Abstract
Starch composition which is dependent on the waxy protein, the enzyme responsible for amylose synthesis in the grain, is an important aspect of the wheat quality. In this report, we describe the characterization of a novel Wx-A1 allele (Wx-A1g formerly known as -Wx-A1a) in Spanish spelt wheat lines which is responsible for a remarkable decline in the concentration of Wx-A1 protein found in the endosperm. Comparison of the DNA sequences in the Wx-A1a and Wx-A1g alleles showed the presence of a 160-bp insertion within the fourth intron in the latter. This insertion had some characteristics of a transposable-like element. RT-PCR analysis showed the presence of normal and aberrant mRNA transcripts in the Wx-A1g lines, indicating that the aberrant transcripts are un-spliced and contained the longer fourth intron. This may be related to the low level of Wx-A1 protein in these lines. In addition, a simple and fast PCR assay was designed for differentiating among different Wx-A1 alleles (a, b, f and g). The mutation described here is not related to either of the Wx-A1 mutations identified previously in common and durum wheats and could help to extend the range of amylose content of wheats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One important component of wheat grain is starch, which accounts for between 65 and 75% of its dry weight. This macromolecule is composed of two types of glucose polymer: amylose and amylopectin (James et al. 2003). The ratio of the polymers (usually 22–35% amylose, 68–75% amylopectin) is important as it affects properties of the starch such as gelatinization, pasting and gelation which are determinants of the end-uses of the flour or starch (Zeng et al. 1997).
Recently, wheat lines with varying ratios of amylose/amylopectin have been investigated, and these lines have then been used in breeding programmes to develop new wheat lines with a modified ratio of these components (Nakamura et al. 1995; Kiribuchi-Otobe et al. 1997; Yamamori et al. 2000). As amylose is digested more slowly than amylopectin, and slow digestion is beneficial to human health, wheat lines with high amylose content may be used in the manufacture of healthy food (Higgins et al. 2004; Behall and Scholfield 2005). On the other hand, wheat lines containing lower or zero amount of amylose may have commercial importance since starch with reduced levels of amylose was reported to produce noodles of better quality (Oda et al. 1980) and to extend the shelf life of various baked products (Lee et al. 2001).
Although several enzymes are involved in starch synthesis in cereal endosperms, GBSSI or waxy protein is the sole enzyme responsible for amylose synthesis (Denyer et al. 2001). Three waxy proteins are present in common wheat (Triticum aestivum L. ssp. aestivum) with molecular weights ranging from 59 to 60 kDa and are encoded by three genes: Wx-A1 located on chromosome 7AS, Wx-B1 on chromosome 4AL, and Wx-D1 on chromosome 7DS (Yamamori et al. 1994). Each gene has eleven exons and ten introns (Murai et al. 1999). Several studies have shown that variation in amylose content is associated with the status of the Wx proteins expression (Yamamori et al. 1994; Rodriguez-Quijano et al. 1998; Urbano et al. 2002); particularly important is the occurrence of null alleles (absence of protein) of any of the three Wx genes.
In a worldwide survey of common wheat, Yamamori et al. (1994) found that the null Wx-A1 allele frequently occurred in Japanese, Korean and Turkish cultivars, whereas the null Wx-B1 allele was present in many cultivars from Australia to India. Other authors also found these null variants in materials from all over the world (Graybosch et al. 1998; Rodriguez-Quijano et al. 1998; Urbano et al. 2002). The null Wx-D1 allele is, however, extremely rare and has only been described in few cultivars (Yamamori et al. 1994; Urbano et al. 2002; Guzman et al. 2010).
These null alleles have been subjected to molecular characterization to identify the nature of their inactivation. Vrinten et al. (1999) analysed the null alleles present in cv. Kanto 107 (null for the Wx-A1 and Wx-B1 genes) and cv. Bai Huo (null for the Wx-D1 gene), both of which had previously been crossed by Nakamura et al. (1995) to develop waxy wheat. They found that the inactivation of these genes is caused by different deletions and insertions in each of the waxy genes. However, other types of the null Wx-A1 allele have been also identified, for example Saito et al. (2004) detected an insertion of 173 bp inside the fourth exon of Wx-A1f that changes the ORF and leads to a premature stop codon. Monari et al. (2005) described both deletions and insertions in different materials. In addition, Saito and Nakamura (2005) found an insertion and a deletion of one nucleotide that changed the ORF of the gene in wild and cultivated emmer (T. turgidum ssp. dicoccoides Körn. ex Asch. and Graebner em. Thell. and T. turgidum ssp. dicoccum Schrank, respectively) that lacked the Wx-A1 protein.
The ancient wheats, along with their relatives, have showed to be an important source of variability for the bread- and pasta-quality traits and could be used in the breeding of the modern wheat (Sharma et al. 1981). Recently, diverse studies on the waxy genes have been carried out in three ancient wheat species of Spanish origin: einkorn (T. monococcum L. ssp. monococcum), emmer and spelt (T. aestivum ssp. spelta L. em. Thell). These species have been associated with a certain degree of variation including null alleles (Guzman et al. 2009, 2010, 2011). In a previous study on one broad collection of Spanish spelt (Guzman et al. 2010), we detected different null alleles, including Wx-A1 null (Wx-A1g formerly known as Wx-A1a′) in 36 out of 420 accessions evaluated.
The aim of the current study was the molecular characterization of this null Wx-A1g allele found in spelt in order to determine the cause of its reduced expression.
Materials and methods
Plant material
All spelt (T. aestivum ssp. spelta L. em. Thell) accessions that presented the novel Wx-A1g allele (36), together with a representative sample of accessions carrying the wild allele (Wx-A1a), classified in a previous survey by SDS-PAGE means (Guzman et al. 2010), were analysed in this study. The common wheat (Triticum aestivum L. ssp. aestivum) cultivar Chinese Spring (National Small Grain Collection, Aberdeen, ID, USA) that carries the Wx-A1a allele was used as standard as well as a Wx-A1b line (9906-155) generated from the cross of cvs. Bai Huo and Kanto 107. The cultivar Kanto 107 (Wx-A1b allele) and the Turkey-124 and Turkey-140 accessions (Wx-A1f allele), kindly supplied by the National Small Grain Collection (Aberdeen, ID,USA) and NIAS (Tsukuba, Japan), respectively, were included for comparison in DNA analysis. All plants were grown to maturity in a greenhouse.
Starch extraction and electrophoretic analysis
Preparation of starch granules and separation of waxy proteins by low-bis acrylamide SDS-PAGE were performed as described by Echt and Schwartz (1981) and Zhao and Sharp (1996).
For two-dimensional polyacrylamide-gel electrophoresis (2D-PAGE), 8.0 mg of starch was puffed up at room temperature in 300 μl of lysis buffer [8 M urea, 2% Nonidet-P40, 2% ampholine pH 3.5–10 (Pharmacia LKB) and 5% 2-mercaptoethanol]. After centrifugation, the supernatant containing the solubilised proteins was subjected to 2D-PAGE using isoelectric focusing (IEF) for the first dimension and modified SDS-PAGE for the second (Nakamura et al. 1993). IEF gels contained 2.5% (v/v) ampholines (pH 3.5–10/5–8, 1:1). Focusing was started from the acidic end (0.01 M H3PO4) and continued at 400 V for 15 h, then 800 V for 60 min at room temperature. Proteins were revealed by silver staining according to stain kit (Wako Pure Chemical Industries).
DNA extraction and PCR amplification
For DNA extraction, about 100 mg of young leaf tissue was excised, immediately frozen in liquid nitrogen and stored at −80°C. DNA was isolated using the DNAzol® method (Invitrogen).
The primers designed by Monari et al. (2005) were used to amplify the waxy genes in three regions. The region spanning the first to the third exon was amplified using the primers WxF3 (5′-TCTGGTCACGTCCCAGCTCGCCACCT-3′) and WxVT1R (5′-ACCCCGCGCTTGTAGCAGTGGAAGT-3′); the pair WxBAF (5′-ACTTCCACTGCTACAGCGCGGGGT-3′) and WxBAR (5′-GCTGACGTCCATGCCGTTGACGATG-3′) was used to amplify the region spanning the third to the sixth exon; and the pair WxVT1F (5′-CATCGTCAACGGCATGGACGTTCAGC-3′) and WxVTR (5′-CCAGAAGCACGTCCTCCCAGTTCTTG-3′) for the region spanning the sixth to the eleventh exon. Each 15 μl reaction included 50 ng DNA, 1.5 mM MgCl2, 0.2 µM of each primer, 0.2 mM dNTPs, 1.5 μl 10× PCR buffer and 0.75 U DNA polymerase (Promega). The PCR conditions included an initial denaturation step of 3 min at 94°C followed by 35 cycles as follows: for WxF3/WxVT1R, 40 s at 94°C, 1 min at 62°C, then 1 min at 72°C; for WxBAF/WxBAR, 45 s at 94°C, 2 min at 62°C, then 1 min 5 s at 72°C; and for WxVT1F/WxVTR, 40 s at 94°C, 1 min at 62°C, then 1 min 30 s at 72°C. After the 35 cycles all reactions included a final extension of 5 min at 72°C.
Amplification products were fractionated in vertical PAGE gels with 8% polyacrylamide concentration (w/v, C: 1.28%), and the bands were visualized by ethidium bromide staining.
RNA extraction, cDNA synthesis and analysis
Developing seeds at 10 days post-anthesis (10-DPA) for RNA extractions were collected, immediately frozen by immersion in liquid nitrogen and stored at −80°C. RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions and treated with TURBO DNase (RNase-Free; Ambion, Warrington, UK) to eliminate any DNA contamination. The cDNA was generated from total RNA and random nonamers using Superscript II Reverse Transcriptase (Invitrogen), according to the company’s instructions. One control PCR amplification was routinely performed to ensure that the specific PCR products were from RNA and not from DNA contamination.
Four pairs of specific primers were designed to amplify different regions of the cDNA surrounding the fourth intron in Wx-A1 gene (Table 1). Each 15-μl reaction included 50 ng cDNA, 1.5 mM MgCl2, 0.4 μM of each primer, 0.2 mM dNTPs, 1.5 μl 10× PCR buffer and 0.75 U DNA polymerase. The PCR conditions for each pair are described in Table 1. Amplification products were fractionated as described before.
Cloning of PCR products and sequencing analysis
PCR products were excised from polyacrylamide gel and cloned into pGEM T-easy vector (Promega) for sequencing. Inserts were sequenced using an ABI Prism 310 Genetic Analyzer (Perkin Elmer). The sequences were compared with the sequences of spelt (Wx-A1; HQ338723) and cv. Chinese Spring (Wx-A1a; AB019622) available in the NCBI databases.
PCR marker for differentiation among null and partial-null Wx-A1 alleles
For detection and differentiation of different Wx-A1 alleles (a, b, f and g), a pair of primers that flanked the region where these alleles showed differences was designed: Wx-A1 Diag Forward (5′-GTAAGCTTGCGCCACTGCCT-3′) and Wx-A1 Diag Reverse (5′-TGTGCCAGTCGTTGCACACA-3′). Each 15-μl reaction included 50 ng DNA, 1.5 mM MgCl2, 0.2 μM of each primer, 0.2 mM dNTPs, 1.5 μl 10× PCR buffer and 0.75 U DNA polymerase (Promega). The PCR conditions included an initial denaturation step of 3 min at 94°C followed by 35 cycles as follows: 40 s at 94°C, 30 s 66°C and then 1 min 55 s at 72°C. After the 35 cycles a final extension of 5 min at 72°C. Amplifications products were fractionated as described above. Additionally, the PCR products were restricted with the ApoI endonuclease (Fermentas) following the supplier’s instructions.
Amplification and digestion products were fractionated in vertical PAGE gels with 8% polyacrylamide concentration (w/v, C: 1.28%), and the bands were visualized by ethidium bromide staining.
Results
SDS-PAGE and 2D-PAGE electrophoresis
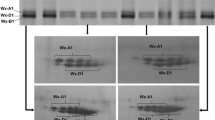
The separation of the waxy proteins in several representative lines of the variation detected is shown in Fig. 1. In a previous study (Guzman et al. 2010) these lines were classified as null for Wx-A1 protein (Wx-A1b allele). However, in the current study, when the SDS-PAGE gels were stained for an extended period, a faint band was detected in the Wx-A1 position (lanes 1, 3, 4, 5, 7, and 8), although it was much less intense than in the lines carrying the Wx-A1a allele (lanes 2 and 6).
SDS-PAGE and 2D-PAGE electrophoresis patterns of waxy proteins. Lanes are as follows: 1, Line 9906-155 (Wx-A1b); 2, PI-348437 (Wx-A1g); 3, BGE-012911 (Wx-A1g); 4, PI-469029 (Wx-A1a); 5, PI-348518 (Wx-A1g); 6, PI-348741 (Wx-A1g); 7, PI-348728 (Wx-A1g); and 8, cv. Chinese Spring (Wx-A1a. All lines present the Wx-B1a and Wx-D1a proteins. Faint Wx-A1g proteins are indicated by arrows. In 2D-PAGE electrophoresis, Wx-A1b (a), Wx-A1a (b) and Wx-A1g (c) are shown
One similar result was indicated by Caballero et al. (2008) that provisionally named the partially null allele found as Wx-A1a′. Although following this discovery, this new allele was designated Wx-A1g by McIntosh et al. (2009). The comparison between the three Spanish spelt lines used by Caballero et al. (2008) and that ones used in the current study suggested that both alleles could be the same. Consequently, these lines were reassigned as partially null for the Wx-A1 protein (Wx-A1g allele). This result was confirmed with 2D-PAGE, which showed three faint spots in the expected position of the Wx-A1 protein (Fig. 1b) but with less intensity than that shown in the spelt reference sample (Fig. 1a).
Molecular characterization of Wx-A1g allele
The PCR amplification of the Wx-A1 gene was carried out in genomic DNA with the specific primers designed by Monari et al. (2005). This permitted the simultaneous amplification of the three waxy genes (A, B, and D) as three fragments. In the first and third fragments of the Wx-A1 gene no differences were detected between the amplicons of the lines carrying the Wx-A1g and Wx-A1a alleles (data not shown). However, amplification of the central region of these genes revealed a remarkable difference between the spelt and common wheat lines (Fig. 2, lanes 2 and 6, respectively) which were used as standards. Spelt showed two bands due to the Wx-A1 and Wx-B1 which co-migrated, while the common wheat presented three conspicuous bands. When the amplicons were sequenced, three different sequences for each species were identified. The size of the Wx-A1 (953-bp) and Wx-D1 (1017-bp) was similar in both species, while that of the Wx-B1 bands was different: 973-bp for spelt and 935-bp for common wheat.
Diagrammatic representation of the waxy gene structure and amplification of the central region (Fragment 2) in several representative lines. Lanes are as follows: 1, PI-348437 (Wx-A1g); 2, PI-469029 (Wx-A1a); 3, PI-348518 (Wx-A1g); 4, PI-348741 (Wx-A1g); 5, PI-348728 (Wx-A1g); 6, cv. Chinese Spring (Wx-A1a); 7, PI-469029 (Wx-A1g); and 8, BGE-012911 (Wx-A1g). Wx-A1g bands are indicated with stars
On the other hand, the main difference between spelt lines containing the Wx-A1a and Wx-A1g alleles was the presence of an extra band larger than the Wx-D1a band (Fig. 2, lanes 1, 3, 4, 5, 7 and 8) in Wx-A1g lines. This larger band was purified and sequenced. It showed 100% homology with respect to the Wx-A1a allele from spelt reference line and cv. Chinese Spring, with the exception of one insertion of 160 bp located in the fourth intron (Fig. 3). This fragment had the characteristics of a transposable-like element, with target site duplications and inverted repeats, although the inverted repeats were not located in the terminal regions of the insertion and did not share 100% homology.
The insertion in the partial null Wx-A1g allele of spelt wheat. a Diagrammatic representation of the central region in the Wx-A1a and Wx-A1g alleles. b Comparison of genomic DNA sequences of Wx-A1 alleles between the primers Wx-BAF and Wx-BAR. The exons are indicated by capital letters. Target site duplications are indicated by arrows and terminal inverted repeats like sequences are shown in grey boxes
The rest of the Wx-A1 gene (first and third fragment) was also sequenced in two Wx-A1g lines (PI-348437, BGE-012911) in order to identify other differences that had not been detected by PCR analysis. In this case no variation was found with respect to the Wx-A1a allele; however, there was 100% homology with the spelt reference sample. The entire Wx-A1g sequences of both lines are available in GenBank (HQ625382, HQ625383).
RT-PCR analysis of the Wx-A1 gene
Because low levels of Wx-A1 protein were detected in lines containing the Wx-A1g allele in comparison with the lines containing Wx-A1a allele, it was important to determine whether the insertion found in the fourth intron of the Wx-A1g allele affected mRNA processing and, consequently, the expression of the protein. Total RNA was extracted from the 10-DPA endosperm for use in the synthesis of cDNA and RT-PCR analysis. Four pairs of specific primers were designed on the exons flanking the fourth intron which was expected to be removed during the splicing as was the remainder of the introns (Fig. 4a). Both the Wx-A1a and Wx-A1g alleles showed transcripts containing only the exons, all the introns having been removed (Fig. 4b; and Fig. 4d lanes 1-4.694 bp bands). In addition, lines carrying the Wx-A1g allele showed larger transcripts that were not present in the Wx-A1a lines with each pair of primes used (Fig. 4c; and Fig. 4d lanes 1, 2 975 bp bands). Sequencing revealed that these products consisted of Wx-A1 sequences carrying the fourth intron which had not been spliced. This gave a clear indication that splicing of part of the mRNA was not successful in the Wx-A1g genotypes. In addition, it was found with one pair of primers that part of the first exon was also deleted in some fragments of mRNA of Wx-A1g lines during the splicing, resulting in an incomplete Wx-A1 coding sequence (Fig. 4c).
a Schematic representation of the Wx-A1a and Wx-A1g alleles. b Fragments amplified from total RNA of Wx-A1a and Wx-A1g lines using RT-PCR. c Fragments amplified exclusively from total RNA of Wx-A1g lines. Four pairs of primers were used: 1, WxA1DFw/WxA1B2Rv; 2, WxA1AFw/WxA1A3Rv; 3, WxA1BFw/WxA1B1Rv; and 4, WxA1BFw/WxA1B2Rv. Small deletion in first exon was detected (star). d Representative gel of amplicons obtained from cDNA with primers WxA1AFw/Wx-A1A3Rv. Bands of 974 bp contained fourth intron unspliced with insertion. Lanes are as follows: 1, PI-348437 (Wx-A1g); 2, BGE-012911 (Wx-A1g); 3, PI-469029 (Wx-A1a); and 4, Pi-348515 (Wx-A1a)
Development of a molecular marker to differentiate Wx-A1 alleles
Two null Wx-A1 alleles (Wx-A1b and Wx-A1f) together with the wild allele (Wx-A1a) were compared with the Wx-A1g alleles by the amplification of the gene region between the first and fifth introns (Fig. 5a). This area include the In/Del sequences that differentiate the three mutant alleles and the wild ones. In addition, the primers were designed in areas of the Wx-A1 gene that have several mismatches with respect to Wx-B1 and Wx-D1 ones, which avoid the simultaneous amplification of the Wx-B1 and Wx-D1 homoeologous genes when the annealing temperature increases.
Molecular marker to differentiate Wx-A1 alleles. a Schematic representation of the Wx-A1a, Wx-A1b, Wx-A1f and Wx-A1g alleles. Primers Wx-A1 Diag forward and reverse are indicated in their annealing area. b PCR products with Wx-A1 Diag primers fractionated in polyacrylamide gels. c Digestion with ApoI of PCR products. Lanes are as follow: 1, cv. Chinese Spring (Wx-A1a); 2, spelt PI-469029 (Wx-A1a); 3, cv. Kanto 107 (Wx-A1b); 4, Turkey-124 (Wx-A1f); 5, Turkey-140 (Wx-A1f); and 6, spelt PI-348437 (Wx-A1g)
The Wx-A1a allele showed a 770-bp amplicon (Fig. 5b, lanes 1 and 2). However, as it was expected, the Wx-A1b allele present in cv. Kanto 107 did not show any PCR product, since the forward primer was designed on a region of the Wx-A1 sequence that was deleted in this allele, exactly at the first exon–intron junction. The other two alleles (Wx-A1f and Wx-A1g) showed amplicons with a higher size of the Wx-A1a allele (Fig. 5b, lanes 4–7), but with slight differences between them (943 and 930 bp, respectively). To make a more clear differentiation between Wx-A1g and Wx-A1f alleles, the PCR product was digested with ApoI endonuclease that has a sequence target in Wx-A1f allele (Fig. 5c). Wx-A1a and Wx-A1g products were not affected by this enzyme. Consequently, the use of this PCR assay permitted to discriminate among all alleles evaluated here, with special interest between both null allele type (Wx-A1b and Wx-A1f) and the new Wx-A1g allele that shows a partial expression.
Discussion
Waxy protein (GBSSI) is the key enzyme in amylose synthesis, whose variation has a considerable influence on the amylose content, mainly due to the presence of null alleles (Yamamori et al. 1994; Rodriguez-Quijano et al. 1998). Numerous studies have been carried out to characterize the mutations that cause these null Wx variants (Vrinten et al. 1999; Saito et al. 2004; Monari et al. 2005).
In the case of the Wx-A1 locus, several In/Del mutations that caused a change in the ORF which led to a premature stop codon have been found. Vrinten et al. (1999) described a 23-bp deletion at an exon–intron junction in cv. Kanto 107 that cause the lack of Wx-A1 protein. In the same way, Saito et al. (2004) described one mutation in Turkish common wheat accessions lacking Wx-A1 protein that consisted of a 173-bp insertion in the fourth exon of the Wx-A1 locus (Wx-A1f allele). They also found that the insertion had the characteristics of a transposable element of class II which affected RNA maturation; this element was designated Hikkoshi (Saito et al. 2004). In fact, Wessler and Varagona (1985) indicated that some spontaneous waxy mutations in maize were associated with the presence of these transposable elements. Monari et al. (2005) also identified a Turkish durum wheat accession that lacked the Wx-A1 protein as consequence of an insertion of 89-bp in the sixth exon.
Although Debiton et al. (2010) found a non-functional truncated protein for one null Wx allele (Wx-D1b), which presented a smaller size than the wild protein, the possible proteins of the abovementioned null Wx-A1 alleles have not been detected yet. On the contrary, the corresponding protein in the Wx-A1g allele detected in spelt by Caballero et al. (2008) and in the current study was found at very low levels. The analysis of its sequence indicated the presence of a 160-bp insertion in the fourth intron which showed some characteristics typical of transposable-like elements such as target site duplications and inverted repeats. Due to the similarity of the remaining sequence in the Wx-A1a and Wx-A1g alleles, it is likely that this insertion is related to the reduced expression of the Wx-A1 protein. The RT-PCR analysis of developing seeds showed that—although the correct processed form of Wx-A1 transcript was present in the Wx-A1a and Wx-A1g lines—transcripts with the un-spliced fourth intron were also found in all Wx-A1g lines. Saito et al. (2004), who also found un-spliced transcripts in null mutant lines, suggested that these aberrant mRNA products were rapidly degraded by a process referred to as nonsense-mediated mRNA decay (NMD), or formed a truncated protein that would be promptly eliminated because of its lack of function. In the current case, the 160-bp insertion found in this intron appeared to prevent the complete splicing of all the RNA molecules, thus reducing the yield of correctly processed RNA leading to a remarkable decrease in the final concentration of Wx-A1 protein.
The presence of three Wx loci in hexaploid wheat has made it difficult to develop wheats with low or null amylose content. Yamamori et al. (1994) classified these wheats in eight types by combining wild and null alleles at the Wx-A1, Wx-B1 and Wx-D1 loci. The wild type was named as type 1, waxy wheat as type 8 and the partial waxy lines as types 2–6. In general, the effect of any active Wx gene makes that the mutations of any of the other Wx genes have scarce impact on the amylose content due to one dosage effect, as it was showed again later by Yamamori and Quynh (2000). Consequently, the partial waxy lines show amylose contents almost always over 20%, and only the triple null mutant (type 8) present values less than 1% (Nakamura et al. 1995). Although further studies, similar to that made by Yamamori (2009) and Yamamori and Yamamoto (2011) with other Wx alleles, should be carried out, data suggest that the allele describe in the current survey (Wx-A1g) could have one different behaviour in combination with the null alleles for the Wx-B1 and Wx-D1 loci, thus opening the possibility to obtain quasi-waxy wheats with amylose content above 1% but less than 20%.
The main difficulty is the correct and unequivocal identification of the Wx-A1 alleles (a, b, f and g). The evaluation by SDS-PAGE (uni- or bi-dimensional) is low efficient for its use in breeding programmes. Consequently, the development of an easy and fast PCR marker associated with these mutations, as the designed one in the current study, is an important key for the successfully development of new waxy, partial waxy or quasi-waxy wheats.
In conclusion, a new waxy allele (Wx-A1g) has been characterized at a molecular level. The mutation described here is not related to either of the Wx-A1 mutations identified previously in common and durum wheats and could help to extend the range of amylose content of wheats or to breed wheats with very low amylose content.
Abbreviations
- GBSSI:
-
Granule-bound starch synthase I
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- RT-PCR:
-
Reverse transcription polymerase chain reaction,
- SDS-PAGE:
-
Sodium dodecyl sulphate polyacrylamide gel electrophoresis
References
Behall KM, Scholfield DJ (2005) Food amylose content affects postprandial glucose and insulin responses. Cereal Chem 82:654–659
Caballero L, Bancel E, Debiton C, Branlard G (2008) Granule-bound starch synthase (GBSS) diversity of ancient wheat and related species. Plant Breed 127:548–553
Debiton C, Bancel E, Chambon C, Rhazi L, Branlard G (2010) Effect of the three waxy null alleles on enzymes associated to wheat starch granules using proteomic approach. J Cereal Sci 52:466–474
Denyer K, Johnson P, Zeeman S, Smith AM (2001) The control of amylose synthesis. J Plant Physiol 158:479–487
Echt CS, Schwartz D (1981) Evidence for the inclusion of controlling elements within structural gene at the waxy locus in maize. Genetics 99:275–284
Graybosch RA, Peterson CJ, Hansen LE, Rahman S, Hill A, Skerritt JH (1998) Identification and characterization of US wheats carrying null alleles at the wx loci. Cereal Chem 75:162–165
Guzman C, Caballero L, Alvarez JB (2009) Variation in Spanish cultivated einkorn wheat (Triticum monococcum L ssp. monococcum) as determined by morphological traits and waxy proteins. Genet Resour Crop Evol 56:601–604
Guzman C, Caballero L, Moral A, Alvarez JB (2010) Genetic variation for waxy proteins and amylose content in Spanish spelt wheat (Triticum spelta L.). Genet Resour Crop Evol 57:721–725
Guzman C, Caballero L, Alvarez JB (2011) Molecular characterization of the Wx-B1 allelic variants identified in cultivated emmer wheat and comparison with those of durum wheat. Mol Breeding 28:403–411
Higgins JA, Higbee DR, DonahooWT BrownIL, Bell ML, Bessesen DH (2004) Resistant starch consumption promotes lipid oxidation. Nutr Metabol 1:8
James MG, Denyer K, Myers AM (2003) Starch synthesis in the cereal endosperm. Curr Opin Plant Biol 6:215–222
Kiribuchi-Otobe C, Nagamine T, Yanagisawa M, Ohnishi M, Yamaguchi I (1997) Production of hexaploid wheats with waxy endosperm character. Cereal Chem 74:72–74
Lee MR, Swanson BG, Baik BK (2001) Influence of amylose content on properties of wheat starch and breadmaking quality of starch and gluten blends. Cereal Chem 78:701–706
McIntosh RA, Dubcovsky J, Rogers WJ, Morris C, Appels R, Xia XC (2009) Catalogue of gene symbols for wheat: 2009 Supplement (http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2009.pdf)
Monari AM, Simeone MC, Urbano M, Margiotta B, Lafiandra D (2005) Molecular characterization of new waxy mutants identified in bread and durum wheat. Theor Appl Genet 110:1481–1489
Murai J, Taira T, Ohta D (1999) Isolation and characterization of the three Waxy genes encoding the granule-bound starch synthase in hexaploid wheat. Gene 234:71–79
Nakamura T, Yamamori M, Hirano H, Hidaka S (1993) Identification of three Wx proteins in wheat (Triticum aestivum L.). Biochem Genet 31:75–86
Nakamura T, Yamamori M, Hirano H, Hidaka S (1995) Production of waxy (amylose-free) wheats. Mol Gen Genet 248:253–259
Oda M, Yasuda Y, Okazaki S, Yamauchi Y, Yokoyama Y (1980) A method of flour quality assessment for Japanese noodles. Cereal Chem 54:253–254
Rodriguez-Quijano M, Nieto-Taladriz MT, Carrillo JM (1998) Polymorphism of waxy proteins in Iberian hexaploid wheats. Plant Breed 117:341–344
Saito M, Nakamura T (2005) Two point mutations identified in emmer wheat generate null Wx-A1 alleles. Theor Appl Genet 110:276–282
Saito M, Konda M, Vrinten P, Nakamura K, Nakamura T (2004) Molecular comparison of waxy null alleles in common wheat and identification of a unique null allele. Theor Appl Genet 108:1205–1211
Sharma HC, Waines JG, Foster KW (1981) Variability in primitive and wild wheats for useful genetic characters. Crop Sci 21:555–559
Urbano M, Margiotta B, Colaprico G, Lafiandra D (2002) Waxy proteins in diploid, tetraploid and hexaploid wheats. Plant Breed 121:465–469
Vrinten P, Nakamura T, Yamamori M (1999) Molecular characterization of waxy mutations in wheat. Mol Gen Genet 261:463–471
Wessler SR, Varagona MJ (1985) Molecular basis of mutations at the waxy locus of maize: correlation with the fine structure genetic map. Proc Natl Acad Sci USA 82:4177–4182
Yamamori M (2009) Amylose content and starch properties generated by five variant Wx alleles for granule-bound starch synthase in common wheat (Triticum aestivum L.). Euphytica 165:607–614
Yamamori M, Quynh NT (2000) Differential effects of Wx-A1, -B1 and -D1 protein deficiencies on apparent amylose content and starch pasting properties in common wheat. Theor Appl Genet 100:32–38
Yamamori M, Yamamoto K (2011) Effects of two novel Wx-A1 alleles of common wheat (Triticum aestivum L.) on amylose and starch properties. J Cereal Sci 54:229–235
Yamamori M, Nakamura T, Endo R, Nagamine T (1994) Waxy protein deficiency and chromosomal location of coding genes in common wheat. Theor Appl Genet 89:179–184
Yamamori M, Fujita S, Hayakawa K, Matsuki J, Yasui T (2000) Genetic elimination of a starch granule protein, SGP-1, of wheat generates an altered starch with apparent high amylose. Theor Appl Genet 101:21–29
Zeng M, Morris CF, Batey II, Wrigley CW (1997) Sources of variation for starch gelatinization, pasting, and gelation properties in wheat. Cereal Chem 74:63–71
Zhao XC, Sharp P (1996) An improved 1-D SDS-PAGE method for the identification of three bread wheat ‘waxy’ proteins. J Cereal Sci 23:191–193
Acknowledgments
This research was supported by grants AGL2007-65685-C02-02 and AGL2010-19643-C02-01 from the Spanish Ministry of Science and Innovation, co-financed with European Regional Development Fund (FEDER) from the European Union. The first author thanks the Spanish Ministry of Education and Science (FPU programme) for a predoctoral fellowship. In the same way he expresses his appreciation to Alessandra Di Francesco, Andrea Aglieco and Ana Moral for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guzmán, C., Caballero, L., Yamamori, M. et al. Molecular characterization of a new waxy allele with partial expression in spelt wheat. Planta 235, 1331–1339 (2012). https://doi.org/10.1007/s00425-011-1577-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-011-1577-7