSs seem to exist before the split of monocots and dicots and expand independently after this split. Both H. coronarium mono-TPS genes have six introns that are common in angiosperms (Fig. 2b) (Aubourg et al. 2002; Trapp and Croteau 2001). However, exceptional number of intron due to loss and gain of intron in the evolution was also observed in angiosperms (Falara et al. 2011; Lee and Chappell 2008).
In conclusion, we have first identified two monoterpene synthases involved in the biosynthesis of sabinene and linalool, which are two important constituents of floral scent in H. coronarium. Wherein, HcTPS7 is a multiproduct monoterpene synthase and HcTPS8 is a bifunctional TPS with special catalytic activity on sesquiterpene. Our results provide new insights into molecular mechanisms of terpene biosynthesis in H. coronarium and also provide the opportunities for breeding and genetic manipulation of scent-associated traits in Hedychium to enhance their commercial values. In the future it will be interesting to unravel the transcriptional regulatory network of floral scent formation and emission in H. coronarium.
Abbreviations
- DMAPP:
-
Dimethylallyl pyrophosphate
- FPP:
-
Farnesyl diphosphate
- GC-MS:
-
Gas chromatography-mass spectrometry
- GFP:
-
Green fluorescence protein
- GGPP:
-
Geranylgeranyl diphosphate
- GPP:
-
Geranyl diphosphate
- IPP:
-
Isopentenyl pyrophosphate
- LB:
-
Luria–Bertani
- ORF:
-
Open reading frame
- PDMS:
-
Polydimethylsiloxane
- RACE:
-
Rapid amplification of cDNA ends
- RT-PCR:
-
Reverse transcription PCR
- TPS:
-
Terpene synthase
References
Aharoni A, Giri AP, Deuerlein S, Griepink F, de Kogel WJ, Verstappen FW, Verhoeven HA, Jongsma MA, Schwab W, Bouwmeester HJ (2003) Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15:2866–2884
Aharoni A, Giri AP, Verstappen F, Bertea CM, Sevenier R, Sun ZK, Jongsma MA, Schwab W, Bouwmeester HJ (2004) Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell 16:3110–3131
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Aros D, Gonzalez V, Allemann RK, Muller CT, Rosati C, Rogers HJ (2012) Volatile emissions of scented Alstroemeria genotypes are dominated by terpenes, and a myrcene synthase gene is highly expressed in scented Alstroemeria flowers. J Exp Bot 63:2739–2752
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267:730–745
Baez D, Pino JA, Morales D (2011) Floral scent composition in Hedychium coronarium J. Koenig analyzed by SPME. J Essent Oil Res 23:64–67
Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. P Natl Acad Sci USA 95:4126–4133
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chen F, Tholl D, D’Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15:481–494
Chen F, Tholl D, Bohlmann J, Pichersky E (2011a) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66:212–229
Chen L, Zhong H, Kuang J, Li J, Lu W, Chen J (2011b) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234:377–390
Christianson DW (2006) Structural biology and chemistry of the terpenoid cyclases. Chem Rev 106:3412–3442
Crowell AL, Williams DC, Davis EM, Wildung MR, Croteau R (2002) Molecular cloning and characterization of a new linalool synthase. Arch Biochem Biophys 405:112–121
Cseke L, Dudareva N, Pichersky E (1998) Structure and evolution of linalool synthase. Mol Biol Evol 15:1491–1498
Danner H, Boeckler GA, Irmisch S, Yuan JS, Chen F, Gershenzon J, Unsicker SB, Koellner TG (2011) Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 72:897–908
Davidovich-Rikanati R, Lewinsohn E, Bar E, Iijima Y, Pichersky E, Sitrit Y (2008) Overexpression of the lemon basil alpha-zingiberene synthase gene increases both mono- and sesquiterpene contents in tomato fruit. Plant J 56:228–238
Degenhardt J, Kollner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70:1621–1637
Dos Santos BCB, Barata LES, Marques FA, Baroni ACM, Karnos BAC, de Oliveira PR, Guerrero PG Jr (2010) Composition of leaf and rhizome essential oils of Hedychium coronarium Koen. from Brazil. J Essent Oil Res 22:305–306
Dudareva N, Pichersky E (2000) Biochemical and molecular genetic aspects of floral scents. Plant Physiol 122:627–633
Dudareva N, Cseke L, Blanc VM, Pichersky E (1996) Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8:1137–1148
Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Faldt J, Miller B, Bohlmann J (2003) (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15:1227–1241
Dudareva N, Pichersky E, Gershenzon J (2004) Biochemistry of plant volatiles. Plant Physiol 135:1893–1902
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Faehnrich A, Krause K, Piechulla B (2011) Product variability of the ‘Cineole Cassette’ monoterpene synthases of related Nicotiana Species. Mol Plant 4:965–984
Falara V, Akhtar TA, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini ME, Schilmiller AL, Last RL, Schuurink RC, Pichersky E (2011) The tomato terpene synthase gene family. Plant Physiol 157:770–789
Fan YP, Yu RC, Huang Y, Chen YF (2003) Studies on the essential constituent of Hedychium flavum and H. coronarium. Acta Horticulturae Sinica 30:475 (in Chinese)
Fan YP, Wang XR, Yu RC, Yang P (2007) Analysis on the aroma components in several species of Hedychium. Acta Horticulturae Sinica 34:231–234 (in Chinese)
Green S, Baker EN, Laing W (2011) A non-synonymous nucleotide substitution can account for one evolutionary route to sesquiterpene synthase activity in the TPS-b subgroup. FEBS Lett 585:1841–1846
Green SA, Chen X, Nieuwenhuizen NJ, Matich AJ, Wang MY, Bunn BJ, Yauk Y, Atkinson RG (2012) Identification, functional characterization, and regulation of the enzyme responsible for floral (E)-nerolidol biosynthesis in kiwifruit (Actinidia chinensis). J Exp Bot 63:1951–1967
Gutensohn M, Orlova I, Nguyen TTH, Davidovich-Rikanati R, Ferruzzi MG, Sitrit Y, Lewinsohn E, Pichersky E, Dudareva N (2013) Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits. Plant J 75:351–363
Guterman I, Shalit M, Menda N, Piestun D, Dafny-Yelin M, Shalev G, Bar E, Davydov O, Ovadis M, Emanuel M, Wang J, Adam Z, Pichersky E, Lewinsohn E, Zamir D, Vainstein A, Weiss D (2002) Rose scent: genomics approach to discovering novel floral fragrance-related genes. Plant Cell 14:2325–2338
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre J, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G, Huang L, Peters RJ (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J 68:1051–1060
Hsiao Y, Jeng M, Tsai W, Chuang Y, Li C, Wu T, Kuoh C, Chen W, Chen H (2008) A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)(2-4)D motif. Plant J 55:719–733
Jaakola L, Pirttila AM, Halonen M, Hohtola A (2001) Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol 19:201–203
Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J (2011) Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS)-a and TPS-b subfamilies, including santalene synthases. J Biol Chem 286:17445–17454
Joy B, Rajan A, Abraham E (2007) Antimicrobial activity and chemical composition of essential oil from Hedychium coronarium. Phytother Res 21:439–443
Kampranis SC, Ioannidis D, Purvis A, Mahrez W, Ninga E, Katerelos NA, Anssour S, Dunwell JM, Degenhardt J, Makris AM, Goodenough PW, Johnson CB (2007) Rational conversion of substrate and product specificity in a Salvia monoterpene synthase: structural insights into the evolution of terpene synthase function. Plant Cell 19:1994–2005
Kohzaki K, Gomi K, Yamasaki-Kokudo Y, Ozawa R, Takabayashi J, Akimitsu K (2009) Characterization of a sabinene synthase gene from rough lemon (Citrus jambhiri). J Plant Physiol 166:1700–1704
Koo HJ, Gang DR (2012) Suites of terpene synthases explain differential terpenoid production in ginger and turmeric tissues. PLoS One 7:e51481
Lan JB, Yu RC, Yu YY, Fan YP (2013) Molecular cloning and expression of Hedychium coronarium farnesyl pyrophosphate synthase gene and its possible involvement in the biosynthesis of floral and wounding/herbivory induced leaf volatile sesquiterpenoids. Gene 518:360–367
Landmann C, Fink B, Festner M, Dregus M, Engel KH, Schwab W (2007) Cloning and functional characterization of three terpene synthases from lavender (Lavandula angustifolia). Arch Biochem Biophys 465:417–429
Lee S, Chappell J (2008) Biochemical and genomic characterization of terpene synthases in Magnolia grandiflora. Plant Physiol 147:1017–1033
Li R, Fan Y (2011) Molecular cloning and expression analysis of a terpene synthase gene, HcTPS2, in Hedychium coronarium. Plant Mol Biol Rep 29:35–42
Li G, Koellner TG, Yin Y, Jiang Y, Chen H, Xu Y, Gershenzon J, Pichersky E, Chen F (2012) Nonseed plant Selaginella moellendorffii has both seed plant and microbial types of terpene synthases. P Natl Acad Sci USA 109:14711–14715
Martin DM, Faldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135:1908–1927
Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10:226
Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J 55:224–239
Pichersky E, Dudareva N (2007) Scent engineering: toward the goal of controlling how flowers smell. Trends Biotechnol 25:105–110
Pichersky E, Lewinsohn E, Croteau R (1995) Purification and characterization of S-linalool synthase, an enzyme involved in the production of floral scent in Clarkia breweri. Arch Biochem Biophys 316:803–807
Pichersky E, Noel JP, Dudareva N (2006) Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311:808–811
Raguso RA (2008) Wake up and smell the roses: the ecology and evolution of floral scent. Annu Rev Ecol Evol Syst 39:549–569
Schnee C, Kollner TG, Gershenzon J, Degenhardt J (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)-beta-farnesene, (E)-nerolidol, and (E, E)-farnesol after herbivore damage. Plant Physiol 130:2049–2060
Tholl D (2006) Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol 9:297–304
Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E (2005) Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42:757–771
Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158:811–832
Vainstein A, Lewinsohn E, Pichersky E, Weiss D (2001) Floral fragrance. New inroads into an old commodity. Plant Physiol 127:1383–1389
van Schie C, Haring MA, Schuurink RC (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol Biol 64:251–263
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: research0034.1–0034.11. doi:10.1186/gb-2002-3-7-research0034
Wang W, Zhou W (2008) Efficient extraction and application of total RNA in sisal. Chin J Trop Crops 29:725–729
Williams DC, McGarvey DJ, Katahira EJ, Croteau R (1998) Truncation of limonene synthase preprotein provides a fully active ‘Pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37:12213–12220
Wise ML, Savage TJ, Katahira E, Croteau R (1998) Monoterpene synthases from common sage (Salvia officinalis): cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J Biol Chem 273:14891–14899
Wu ZY, Raven HP (2001) Flora of China 24. Missouri Botanical Garden Press, Saint Louis, pp 370–377
Wu SQ, Schalk M, Clark A, Miles RB, Coates R, Chappell J (2006) Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol 24:1441–1447
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protoc 2:1565–1572
Yu F, Utsumi R (2009) Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell Mol Life Sci 66:3043–3052
Zhuang X, Koellner TG, Zhao N, Li G, Jiang Y, Zhu L, Ma J, Degenhardt J, Chen F (2012) Dynamic evolution of herbivore-induced sesquiterpene biosynthesis in sorghum and related grass crops. Plant J 69:70–80
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (Grant No. 30972026 and 31370694), the Guangdong-industry Technology Research System (Grant No. 2009-356), Key Laboratory of Innovation and Utilization for Germplasm Resources in Horticultural Crops in Southern China of Guangdong Higher Education Institutes, South China Agricultural University (No. KBL11008), and a Specialized Research Fund for the Doctoral Program of Higher Education of China to Yanping Fan (Grant No. 20134404110016).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yue, Y., Yu, R. & Fan, Y. Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium . Planta 240, 745–762 (2014). https://doi.org/10.1007/s00425-014-2127-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-014-2127-x