Abstract
Main conclusion
CaTPS2 and CaTPS3 were significantly expressed in flowers of Curcuma alismatifolia ‘Shadow’ and demonstrated bifunctional enzyme activity, CaTPS2 generated linalool and nerolidol as products, and CaTPS3 catalyzed β-myrcene and β-farnesene formation.
Abstractsss
This study presents the discovery and functional characterization of floral terpene synthase (TPS) genes in Curcuma alismatifolia ‘Shadow’, a cultivar renowned for its unique fragrance. Addressing the gap in understanding the genetic basis of floral scent in this species, we identified eight TPS genes through comprehensive transcriptome sequencing. Among these, CaTPS2 and CaTPS3 were significantly expressed in floral tissues and demonstrated bifunctional enzyme activity corresponding to the major volatile compounds detected in ‘Shadow’. Functional analyses, including in vitro assays complemented with rigorous controls and alternative identification methods, elucidated the roles of these TPS genes in terpenoid biosynthesis. In vitro studies were conducted via heterologous expression in E. coli, followed by purification of the recombinant protein using affinity chromatography, enzyme assays were performed with GPP/FPP as the substrate, and volatile products were inserted into the GC–MS for analysis. Partially purified recombinant protein of CaTPS2 catalyzed GPP and FPP to produce linalool and nerolidol, respectively, while partially purified recombinant protein of CaTPS3 generated β-myrcene and β-farnesene with GPP and FPP as substrates, respectively. Real-time quantitative PCR further validated the expression patterns of these genes, correlating with terpenoid accumulation in different plant tissues. Our findings illuminate the molecular mechanisms underpinning floral fragrance in C. alismatifolia and provide a foundation for future genetic enhancements of floral scent in ornamental plants. This study, therefore, contributes to the broader understanding of terpenoid biosynthesis in plant fragrances, paving the way for biotechnological applications in horticulture plant breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The realm of plant fragrances, a complex interplay of volatile organic compounds (VOCs), is dominated by terpenoids, notable for their structural diversity and significant contributions to floral aroma profiles (Dudareva et al. 2013). Curcuma alismatifolia, also known as the Siam Tulip or ‘Shadow’, is celebrated not just for its visually striking flowers but also for its unique fragrance, making it a subject of both commercial interest and scientific inquiry (Brown and Patel 2019). At present, studies about the C. alismatifolia traits have mainly concentrated on vase life, bract color, and flower initiation and development (Kjonboon and Kanlayanarat 2005; Koshioka et al. 2015). As one of the main ornamental attributes of C. alismatifolia, bract color has drawn in expanding consideration among scientists, and different species with bright and beautiful inflorescences have been developed. For plants, the color and scent of flowers are equally important in attracting consumer attention, and these two traits are crucial for attracting pollinating insects and for the success of plant evolution (Zuker et al. 2002; Parachnowitsch et al. 2012). Despite its appeal, large amounts of Curcuma species have flashy bracteate inflorescences but no floral fragrance, and the genetic underpinnings and enzymatic pathways shaping its scent have remained largely uncharted territory.
At the heart of terpenoid biosynthesis lie terpene synthases (TPSs), enzymes catalyzing the transformation of prenyl diphosphate substrates into a plethora of terpenoids, integral for plants’ interactions with their environment, such as attracting pollinators and deterring herbivores (Sheehan et al. 2012; Lin et al. 2021). Volatile terpenes can serve as long-distance attractants in Mimulus lewisii, predominantly attracting bumblebee for pollination (Byers et al. 2014). In addition, volatile terpenes function over shorter distances, such as within-flower nectar guides in monkeyflowers (Liang et al. 2023), or act directly as chemical defense against herbivores or pathogens in Petunia hybrida (Boachon et al. 2019). Despite the pivotal roles of terpenes in attracting pollinators and plant evolution, the correlation between volatile terpenes and pollinator attraction remains largely unknown for most flowering plant species (Shen et al. 2024). Identifying and understanding the function of TPS genes responsible for the floral scent in C. alismatifolia could pave the way for genetic enhancements, improving both ornamental and aromatic traits in this species and others (Nguyen et al. 2023).
The advent of transcriptome sequencing technologies has revolutionized our ability to uncover the genetic bases of complex traits, including floral fragrance providing insights into the specific genes involved in these pathways (Fisher and Kramer 2019). Nonetheless, functional validation of these genes is crucial for a comprehensive understanding of their roles in terpenoid biosynthesis (Williams and Jackson 2021a, b). This study seeks to fill this gap by identifying and functionally characterizing TPS genes involved in volatile terpene production in C. alismatifolia ‘Shadow’. Employing an integrative approach that combines transcriptome analysis, gene cloning, in vitro functional assays, and expression profiling, we aim to decode the molecular basis of floral scent in this species.
We selected a scented cultivar (‘Shadow’) as materials to explore the regulatory mechanism of terpene formation in C. alismatifolia. The emission pattern of terpenes in different developmental stages and various tissues of ‘Shadow’ will be studied. Moreover, we investigated the key terpene synthase genes by transcriptome sequencing and identified their functions. The expression patterns of candidate CaTPS genes were also analyzed to determine their important roles in the biosynthesis pathway of terpenes. Our research not only focuses on the identification of TPS genes but also delves into their functional implications in terpene biosynthesis. This dual focus allows us to contribute to the expanding field of plant fragrance biosynthesis and opens avenues for the genetic improvement of floral characteristics in ornamental plants, aligning with the study’s goals and addressing the gaps identified in previous research.
Materials and methods
Plant materials
Curcuma alismatifolia was planted in the Ginger Garden of Flower Research Center, South China Agricultural University (Guangzhou, China) under natural light. The different tissues of C. alismatifolia are divided into flowers (F), fertile bracts (Fb), sterile bracts (Sb) and leaves (Le) (Fig. 1a). The flowers of C. alismatifolia contain petals (Pe), labella (L), lateral petals (Lp) and gynandrium (Gy) (Fig. 1b). In our study, the flowering process was divided into four stages (S1–S4) (Fig. 1c). The stages (S1–S3) constitute the process of floral buds to the full-openness stage (S3). Afterward, the flower undergoes early wilting (S4). All samples were collected during the blooming period from July to August. The material samples for RNA extraction were frozen in liquid nitrogen immediately after picking and stored in a – 80 °C freezer.
GC–MS analysis of floral volatiles
Approximately 0.3 g of sample was enclosed in a 50-ml glass bottle with 0.432 μg of ethyl caprate as an internal standard. After 30 min, a solid-phase microextraction (SPME) fiber was used to adsorb floral volatiles for 30 min and then inserted into a gas chromatography‒mass spectrometer (GC‒MS system) (Agilent) for volatile analysis as described previously (Zhou et al. 2022). Three replicates of each sample of C. alismatifolia tissues in the floral fragrance collection were included. VOCs were then identified by matching the mass spectra to the NIST Mass Spectral Library (NIST 08) based on an 80% match factor. The relative quantification of floral volatile compounds was calculated by Agilent ChemStation Data Analysis Application according to the quantity of ethyl caprate and the peak area ratio. SPSS software was used to analysis of variance by Tukey’s test (P = 0.05).
Transcriptome sequencing and analysis
Total RNA of ‘Shadow’ flowers and sterile bracts in the blooming stage were extracted using TRIzol® Reagent following the manufacturer’s instructions (Invitrogen). RNA-seq transcriptome libraries were prepared according to the TruSeqTM RNA sample preparation Kit from Illumina (San Diego, CA, USA) and sequenced by Illumina NovaSeq 6000 sequencing (150 bp) with 1 G total bases. Raw reads were filtered using Trimmomatic software (Bolger et al. 2014) to obtain clean reads by removing the adapters and low-quality reads. Transcriptome de novo assembly was implemented by Trinity software (Grabherr et al. 2011) without reference genome. Based on sequence similarity, all the assembled transcripts were compared to the NCBI protein nonredundant (NR), String, and KEGG databases using BLASTX algorithm with a typical cut-off E-value ≤ 10−5. BLAST2GO software was used to get GO annotations of unique assembled transcripts for describing biological processes, molecular functions and cellular components (Götz et al. 2008). KEGG pathway analysis was performed using BLASTX algorithm with an E-value cut-off of 10−5. Annotated protein output was used to extract TPSs by using the hidden Markov model (HMM) profiles of PF03936 and PF01397 from Pfam database as a template. To obtain differential expression genes (DEGs) between flowers and sterile bracts, the expression level of each readcount was normalized to reads per kilobase of exon model per million mapped reads (RPKM) (Mortazavi et al. 2008). DEGseq R package was used for differential expression analysis (Wang et al. 2010), and FDR (False Discovery Rate) ≤ 0.05 and FC (Fold Change) ≥ 2 were used as the cut-off values. GO term enrichment analysis and KEGG pathway enrichment analysis of DEGs were performed using GOseq R package (Young et al. 2010) and KOBAS software (Mao et al. 2005), respectively.
Terpenoid synthase genes identification and phylogenetic analysis
To identify putative floral terpenoid synthase genes, we utilized the transcriptome of ‘Shadow’ as a query by HMMER domain (PF03936, PF01397) screening and the NCBI BLAST function. The expression patterns of candidate CaTPS genes in flowers and sterile bracts were analyzed using RPKM data and visualized with TBtools (Chen et al. 2020). The highly expressed genes in flowers (CaTPS2 and CaTPS3) were selected for further functional studies. The sequences of CaTPSs were obtained from RNA-seq database of C. alismatifolia flower, and the relevant protein sequences from other plant species were downloaded from the NCBI database. The phylogenetic tree was constructed using MEGA X software (Kumar et al. 2018) with the neighbor-joining method after the multiple protein sequences were aligned with Clustal (Sievers et al. 2011).
In vitro TPS enzyme assays
The coding sequences (CDSs) of CaTPS2 and CaTPS3 from the flowers were subcloned and inserted into vector pET-32a with an N-terminal His-tag through Bamh I and Hind III sites. Escherichia coli Rosetta (DE3) was utilized as an expression host for the CaTPS2 and CaTPS3 genes, and the empty vector pET-32a was used as a negative control. Positive colonies verified by sequencing were incubated 10–14 h at 37 °C with shaking at 180 rpm in 1 ml Luria–Bertani (LB) medium added Ampicillin (100 µg/ml). About 100 µl of overnight grown cultures were transferred to 100 ml LB media and were incubated under the same condition as above until OD600 of 0.4–0.6 was achieved. The formation of recombinant proteins with isopropyl-β-D-thiogalactopyranoside (IPTG) and the fusion proteins purification with Ni–NTA His·Bind Resins were implemented as described previously (Yue et al. 2014). Partially purified CaTPS2 and CaTPS3 proteins were dialyzed twice to three times in a buffer containing 5 mM dithiothreitol (DTT) and 30 mM HEPES (pH 7.5). Protein concentrations were performed on Ultra-micro Ultraviolet–visible Spectrophotometer (MIULAB) using the ultraviolet spectrophotometry method. Standard assays were performed in a total volume of 100 µl containing buffer (5 mM DTT, 25 mM MgCl2, 30 mM HEPES, pH 7.5), 20 μM substrate geranyl diphosphate/farnesyl diphosphate (GPP/FPP, Sigma) and 10–20 µg partially purified protein. The mixture was incubated into the reaction bottle at 30 °C. After 1 h, an SPME fiber was used to adsorb volatiles for 1 h and then inserted into the GC‒MS instrument for volatile analysis.
Real-time PCR analysis
Total RNA was extracted from different floral developmental stages and various plant tissue samples using the HiPure Plant RNA Kit (Magen) following the manufacturer’s protocol instructions, and then reverse transcribed using an Evo M-mLV premix kit (Accurate Biology, Carle Place, NY, USA) following the manufacturer’s suggestions. Real-time PCR analysis was performed on a 7500 Real Time PCR System using Hieff qPCR SYBR Green Master Mix (low Rox) (Yeasen, Shanghai, China) following the protocol. Each sample were tested with triplicate biological replicates, and the relative expression levels of CaTPS2 and CaTPS3 were calculated using the formula 2−ΔΔCт method (Livak and Schmittgen 2001). The sequence-specific primers for CaTPS2 and CaTPS3 are listed in Supplementary Table S1. SPSS software was used to analysis of variance by Tukey’s test (P = 0.05).
Results
Emission patterns of floral terpenoids of C. alismatifolia
To investigate the composition and content of volatiles in C. alismatifolia, we chose different varieties of C. alismatifolia for measurement, and among these, C. alismatifolia ‘Shadow’ released the most abundant volatiles’ compounds (Supplementary Fig. S1). Therefore, we selected ‘Shadow’ as the model variety, and seven different tissues were used for volatiles collection and GC‒MS analysis. The results showed that the fertile bracts contained 14 terpenoids, and the total concentrations of monoterpenes and sesquiterpenes represented roughly 43.76% and 56.24% of the total volatile terpenoids (20.52 µg g−1FW h−1), respectively. Caryophyllene, β-pinene, and α-pinene were the dominant volatile compounds (Table 1). A total of nine terpenoids (8.64 µg g−1FW h−1) were detected in the sterile bracts, and the emission rates of β-pinene and α-curcumene were higher. Leaves only contained eight terpenoids, with a total amount of 4.298 µg g−1FW h−1.
In the various parts of the ‘Shadow’ flower, a total of 12 floral terpenoids were detected and analyzed, and these comprised six monoterpenes and six sesquiterpenes (Table 2). The total amount of terpenoids in petals was 53.98 µg g−1FW h−1, of which monoterpenes and sesquiterpenes accounted for approximately 43.02% and 56.98%, respectively, and caryophyllene, and linalool were the main volatile compounds. Twelve types of terpenoids were also detected in the labella, the concentration of the total volatiles was 13.49 µg g−1FW h−1, and the amounts of linalool, caryophyllene, nerolidol and β-myrcene were higher. Only four terpenoids were detected in the lateral petals (11.01 µg g−1FW h−1), and linalool has the highest concentration (9.42 µg g−1FW h−1). The concentration of terpenoids was lowest in the gynandrium (4.35 µg g−1FW h−1), and the dominant compounds were nerolidol, caryophyllene and linalool. In summary, the total mono- and sesquiterpenes were observed to be firmly discharged from the petals, indicating that genuine blossoms could be a vital site for the dispersion of botanical fragrance in C. alismatifolia.
Transcriptome analysis of C. alismatifolia ‘Shadow’
We performed a transcriptome analysis of C. alismatifolia ‘Shadow’ to identify the genes involved in the biosynthesis of floral terpenoids. An overview of the sequencing and assembly data is listed in Supplementary Table S2. The RNA-seq data yielded 53,655 unigenes with an N50 value of 768 bp, and the annotation of these unigenes is outlined in Supplementary Table S3. Briefly, the data suggested that the sequences provide a valuable resource for further analysis. Using HMMER domain (PF03936, PF01397) screening and the NCBI BLAST function against the currently assembled ‘Shadow’ transcriptome, we identified a total of eight TPS genes, named CaTPS1 to CaTPS8, possibly involved in terpenoid biosynthesis. Detailed information of the eight genes is shown in Supplementary Table S4, the full-length sequences of CaTPS1-8 encoding 546, 579, 590, 562, 775, 550, 772, and 593 amino acids (aa) with predicted molecular mass of 64.2, 66.96, 68.23, 65.16, 89.03, 64.65, 82.99, and 68.38 kDa, respectively. In ‘Shadow’, the differential expressions of eight CaTPS genes in flowers and sterile bracts were analyzed based on the RPKM data of transcriptome and visualized by TBtools (Fig. 2b). The result shows that high expression of CaTPSs was observed in the flowers, among which CaTPS2 and CaTPS3 displayed the highest expression (Fig. 2b), suggesting that these two TPS genes are the top candidate genes for floral terpenoid formation.
Expression profile of TPS genes in C. alismatifolia ‘Shadow’. a Photographs of sterile bracts and flowers of C. alismatifolia ‘Shadow’. b Expression profile of CaTPSs in sterile bracts and flowers. Heatmap are drawn based on the relative expression levels of CaTPSs genes RNA-Seq data. Red and blue represent the log2-transformed expression values
Phylogenetic analysis of CaTPS2 and CaTPS3
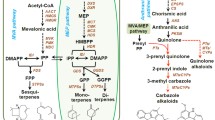
The full-length amino acid sequences of CaTPS2 and CaTPS3 are highly conserved with TPS proteins from other plant species (Fig. 3a). The sequence alignment of CaTPS2 and CaTPS3 proteins revealed conserved DDXXD regions and two conserved sequences of the N-terminal domain (PF01397) and the C-terminal active domain (PF03936), and the motif DDXXD is essential for metal ion binding and consists in almost all TPSs. Moreover, NSE/DTE motif is also found in CaTPS2 and CaTPS3 protein sequences, which function similarly to DDXXD motif, while RRX8W motif is found in CaTPS2 but not present in CaTPS3 (Fig. 3a). A phylogenetic analysis of CaTPS2 and CaTPS3 amino acid sequences with those of other species was performed to further explore their potential biochemical function. The results showed that CaTPS3 was assigned to the TPS-b subfamily and mainly encoded monoterpene synthase. CaTPS2 belongs to the TPS-g subfamily encoding acyclic terpene synthases (Fig. 3b). In summary, the cloned CaTPS2 and CaTPS3 are considered as the typical members of TPS gene family based on the kinship and similarity analysis, indicating that they may have functional properties as monoterpene and sesquiterpene synthases.
Sequence analysis and phylogenetic tree of CaTPS2 and CaTPS3 with their homologous genes. a Alignment of the amino acid sequences of CaTPS2 and CaTPS3 and other TPS proteins originated from three species, in which the conserved RRx8W, DDXXD and NSE/DTE regions were underlined with red. b Phylogenetic analysis between CaTPSs and homologous proteins from other plant species. The detailed information of comparative plant species is provided in Supplementary Table S5. The scale bar indicates 10% sequence divergence
Functional analysis of the CaTPS gene in vitro
To further study the enzymatic activity of CaTPS2 and CaTPS3 in vitro, the coding regions of these two genes were expressed in the pET32a vector using E. coli Rosetta (DE3), and their activity was tested with the substrates GPP and FPP. The results of bacterial expression and purification of recombinant CaTPS2 and CaTPS3 proteins are shown in Supplementary Fig. S2, and the concentration of partially purified CaTPS2 and CaTPS3 protein were 0.982 mg/ml and 1.084 mg/ml, respectively. Our findings indicated that CaTPS2 can catalyze GPP to produce a single monoterpene (linalool) (Fig. 4c), while incubated with FPP, CaTPS2 only synthesized nerolidol (Fig. 4d). With GPP as the substrate, CaTPS3 catalyzed β-myrcene as the only product (Fig. 4g) and transformed FPP into a single sesquiterpene (β-farnesene) (Fig. 4h). In contrast, no product was obtained by incubating the recombinant empty vector with GPP and FPP, respectively (Fig. 4a, b). These results showed that the products formed by the two CaTPSs were consistent with the main volatile compounds released from the flowers, indicating that CaTPS2 and CaTPS3 proteins are the key determinants of the production of monoterpenes and sesquiterpenes in C. alismatifolia ‘Shadow’ flowers.
Characterization of CaTPS2 and CaTPS3 in vitro. a–b Total ion chromatogram (TIC) of the products obtained by recombinant empty vector incubated with geranyl pyrophosphate (a) and farnesyl diphosphate (b). c–d TIC of the CaTPS2 enzyme with GPP (c) and FPP (d). e–f TIC of linalool (e) and nerolidol (f) authentic standards. Insets in panels (c–f) represent the mass spectra of the corresponding main peaks. g–h TIC of the CaTPS3 enzyme with GPP (g) and FPP (h). i Mass spectrum of the peak in (g) and β-myrcene in the NIST08 library. j Mass spectrum of the peak in (h) and β-farnesene in the NIST08 library
Expression pattern of CaTPS genes and their major products
To clarify the temporal and spatial expression patterns of CaTPS2 and CaTPS3, we analyzed the expression of these two genes in diverse flower development stages and plant tissues by real-time PCR (Fig. 5). The correlation between gene expression level and emission of major products was established. The analysis of the expression of four different plant tissues indicated that CaTPS2 showed significantly higher expression levels in the flower, followed by fertile bracts, and almost no expression in sterile bracts and leaves (Fig. 5a), which was correlated with the release of linalool and nerolidol (Fig. 5b). In different floral tissues, CaTPS2 was expressed mainly in the petals and labella, with relative low expression level in lateral petals and gynandrium (Fig. 5c), the expression pattern was basically consistent with that of linalool but slightly different from that of nerolidol in gynandrium (Fig. 5d). Consistent with the accumulation pattern of linalool and nerolidol, the expression levels of CaTPS2 increased during the process of flower development, reached the highest level at the full-openness stage, and sharply decreased at the aging stage (Fig. 5e, f).
CaTPS2 and CaTPS3 expression analyses, and the emission of their major products in different floral developmental stages and plant tissues of C. alismatifolia ‘Shadow’. a–d Expression analysis of CaTPS2 (a, c), and the emission of linalool and nerolidol (b, d) in diverse plant tissues. e–f Expression analysis of CaTPS2 (e), and the emission of linalool and nerolidol (f) in flowers at four floral developmental stages. g–j Expression analysis of CaTPS3 (g, i), and the emission of β-myrcene and β-farnesene (h, j) in different plant tissues. k–l Expression analysis of CaTPS3 (k), and the emission of β-myrcene and β-farnesene (l) at four floral developmental stages. Error bars demonstrate standard deviation of three biological replicates. Various letters marked on bars demonstrate significant differences (P < 0.05)
In different organs of C. alismatifolia, high expression of CaTPS3 was detected in the flowers, and its main product, β-myrcene, was detected only in the same tissue (Fig. 5g, h). However, the contents of another product, β-farnesene, were highest in fertile bracts (Fig. 5h), indicating that other unknown factors are also involved in the production of β-farnesene. Compared with flowers, this gene showed relative low expression in fertile bracts and sterile bracts, and nearly no expression in leaves (Fig. 5g). The expression pattern of CaTPS3 tended to be similar to the emission of its corresponding products (β-myrcene and β-farnesene) in different floral organs and developmental stages, and highest expression level was discovered in petals at the full-openness stage (Fig. 5i–l). Otherwise, the expression level of CaTPS3 was higher in labella than in gynandrium, and the lowest expression in lateral petals (Fig. 5i). These results indicated that CaTPS2 and CaTPS3 were specifically expressed in floral tissues and regulated by flower development corresponding to their protein catalytic products detected in ‘Shadow’.
Discussion
This investigation into the accumulation and release of floral volatiles from diverse tissues of Curcuma alismatifolia ‘Shadow’ indicated the emission pattern of terpenes in the flowers. We found that the main volatile compounds released by ‘Shadow’ were monoterpenes and sesquiterpenes, mainly including linalool, caryophyllene, nerolidol, α-pinene, α-humulene, β-farnesene, and β-myrcene (Table 2), similar to the findings in other Zingiberaceae species, such as Hedychium coronarium (Yue et al. 2015) and C. alismatifolia ‘Chiang Mai Pink’ (Liao et al. 2022), with the main volatile components being monoterpenes and sesquiterpenes. In plants, the emission of volatile compounds is spatiotemporally regulated (Dudareva et al. 2013), a majority of floral scents usually occurs in a flower-specific, and the emission levels regulate with flower development. In H. coronarium, the flower is a source of floral volatile compounds, and floral scent emission increased as flower buds approach the full blooming stages (Yue et al. 2015). In Osmanthus fragrans, the largest number of volatile terpenes are released during the full booming stages (Li et al. 2020). Here, we found that volatile terpenes firmly transmitted from the petals in ‘Shadow’, demonstrating that the true flowers could be the critical wellsprings of flower volatiles as opposed to its colorful bracts. Moreover, the emission levels of its main volatile compounds increase with the development of flowers and decrease during the aging stage (Fig. 5). This pattern of volatile terpenes release is common in flowering plants and may be related to environmental interactions and self-development.
Volatile terpenes may serve as chemical signals to mediate communications between flowering plants and other organisms, and therefore, the TPS family have an important role in plant adaptation (Pichersky and Gershenzon 2002). A majority of orchids emit a charming fragrance, and researchers have demonstrated that the number of genes in the TPS gene family of orchids ranges from 20 to 150, belonging to a mid-size family (Chen et al. 2011). In H. coronarium, its flowers release a fresh and inviting fragrance, with the main floral aroma compounds being volatile terpenes (Yue et al. 2015), and 55 TPS genes were found in the H. coronarium (unpublished). A total of 111 RoTPS genes found in Rhododendron ovatum, the gene number is significantly larger than that of other Rhododendron species without floral scents (Wang et al. 2021). Through the analysis of transcriptome data of C. alismatifolia ‘Shadow’, we identified a total of eight TPS genes (CaTPS1 to CaTPS8) (Fig. 2), which belong to a small-scale family. The identification of TPS genes in C. alismatifolia ‘Shadow’ elucidates key aspects of the genetic basis of floral fragrance. These findings further demonstrated that the diversity and abundance of terpenes are largely related to the action of multiproduct TPSs in aromatic plants. This investigation into the TPS genes of C. alismatifolia ‘Shadow’ not only enriches our knowledge of plant fragrance biosynthesis but also heralds new possibilities for the genetic modification of floral traits.
TPS genes are the key enzymes for producing various terpenes (Chen et al. 2003). Our findings, focusing on the prominent expression and bifunctional activity of CaTPS2 and CaTPS3 in floral tissues, shed light on the complex pathways governing terpenoid biosynthesis. CaTPS2 was shown to be able to catalyze linalool and nerolidol using GPP and FPP, respectively, and the biochemical activity of CaTPS3 includes catalyzing GPP to β-myrcene and converting FPP to β-farnesene (Fig. 4), similar to the function of CfTPS1/CfTPS2, PamTPS1 and LoTPS12 isolated from Clematis florida, Plectranthus amboinicus, and Lathyrus odoratus, respectively (Ashaari et al. 2020; Jiang et al. 2020; Bao et al. 2020). Developmental regulation of floral scent release occurs at several levels, including orchestrated expression of floral fragrance biosynthesis pathway genes (Colquhoun et al. 2010). In this study, a connection in floral tissues was found between the expression levels of CaTPS2 and CaTPS3 and their major catalytic products, and the articulation levels of CaTPS2 and CaTPS3 in the flower displayed a pattern of simultaneous upregulation with the pattern of their protein products at four formative stages (Fig. 5). Moreover, the emission of floral scent is also affected by enzyme activities, substrate availability, and regulation of transcription factors (Pichersky et al. 1995; Ramya et al. 2017), which may be the factor leading to slight differences between the expression of CaTPS3 and the release of β-farnesene (Fig. 5g, h). In general, the results demonstrated that these two practical TPS might be liable for the temporal and spatial particularity of terpenoid creation in ‘Shadow’. This contributes significantly to our understanding of plant secondary metabolism and its implications for floral ecology and horticulture (Johnson and Ecker 2019).
The bifunctional nature of CaTPS2 and CaTPS3 highlights the evolutionary adapt ability of the TPS gene family, reflecting the nuanced mechanisms plants utilize to produce a diverse array of terpenes (Chen et al. 2003). The presence of bifunctionality, where an enzyme catalyzes the formation of multiple products, can be a result of evolutionary pressures. It suggests a versatile enzymatic machinery that allows for the synthesis of different terpene products, potentially providing adaptive advantages to the organism. The missing motif (RRx8W) in CaTPS3 (Fig. 3) could indicate a divergence from the canonical function of TPS-b enzymes, suggesting either a loss of function, a gain of a new function, or a specialization toward a different substrate or reaction pathway. Overall, the evolutionary relationship between bifunctionality and the missing conserved motif in CaTPS3 likely reflects complex dynamics shaped by selective pressures, genetic drift, and functional diversification over evolutionary time scales. Further biochemical and evolutionary studies are necessary to elucidate the precise mechanisms driving these observations and their implications for plant terpene biosynthesis and evolution (Bohlmann and Keeling 2008; Degenhardt et al. 2009; Tholl 2015). Additionally, the release pattern of volatile terpenes follows a temporal rhythm, which is closely related to the behavior of pollinator and herbivores (Fenske and Imaizumi 2016). Generally, when the flowers are ready for pollination, the release levels of related volatile compound also increase accordingly, while decrease in post-pollination to deter further visits and against herbivorous insects (Martignier et al. 2019). In Quisqualis indica, scent components and emission rates change at different stages of flower development to attract different pollinators and contribute to reproductive fitness (Yan et al. 2016). Here, our result indicated that the volatile terpenes emission of C. alismatifolia ‘Shadow’ firmly transmitted from the petals, and the expression levels of CaTPS2 and CaTPS3 with their protein catalytic products increase with the flowering process, implies a potential attractive function for subsequent pollination. These findings underscore the ecological role of volatile terpene compounds in mediating interactions with pollinators, a critical aspect of plant biology with far-reaching implications for biodiversity and ecosystem sustainability (Peterson and Harrington 2018; Williams and Jackson 2021a, b).
In conclusion, employing a comprehensive methodology that includes transcriptome sequencing, gene cloning, and in vitro functional assays, this study underscores the importance of a multifaceted approach in deciphering the genetic and biochemical foundations of scent production in plants (Lee et al. 2021; Green and Brown 2022). The conservation and ecological examination of C. alismatifolia and related species stand to benefit from our insights into scent production genetics. These efforts contribute not only to the preservation of genetic diversity but also to our understanding of plant-environment interactions, offering strategies for integrated pest management and pollinator support (Li and Zhang 2019; Kumar and Sharma 2020). Future research will undoubtedly leverage these insights, exploring the intersection of genetic engineering, horticulture, and ecology in the quest to enhance plant-pollinator interactions and floral aesthetic appeal.
Data availability
RNA-seq data and have been deposited in the GenBank of the National Center for Biotechnology Information (NCBI) (PRJNA1049307). The CaTPS2 and CaTPS3 gene sequences data generated in this study are available in NCBI under the access numbers: OR921088 and OR921089. Other data supporting the findings of this study are available within the paper or its supplementary data.
Abbreviations
- FPP:
-
Farnesyl diphosphate
- GPP:
-
Geranyl diphosphate
- TPS:
-
Terpene synthase
- VOCs:
-
Volatile organic compounds
References
Ashaari NS, Ab Rahim MH, Sabri S, Lai KS, Song AA, Abdul Rahim R, Wan Abdullah W, Ong Abdullah J (2020) Functional characterization of a new terpene synthase from Plectranthus amboinicus. PLoS ONE 15(7):e0235416
Bao T, Shadrack K, Yang S, Xue X, Li S, Wang N, Wang Q, Wang L, Gao X, Cronk Q (2020) Functional characterization of terpene synthases accounting for the volatilized-terpene heterogeneity in Lathyrus odoratus cultivar flowers. Plant Cell Physiol 61(10):1733–1749
Boachon B, Lynch JH, Ray S, Yuan J, Caldo KMP, Junker RR, Kessler SA, Morgan JA, Dudareva N (2019) Natural fumigation as a mechanism for volatile transport between flower organs. Nature Chem Biol 15(6):583–588
Bohlmann J, Keeling CI (2008) Terpenoid biomaterials. Plant J 54(4):656–669
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120
Brown DS, Patel KR (2019) Curcuma alismatifolia: a review on its botanical aspects, cultivation, and phytochemistry. Bot J 47(2):123–137
Byers KJRP, Bradshaw HD, Rifell JA (2014) Three floral volatiles contribute to differential pollinator attraction in monkeyflowers (Mimulus). J Exp Bot 217:614–623
Chen F, Tholl D, D’Auria JC, Farooq A, Pichersky E, Gershenzon J (2003) Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15(2):481–494
Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66(1):212–229
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Colquhoun TA, Verdonk JC, Schimmel BCJ, Tieman DM, Underwood BA, Clark DG (2010) Petunia floral volatile benzenoid/phenylpropanoid genes are regulated in a similar manner. Phytochemistry 71(2–3):158–167
Degenhardt J, Köllner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70(15–16):1621–1637
Dudareva N, Klempien A, Muhlemann JK, Kaplan I (2013) Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198(1):16–32
Fenske MP, Imaizumi T (2016) Circadian rhythms in floral scent emission. Front Plant Sci 7:462
Fisher RA, Kramer EM (2019) Advances in transcriptome sequencing and their application to the study of plant development. J Exp Bot 70(8):2075–2084
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–3435
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644–652
Green T, Brown S (2022) Transcriptomic insights into terpenoid biosynthesis in ornamental plants. J Exp Bot 73(5):1423–1437
Jiang Y, Qian R, Zhang W, Wei G, Ma X, Zheng J, Köllner TG, Chen F (2020) Composition and biosynthesis of scent compounds from sterile flowers of an ornamental plant Clematis florida cv. ‘Kaiser.’ Molecules 25(7):1711
Johnson A, Ecker JR (2019) The role of TPS genes in plant terpenoid synthesis: a review. Biochem J 476(3):4505–4520
Kjonboon T, Kanlayanarat S (2005) Effects of gibberellic acid on the vase life of cut patumma (Curcuma alismatifolia Gagnep.) ‘Chaing Mai’ flowers. Acta Hort 673:525–529
Koshioka M, Umegaki N, Boontiang K, Pornchuti W, Thammasiri K, Yamaguchi S, Tatsuzawa F, Nakayama M, Tateishi A, Kubota S (2015) Anthocyanins in the bracts of Curcuma species and relationship of the species based on anthocyanin composition. Nat Prod Commun 10(3):453–456
Kumar P, Sharma A (2020) Integrated pest management strategies in the era of climate change. Environ Sci Policy 107:54–62
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Lee SH, Morgan JP, Carter SG (2021) Terpene synthases and their role in plant-environment interactions. Environ Chem Lett 19:295–304
Li M, Zhang Y (2019) Genetic engineering for the conservation of biodiversity: opportunities and challenges. New Phytol 222(3):1173–1177
Li HY, Yue YZ, Ding WJ, Chen GW, Li L, Li YL, Shi TT, Yang XL, Wang LG (2020) Genome-wide identification, classification, and expression profiling reveals R2R3-MYB transcription factors related to monoterpenoid biosynthesis in Osmanthus fragrans. Genes 11(4):353
Liang M, Chen W, LaFountain AM, Liu Y, Peng F, Xia R, Bradshaw HD, Yuan YW (2023) Taxon-specific, phased siRNAs underlie a speciation locus in monkeyflowers. Science 379(6632):576–582
Liao X, Ye Y, Zhang X, Peng D, Hou M, Fu G, Tan J, Zhao J, Jiang R, Xu Y, Liu J, Yang J, Liu W, Tembrock LR, Zhu G, Wu Z (2022) The genomic and bulked segregant analysis of Curcuma alismatifolia revealed its diverse bract pigmentation. aBIOTECH 3(3):178–196
Lin T, Vrieling K, Laplanche D, Klinkhamer PGL, Lou Y, Bekooy L, Degen T, Bustos-Segura C, Turlings TCJ, Desurmont GA (2021) Evolutionary changes in an invasive plant support the defensive role of plant volatiles. Curr Biol 31(15):3450–3456
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Mao XZ, Cai T, Olyarchuk JG, Wei LP (2005) Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21(19):3787–3793
Martignier T, Labouche A, Pannell JR (2019) Pollination elicits an accelerated reduction in nocturnal scent emission by flowers of the dioecious herb Silene latifolia. Botany 97:495–502
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Nguyen QT, Zhou XL, Li PY (2023) Enhancing ornamental plant traits through genetic engineering: prospects and challenges. Hortic Res 10(1):88–102
Parachnowitsch AL, Raguso RA, Kessler A (2012) Phenotypic selection to increase floral scent emission, but not flower size or colour in bee-pollinated Penstemon digitalis. New Phytol 195(3):667–675
Peterson RA, Harrington MG (2018) Ecological impacts of plant volatiles on pollinator attraction. Ecol Lett 21(4):516–525
Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5(3):237–243
Pichersky E, Lewinsoh E, Croteau R (1995) Purification and characterization of S-linalool synthase: an enzyme involved in the production of floral scent in Clarkia breweri. Arch Biochem Biophys 316(2):803–807
Ramya M, Kwon OK, An HR, Park PM, Baek YS, Park PH (2017) Floral scent: regulation and role of MYB transcription factors. Phytochem Lett 19:114–120
Sheehan H, Hermann K, Kuhlemeier C (2012) Color and scent: how single genes influence pollinator attraction. Cold Spring Harb Symp Quant Biol 77:117–133
Shen Y, Rao Y, Ma M, Li Y, He Y, Wang Z, Liang M, Ning G (2024) Coordination among flower pigments, scents and pollinators in ornamental plants. Hortic Adv 2(1):6
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7:539
Tholl D (2015) Biosynthesis and biological functions of terpenoids in plants. Adv Biochem Eng Biotechnol 148:63–106
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1):136–138
Wang X, Gao Y, Wu X, Wen X, Li D, Zhou H, Li Z, Liu B, Wei J, Chen F, Chen F, Zhang C, Zhang L, Xia Y (2021) High-quality evergreen azalea genome reveals tandem duplication-facilitated low-altitude adaptability and floral scent evolution. Plant Biotechnol J 19(12):2544–2560
Williams BJ, Jackson DP (2021a) Functional analysis of terpene synthases in plants: a review. Plant Methods 17:29
Williams BJ, Jackson DP (2021b) Terpenes and plant-pollinator interactions. Plant Mol Biol 105(3):287–295
Yan J, Wang G, Sui Y, Wang M, Zhang L (2016) Pollinator responses to floral colour change, nectar, and scent promote reproductive fitness in Quisqualis indica (Combretaceae). Sci Rep 6:24408
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Yue Y, Yu R, Fan Y (2014) Characterization of two monoterpene synthases involved in floral scent formation in Hedychium coronarium. Planta 240(4):745–762
Yue Y, Yu R, Fan Y (2015) Transcriptome profiling provides new insights into the formation of floral scent in Hedychium coronarium. BMC Genomics 16(1):470
Zhou Y, Abbas F, He J, Yan F, Wang Q, Yu Y, Yu R, Fan Y (2022) Floral volatile chemical diversity in Hedychium F1 hybrid population. Ind Crops Products 184:115032
Zuker A, Tzfira T, Ben-Meir H, Ovadis M, Shklarman E, Itzhaki H, Forkmann G, Martens S, Neta-Sharir I, Weiss D, Vainstein A (2002) Modification of flower color and fragrance by antisense suppression of the flavanone 3-hydroxylase gene. Mol Breed 9:33–41
Acknowledgements
This research was supported by the Key-Areas Research and Development Program of Guangdong Province (Grant no. 2020B020220007) and the National Agricultural Technology Modernization Pilot County Construction Project (Conghua District).
Author information
Authors and Affiliations
Contributions
YF and ZC conceived of the study. ZC, LW, DH, GW, YYY and XL performed the experiments and analyzed the data. LW and ZC drafted the manuscript. YCY, RY and YF revised the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cao, Z., Wang, L., Huang, D. et al. Identification and functional analysis of floral terpene synthase genes in Curcuma alismatifolia. Planta 260, 26 (2024). https://doi.org/10.1007/s00425-024-04440-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-024-04440-z