Abstract
The association between common neuroradiological markers of multiple sclerosis (MS) and clinical disability is weak, a phenomenon known as the clinico-radiological paradox. Here, we investigated to which degree it is possible to predict individual disease profiles from conventional magnetic resonance imaging (MRI) using multivariate analysis algorithms. Specifically, we conducted cross-validated canonical correlation analyses to investigate the predictive information contained in conventional MRI data of 40 MS patients for the following clinical parameters: disease duration, motor disability (9-Hole Peg Test, Timed 25-Foot Walk Test), cognitive dysfunction (Paced Auditory Serial Addition Test), and the expanded disability status scale (EDSS). It turned out that the information in the spatial patterning of MRI data predicted the clinical scores with correlations of up to 0.80 (p < 10−9). Maximal predictive information for disease duration was identified in the precuneus and somatosensory cortex. Areas in the precuneus and precentral gyrus were maximally informative for motor disability. Cognitive dysfunction could best be predicted using data from the angular gyrus and superior parietal lobe. For EDSS, the inferior frontal gyrus was maximally informative. In conclusion, conventional MRI is highly predictive of clinical disability in MS when pattern-based algorithms are used for prediction. Thus, the so-called clinico-radiological paradox is not apparent when using suitable analysis techniques.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Since the introduction of the McDonald criteria in 2001, conventional magnetic resonance imaging (MRI) has become one of the cornerstones of diagnosing multiple sclerosis (MS) [1]. However, the association between common neuroradiological markers such as T1 or T2 lesion load and clinical disability in patients suffering from MS is weak [2] and was referred to as the ‘clinico-radiological paradox’ [3]. Although various confounders have been discussed [4], the significance of conventional MRI in describing the clinical status of MS patients has been questioned [5, 6]. Please note that the term conventional MRI generally refers to standard MRI sequences such as T1- and T2-weighted images as compared to advanced MRI techniques such as diffusion tensor imaging.
In other neurological diseases such as Huntington or Alzheimer’s disease, however, multivariate pattern recognition methods have been demonstrated to be a sensitive tool predicting the clinical status of individual subjects including the presence of a disease [7, 8] as well as symptom severity [9, 10]. In the present study, we investigated the amount of predictive information contained in local tissue intensity patterns extracted from conventional T1- (magnetization prepared rapid gradient echo, MPRAGE) and T2-weighted (fluid-attenuated inversion recovery sequence, TIRM) MRI data for clinical disability in MS patients. We conducted separate cross-validated canonical correlation analyses to predict several clinical scores, including the disease duration, motor disability (9-Hole Peg Test [9-HPT], Timed-Walk Test [TWT], [11]), cognitive dysfunction (paced auditory serial addition test [PASAT], [11]) and overall clinical disability (expanded disability status scale [EDSS], [12]).
Methods
Patients
Forty-one patients with clinically definite MS (relapsing-remitting type, [1]) recruited for a recent study of our group entered the analysis [13]. Patients were required to be between 18 and 55 years and to have at least one gadolinium (Gd-DTPA)-enhancing lesion on a qualifying T1-weighted brain MRI scan [14]. The patients were scored on the EDSS [12] and the subtests 9-HPT, TWT, and PASAT of the Multiple Sclerosis Functional Composite (MSFC; [11]). One subject was excluded because his EDSS score exceeded the 97th percentile. Demographic and clinical details are listed in Table 1. Importantly, MRI and clinical ratings were performed on the same day and all patients were in remission at this time point (interval to the last relapse/corticosteroid treatment was at least 30 days).
Consent was obtained according to the Declaration of Helsinki, and the study was approved by the research ethics committee of the Charité - Universitätsmedizin Berlin. All subjects gave written informed consent.
MRI acquisition
Whole-brain high-resolution three-dimensional T1-weighted images (MPRAGE, TR 2110 ms, TE 4.38 ms, TI 1,100 ms, flip angle 15°, resolution 1 × 1 × 1 mm) and a T2-weighted fluid-attenuated inversion recovery sequence (TIRM, TR 10,000 ms, TE 108 ms, TI 2,500 ms, resolution 1 × 1 × 3 mm, 44 contiguous axial slices) were acquired using a 1.5-Tesla MRI (Magnetom Sonata, Siemens, Erlangen, Germany) with an eight-channel standard head coil.
Lesion load
Lesion load for MPRAGE and TIRM images were routinely measured using the MedX v.3.4.3 software package (Sensor Systems Inc., Sterling, VA, USA, [15]). Lesion load of TIRM images were additionally measured using in-house software [13].
Preprocessing
Several preprocessing steps were performed. First, a clinician and experienced reader (CP) applied in-house software to conduct lesion mapping based on individual TIRM images. To be as conservative as possible, CP was instructed to mark any hyperintensities visible in the TIRM images and not only oval lesions as it is common in clinical practice. Next, correction of field inhomogeneities, coregistration of high-resolution MPRAGE and TIRM images, and normalization of these high-resolution images to the Montreal Neurological Institute (MNI) brain template (voxel size: 2 × 2 × 2 mm) were conducted using SPM5 (Wellcome Trust Centre for Neuroimaging, Institute of Neurology, UCL, London, http://www.fil.ion.ucl.ac.uk/spm; Fig. 1). The normalization parameters for the MPRAGE images were estimated by the ‘unified segmentation approach’ [16] and then applied to the MPRAGE and co-registered TIRM images as well as to individual lesion masks. Importantly, lesion areas identified by the clinician were excluded to avoid lesion-mediated artifacts in the normalization routine (see SPM5 manual: www.neuro.nl/fmri/docs/SPM5manual.pdf). Finally, we obtained MPRAGE and TIRM images from all subjects as well as their individual lesion masks in MNI space (voxel size: 2 × 2 × 2 mm).
Overview of data processing. Raw MPRAGE and TIRM images were co-registered and normalized to the Montreal Neurological Institute (MNI) template. Individual lesions that were marked beforehand by a clinician were excluded from the estimation of normalization parameters. The search space was reduced to only voxels within the standard brain mask that were not cerebrospinal fluid with a probability of greater than 0.8. For the normalized MPRAGE and TIRM images separately, we aimed to predict the clinical markers’ disease duration, motor disability, cognitive dysfunction, and overall clinical disability
For the pattern-based analyses, only voxels within the SPM standard brain mask that were not cerebrospinal fluid (CSF) with a probability of more than 0.8 (based on SPM CSF prior map) were included. The rather conservative threshold of 0.8 was chosen to avoid misinterpretation of tissue-free voxels as brain tissue. To account for different intensity levels, the images were standardized within subjects based only on normal-appearing (i.e., non-lesional) brain tissue. This was done to ensure that a higher lesion load did not introduce any biases into the standardization. We continued by using a general linear model to regress out the variance contained in voxel intensities that could be explained by local deformation parameters determined during spatial normalization. The deformation (i.e., shift) parameters in x, y, and z direction were calculated for individual MPRAGE images using the deformations toolbox implemented in SPM5. This step was performed in order to rule out that pattern-based prediction could rely on intensity differences induced by the spatial transformation that correlate with clinical disability (e.g., due to a stronger correction of regional atrophy in patients with a higher disability score).
Pattern-based canonical correlation analysis
In order to decode symptom severity from the preprocessed MRI data, we conducted a pattern-based and cross-validated canonical correlation analysis using in-house software [17] independently for MPRAGE and TIRM images and for each of the following clinical variables: disease duration, motor disability (as measured by 9-HPT and TWT), cognitive dysfunction (as measured by PASAT) and overall clinical disability (as measured by EDSS; Figs. 1, 2).
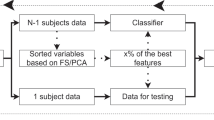
Illustration of pattern-based canonical correlation analysis. In the training phase, local patterns (i.e., so-called searchlights) of n-1 subjects were extracted from the normalized MPRAGE and TIRM images, respectively. The dimension of these feature vectors were reduced to half the samples using principal component analysis (PCA; not shown here). The canonical correlation analysis (CCA) then finds a linear relationship between PCA-projected voxel intensities and clinical scores for all training subjects. In the testing phase, a pattern of a new ‘unseen’ subject is represented and CCA is used to predict the clinical score of this person. For validation, we performed a leave-one-out cross-validation over all subjects. This whole procedure was repeated for each searchlight position, so that each voxel in the brain was the center voxel of the searchlight once
In all analyses, local intensity patterns were extracted from the given MRI data by using a ‘searchlight’ approach [18, 19], which searches across the whole brain for local tissue intensity patterns informative about clinical disability. A searchlight is defined as a spherical cluster of N voxels that is created around a given center voxel v i . Here, we used a searchlight radius of four voxels (i.e., 8 mm, corresponding to at most 257 voxels in one searchlight). For each voxel in a searchlight, we extracted individual tissue intensity values based on MPRAGE and TIRM images, respectively. In the next step, these feature vectors were used to decode disease duration, motor disability, cognitive dysfunction, or overall clinical disability by carrying out a canonical correlation analysis (CCA).
CCA is a standard tool in statistics to study linear relationships between two sets of multidimensional variables [20]. The goal of CCA is to find two bases, one for each set of variables, such that the correlation between the projections of the variables onto these two bases is mutually maximized. Because it is independent of the coordinate system in which the variables were originally described, it can overcome one of the major drawbacks of ordinary correlation analysis.
In our case, the first set of variables is given by the N-dimensional local tissue intensity vector of all subjects; the second set is given by one or two clinical scores of the subjects. Prediction of motor disability comprised two variables, 9-HPT and TWT; all other clinical variables were modeled using one variable.
Since CCA is ill conditioned for redundant sets of variables, we reduced the dimensionality of the feature vectors to 20 (i.e., half the number of subjects) using principal component analysis (PCA, [21]). A fixed number of principal components were chosen to guarantee equal feature vector lengths across all regions.
To assess the generalizability of performance using an independent data set while at the same time avoiding circular inference [22], we performed a leave-one-out cross-validation. This means that we used the feature vectors of all but one subjects as ‘training data’ first to reduce the dimensionality by PCA and second to train the CCA algorithm to learn a relationship between intensity values and clinical scores. We then tested it on the remaining, independent ‘test’ subject. This procedure was repeated so that each subject was the test subject once.
Decoding accuracy is then given by the Pearson’s correlation coefficient between true and predicted clinical scores (i.e., canonical variates) and is assigned to the center voxel of the searchlight. A high correlation implies that the local cluster of voxels surrounding the center voxel spatially encoded information about the clinical variables under investigation. Please note that we extracted only the first canonical variate since it accounts for the largest canonical correlation. After repeating the procedure for each searchlight position, we generated a parametric map indicating the correlation for each searchlight. Corresponding probabilities were calculated using Student’s t distribution. To account for the multiple comparison problem, we report searchlight center coordinates that exhibit a significant correlation on a Bonferroni-corrected level of p < 0.01 (one-tailed). This very conservative threshold was chosen to increase the specificity of the analyses.
To characterize the underlying tissue alterations, we also reported the average grey matter and lesion ratio for each significant cluster. For this purpose, individual grey matter and lesion ratios for all voxels contained in the searchlights of the cluster were calculated and then averaged. The individual grey matter ratio was determined by the number of grey matter voxels (probability of being grey matter >0.5 based on individual grey matter probability maps provided by SPM5 during spatial normalization) divided by the number of all voxels in the cluster. Similarly, individual lesion ratio was determined by the number of lesion voxels (according to the individual lesion masks) divided by the number of all voxels in the cluster.
In a further analysis, we investigated the predictability of clinical disability based on lesion load alone. Here, we conducted a CCA where the first set of variables is given by T1 and/or T2 lesion load and the second set is given by the clinical markers.
Results
Statistical analysis of clinical and volumetric measures
Significant correlations (Spearman’s rho) were found between the univariate variables disease duration and EDSS (r = 0.50, p < 0.01), disease duration, and 9-HPT (r = 0.38, p < 0.05), EDSS and 9-HPT (r = 0.40, p < 0.05), EDSS and TWT (r = 0.33, p < 0.05), 9-HPT and TWT (r = 0.46, p < 0.05), and TWT and PASAT (r = −0.47, p < 0.01). T2 lesion load calculated using in-house software correlated strongly with the T2 lesion load calculated with MedX (r = 0.88, p < 10−12, Pearson), but due to the sensitive lesion mapping performed by the clinician, it resulted in a higher overall mean (in mm3; mean (M) = 12,556, standard deviation (SD) = 9,196 as compared to M = 5,302, SD = 4,142). Decoding accuracies based on T1 and/or T2 lesion load (calculated with MedX) are shown in Table 2.
Pattern-based canonical correlation analysis
Local patterns informative about disease duration, motor disability, cognitive dysfunction, and overall clinical disability are shown in Fig. 3, separately for MPRAGE and TIRM images. Clusters reaching statistical significance after correction for multiple comparisons (p < 0.01, Bonferroni corrected) based on independent test data are listed in Tables 3 and 4. Please note that the decoding accuracy based on tissue intensity patterns was clearly higher than for T1 and T2 lesion load. The anatomical regions were identified using ‘automated anatomical labeling’ (AAL, [23]) and differed between MPRAGE and TIRM images, but included both white matter and task-related grey matter. We found patterns in the somatosensory cortex and posterior parietal cortex linked to disease duration. For motor disability, we identified regions in areas related to motor control such as cerebellum, thalamus, and primary motor cortex as well as in areas related to the planning and coordination of movements such as posterior parietal cortex and middle frontal gyrus. PASAT scores could be accurately predicted from areas known to be involved in working memory [24] such as posterior parietal cortex, but also from inferior temporal lobe, cingulum, and fusiform gyrus. When lowering the threshold to p < 0.05 (Bonferroni corrected), we additionally found clusters in the right prefrontal cortex (inferior frontal gyrus for MPRAGE images and middle frontal gyrus for TIRM images; Fig. 3). For EDSS, one cluster in the inferior frontal gyrus was maximally informative.
Results of pattern-based canonical correlation analysis. Brain regions encoding information about disease duration (pink), motor disability (blue), cognitive dysfunction (red), and overall clinical disability (green) are overlaid on a rendered T1-weighted anatomical template image. For illustrative purposes, searchlight center coordinates are shown at p < 10−5 (uncorrected) with a cluster threshold of five voxels. Peaks of correlation reaching statistical significance after correction for multiple comparisons (p < 0.01) are listed in Tables 3 and 4. AG angular gyrus, CER cerebellum, FFG fusiform gyrus, IFG inferior frontal gyrus, IPL inferior parietal lobe, M1 primary motor cortex, ITL inferior temporal lobe, IOL inferior occipital lobe, MFG middle frontal gyrus, MOL middle occipital lobe, MTL middle temporal lobe, PC paracentral, PCN precuneus, PH parahippocampus, S1 somatosensory cortex, SFG superior frontal gyrus SMA supplementary motor area, SMG supramarginal gyrus, SPL superior parietal lobe, TP temporal pole. All other areas correspond to white matter
A relatively low grey matter ratio (between 16.32 and 79.01 %) in areas classified as grey matter by AAL indicated that most areas were located in the conjunction of white and grey matter. The lesion ratio found was quite low (at most 14.69 %) in all significant clusters, but tended to be higher in white matter.
Maps obtained from MPRAGE and TIRM images indicating the correlation coefficient for each searchlight position were moderately correlated (r = 0.34 for disease duration, r = 0.22 for motor disability, r = 0.29 for cognitive dysfunction, and r = 0.31 for EDSS; Pearson). This means that correlation values between predicted and true clinical scores calculated based on either MPRAGE or TIRM images are to some extent related. The number of significant searchlights (p < 0.01, Bonferroni corrected) was higher for MPRAGE images (n = 36) compared to TIRM images (n = 29). In both modalities, more significant searchlights were found for motor disability and cognitive dysfunction than for disease duration and overall clinical disability.
Discussion
In the present study, we demonstrated that local tissue intensity patterns extracted from conventional MRI of MS patients together with a canonical correlation analysis encode clinically relevant information about symptom severity quantified in terms of disease duration, motor disability, cognitive dysfunction, and overall clinical disability in normal-appearing brain parenchyma. In particular, these patterns were more informative than the global lesion load. Remarkably, decoding accuracy based on local tissue intensity patterns was twice or three times higher than for T2 lesion load.
In recent years, numerous efforts have been made to link clinical disability scores in MS patients and MRI-derived markers. Generally, this association seems to be rather poor and led to the formulation of the so-called clinico-radiological paradox [3]. However, these early studies mostly tried to establish a dependency between T2 lesion load and clinical scores. For T1 lesion load, atrophy and measures based on non-conventional MRI, the results tend to be better [25, 26], but are still not applicable in clinical practice. We used a complex pattern-based decoding approach to establish a relationship between tissue characteristics from conventional MRI and clinical scores. The main advantage over common correlation analysis is that it is capable of detecting subtle tissue alterations in local brain areas that are not distinguishable by the naked eye or measurable by global markers such as lesion load or brain volume. Therefore, it allows for a sensitive mapping of clinically relevant regions. By using a cross-validated procedure, we additionally ensured the generalizability to new data sets.
Although we used a very conservative significance threshold, we found several regions that robustly encode symptom severity. For disease duration, we identified the precuneus and postcentral gyrus as areas with high predictive information. Interestingly, both structures have been linked to significant grey matter loss in MS and seem to be associated with fatigue [27] and sensory disturbances [28]. We hypothesize that this grey matter loss in conjunction with clinical symptoms accumulates over time and therefore correlates with disease duration. For motor disability, we mostly found regions involved in motor control (cerebellum, thalamus, and precentral gyrus) and planning of coordinated movements (posterior parietal cortex). These regions have been linked to various structural and functional abnormalities in MS patients [29, 30]. For cognitive dysfunction, we mainly found working memory areas including posterior parietal cortex and—when lowering the significance threshold—prefrontal cortex. Both areas have been shown to be functionally related to numerical information processing [24]. In MS patients, the PASAT score has been shown to correlate with atrophy [31], reduced diffusivity [32], and a higher functional activation [33] in these areas. Additionally, we have found patterns in the cingulum for which structural abnormalities have actually been reported to correlate with the PASAT score [34]. In line with several studies arguing that atrophy in frontal areas is a good indicator of the EDSS [35, 36], we found one area in the inferior frontal gyrus to accurately predict the EDSS score. However, most other studies did not find a connection between regional brain MRI abnormalities and the EDSS score, which has been explained by the low specificity of the EDSS [37] and a major involvement of the spinal cord in constituting the EDSS score [38].
Although the result maps based on MPRAGE and TIRM images were moderately correlated, peak regions encoding symptom severity differed between MPRAGE and TIRM images. This might be due to the fact that T1- and T2-weighted images measure different tissue and disease properties. For instance, it has been argued that T1 signal intensity is a marker of neuronal density, whereas T2 hypointensity rather correlates with the myelin content [39]. In line with several studies highlighting the importance of T1-relaxation time measurements for disease progression and clinical disability [40, 41], predictions based on MPRAGE images (both lesion load and intensity patterns) were superior to predictions based on TIRM images.
The question regarding the underlying tissue characteristics explaining the high predictability in certain areas is the most challenging one in the present study. This is due to the fact that T1- as well as T2-weighted imaging is relatively unspecific [42]. A hallmark of MS pathology is the development of focal inflammatory lesions. However, since the proportion of lesions was below 5 % in most brain areas listed in Tables 3 and 4, we argue that lesions visualized by TIRM images play a minor role in revealing symptom severity, though. Several studies [3], including our own data, confirmed that the T2 lesion load only partially accounts for the individual clinical deficits. In addition to focal lesions, histological studies [43] and advanced imaging techniques such as diffusion-weighted imaging [34, 44], magnetization transfer imaging [45], and proton spectroscopy [29] revealed that tissue damage in MS is more widespread than previously believed. This tissue damage usually remains undetected on conventional MRI and thus the referring parenchyma is termed normal-appearing brain tissue (NABT). We claim that our algorithm captures not only macroscopic features as presented by lesions, but also subtle signal alterations in the NABT that remain occult to the human eye. We showed that these diffuse abnormalities help to elucidate the extent of clinical disability in MS patients when suitable and more complex analysis techniques are employed. This becomes especially true for brain areas involved in most common symptoms of MS patients, such as visual disturbances and motor as well as sensory pathways, which were revealed in both T1- and T2-weighted sequences.
Several limitations should be pointed out. First, the exact histopathological processes in MS remain unclear, although several possible pathomechanisms are discussed that could account for an accurate prediction. Future studies should correlate prediction accuracy with histopathological findings and additionally assess whether our findings can be confirmed with other quantitative MRI techniques giving an additional insight into tissue integrity such as magnetization transfer imaging or diffusion tensor imaging. Second, our sample size is rather small and might cause effects that are not representative for all MS patients. Therefore, our results need to be confirmed in a larger patient cohort, preferentially including also other forms of MS (e.g., primary or secondary progressive MS) as well as other neurological diseases. Additionally, it would be interesting to assess whether our findings can be confirmed in patients with greater overall clinical disability and impairment in specific clinical domains. Third, the common clinical parameters were criticized to be rather unspecific, in particular the EDSS. Future studies might consider the sub-scores of the EDSS or additional markers of symptom severity. Fourth, the spatial normalization procedure, which is necessary to conduct a group-based analysis, may be influenced by cerebral atrophy. Although we aimed to minimize this error by regressing out the variance induced by the deformations, we cannot guarantee a totally bias-free normalization. Fifth, although we have found a strong association between local brain patterns and the clinical disability markers, the results have to be interpreted with caution. This is due to the fact that we cannot estimate the clinical contribution of spinal cord lesions, in particular to the scores reflecting motor disability, at the current stage of study. Finally, even though our proposed method is not yet applicable in clinical practice, we generally believe in the potential of pattern-recognition methods in complementing macrotexture information already used by neuroradiologists. With the emergence of large databases (such as the Alzheimer’s Disease Neuroimaging Initiative [ADNI] database, www.loni.ucla.edu/ADNI) and more powerful computers, the elaborate methods described in the present study might be transferable to large-scale clinical practices. However, future studies are necessary to evaluate the prognostic information of local intensity patterns in clinical trials and to make this information usable for individual patients.
In conclusion, we have shown that predictive information contained in local brain tissue intensity patterns of MS patients clearly outperforms information contained in conventional neuroradiological markers such as the lesion load when using suitable analysis techniques. Predicting areas were located in the conjunction of white and task-related grey matter and consisted mostly of NABT. We hypothesized that our proposed algorithm uses a mixture of slight intensity changes due to several pathomechanisms, including diffuse T2 hyperintensities with lesions as endpoint, T1 hypointensities, and atrophy. Our findings suggest that local intensity patterns might perform as clinically relevant biomarkers of clinical disability in MS and should be considered in future studies.
References
McDonald WI, Compston A, Edan G et al (2001) Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann Neurol 50:121–127
Bakshi R, Thompson AJ, Rocca MA et al (2008) MRI in multiple sclerosis: current status and future prospects. Lancet Neurol 7:615–625
Barkhof F (1999) MRI in multiple sclerosis: correlation with expanded disability status scale (EDSS). Mult Scler 5:283–286
Barkhof F (2002) The clinico-radiological paradox in multiple sclerosis revisited [Review]. Curr Opin Neurol 15:239–245
Filippi M, Grossman RI (2002) MRI techniques to monitor MS evolution: the present and the future. Neurology 58:1147–1153
Filippi M, Rocca MA (2005) MRI evidence for multiple sclerosis as a diffuse disease of the central nervous system. J Neurol 252(Suppl):16–24
Klöppel S, Chu C, Tan GC et al (2009) Automatic detection of preclinical neurodegeneration: presymptomatic Huntington disease. Neurology 72:426–431
Klöppel S, Stonnington CM, Chu C et al (2008) Automatic classification of MR scans in Alzheimer’s disease. Brain 131:681–689
Stonnington CM, Chu C, Klöppel S et al (2010) Predicting clinical scores from magnetic resonance scans in Alzheimer’s disease. Neuroimage 51:1405–1413
Wang Y, Fan Y, Bhatt P, Davatzikos C (2010) High-dimensional pattern regression using machine learning: from medical images to continuous clinical variables. Neuroimage 50:1519–1535
Cutter GR, Baier ML, Rudick RA et al (1999) Development of a multiple sclerosis functional composite as a clinical trial outcome measure. Brain 122:871–882
Kurtzke JF (1983) Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS). Neurology 33:1444–1452
Weygandt M, Hackmack K, Pfueller C et al (2011) MRI pattern recognition in multiple sclerosis normal-appearing brain areas. PLoS One 6:e21138
Paul F, Waiczies S, Wuerfel J et al (2008) Oral high-dose atorvastatin treatment in relapsing-remitting multiple sclerosis. PLoS One 3:e1928
Wuerfel J, Bellmann-Strobl J, Brunecker P et al (2004) Changes in cerebral perfusion precede plaque formation in multiple sclerosis: a longitudinal perfusion MRI study. Brain 127:111–119
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26:839–851
Weygandt M, Schaefer A, Schienle A, Haynes JD (2011) Diagnosing different binge-eating disorders based on reward-related brain activation patterns. Hum Brain Mapp. doi:10.1002/hbm.21345
Kriegeskorte N, Goebel R, Bandettini P (2006) Information-based functional brain mapping. Proc Natl Acad Sci USA 103:3863–3868
Haynes JD, Sakai K, Rees G et al (2007) Reading hidden intentions in the human brain. Curr Biol 17:323–328
Hardoon DR, Szedmak S, Shawe-Taylor J (2004) Canonical correlation analysis: an overview with application to learning methods. Neural Comput 16:2639–2664
Jolliffe IT (2002) Principal component analysis. Springer, Berlin
Kriegeskorte N, Simmons WK, Bellgowan PS, Baker CI (2009) Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci 12:535–540
Tzourio-Mazoyer N, Landeau B, Papthanassiou D et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289
Dehaene S, Molko N, Cohen L, Wilson AJ (2004) Arithmetic and the brain. Curr Opin Neurobiol 14:218–224
Chard D, Miller D (2009) Grey matter pathology in clinically early multiple sclerosis: evidence from magnetic resonance imaging. J Neurol Sci 282:5–11
Filippi M, Agosta F (2010) Imaging biomarkers in multiple sclerosis. [Review]. J Magn Reson Imaging 31:770–788
Filippi M, Rocca MA, Colombo B et al (2002) Functional magnetic resonance imaging correlates of fatigue in multiple sclerosis. Neuroimage 15:559–567
Prinster A, Quarantelli M, Orefice G et al (2006) Grey matter loss in relapsing-remitting multiple sclerosis: a voxel-based morphometry study. Neuroimage 29:859–867
Wylezinska M, Cifelli A, Jezzard P et al (2003) Thalamic neurodegeneration in relapsing-remitting multiple sclerosis. Neurology 60:1949–1954
Harirchian MH, Rezvanizadeh A, Fakhri M et al (2010) Non-invasive brain mapping of motor-related areas of four limbs in patients with clinically isolated syndrome compared to healthy normal controls. J Clin Neurosci 17:736–741
Morgen K, Sammer G, Courtney SM et al (2007) Distinct mechanisms of altered brain activation in patients with multiple sclerosis. Neuroimage 37:937–946
Bodini B, Khaleeli Z, Cercignani M et al (2009) Exploring the relationship between white matter and gray matter damage in early primary progressive multiple sclerosis: an in vivo study with TBSS and VBM. Hum Brain Mapp 30:2852–2861
Morgen K, Sammer G, Courtney SM et al (2006) Evidence for a direct association between cortical atrophy and cognitive impairment in relapsing-remitting MS. Neuroimage 30:891–898
Dineen RA, Vilisaar J, Hlinka J et al (2009) Disconnection as a mechanism for cognitive dysfunction in multiple sclerosis. Brain 132:239–249
Bakshi R, Benedict RH, Bermel RA, Jacobs L (2001) Regional brain atrophy is associated with physical disability in multiple sclerosis: semiquantitative magnetic resonance imaging and relationship to clinical findings. J Neuroimaging 11:129–136
Sailer M, Fischl B, Salat D et al (2003) Focal thinning of the cerebral cortex in multiple sclerosis. Brain 126:1734–1744
Rudick RA, Lee JC, Nakamura K, Fisher E (2009) Gray matter atrophy correlates with MS disability progression measured with MSFC but not EDSS. J Neurol Sci 282:106–111
Stevenson VL, Leary SM, Losseff NA et al (1998) Spinal cord atrophy and disability in MS: a longitudinal study. Neurology 51:234–238
Schmierer K, Parkes HG, So PW et al (2010) High field (9.4 Tesla) magnetic resonance imaging of cortical grey matter lesions in multiple sclerosis. Brain 133:858–867
Van Walderveen MA, Lycklama A, Nijeholt GJ et al (2001) Hypointense lesions on T1-weighted spin-echo magnetic resonance imaging: relation to clinical characteristics in subgroups of patients with multiple sclerosis. Arch Neurol 58:76–81
Vrenken H, Geurts JJ, Knol DL et al (2006) Whole-brain T1 mapping in multiple sclerosis: global changes of normal-appearing gray and white matter. Radiology 240:811–820
Neema M, Stankiewicz J, Arora A et al (2007) T1- and T2-based MRI measures of diffuse gray matter and white matter damage in patients with multiple sclerosis. J Neuroimaging 17(Suppl 1):16S–21S
Lucchinetti C, Brück W, Noseworthy J (2001) Multiple sclerosis: recent developments in neuropathology, pathogenesis, magnetic resonance imaging studies and treatment. Curr Opin Neurol 14:259–269
Roosendaal SD, Geurts JJ, Vrenken H et al (2009) Regional DTI differences in multiple sclerosis patients. Neuroimage 44:1397–1403
Ge Y, Grossman RI, Babb JS et al (2003) Dirty-appearing white matter in multiple sclerosis: volumetric MR imaging and magnetization transfer ratio histogram analysis. Am J Neuroradiol 24:1935–1940
Acknowledgments
This work was supported by the Max Planck Society, the Bernstein Computational Program of the German Federal Ministry of Education and Research [01GQ0411, 01GQ0851, and 01GQ1001C to J.-D.H., GRK 1,589/1 to K.H.] and the German Research Foundation [Exc 257 to C.P. and F.P., KFO 218/1 to M.W.].
Conflicts of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
K. Hackmack and M. Weygandt contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hackmack, K., Weygandt, M., Wuerfel, J. et al. Can we overcome the ‘clinico-radiological paradox’ in multiple sclerosis?. J Neurol 259, 2151–2160 (2012). https://doi.org/10.1007/s00415-012-6475-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-012-6475-9