Abstract
Motor disability is a dominant and restricting symptom in multiple sclerosis, yet its neuroimaging correlates are not fully understood. We apply statistical and machine learning techniques on multimodal neuroimaging data to discriminate between multiple sclerosis patients and healthy controls and to predict motor disability scores in the patients. We examine the data of sixty-four multiple sclerosis patients and sixty-five controls, who underwent the MRI examination and the evaluation of motor disability scales. The modalities used comprised regional fractional anisotropy, regional grey matter volumes, and functional connectivity. For analysis, we employ two approaches: high-dimensional support vector machines run on features selected by Fisher Score (aiming for maximal classification accuracy), and low-dimensional logistic regression on the principal components of data (aiming for increased interpretability). We apply analogous regression methods to predict symptom severity. While fractional anisotropy provides the classification accuracy of 96.1% and 89.9% with both approaches respectively, including other modalities did not bring further improvement. Concerning the prediction of motor impairment, the low-dimensional approach performed more reliably. The first grey matter volume component was significantly correlated (R = 0.28-0.46, p < 0.05) with most clinical scales. In summary, we identified the relationship between both white and grey matter changes and motor impairment in multiple sclerosis. Furthermore, we were able to achieve the highest classification accuracy based on quantitative MRI measures of tissue integrity between patients and controls yet reported, while also providing a low-dimensional classification approach with comparable results, paving the way to interpretable machine learning models of brain changes in multiple sclerosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis is an autoimmune disease of the central nervous system. With an increase in the age-standardized prevalence, the latest epidemiological studies report 2.2 million cases worldwide, ranking it as the most common demyelinating disease (Wallin et al., 2019; Leray et al., 2016).
Multiple sclerosis presents itself by a wide and heterogeneous spectrum of symptoms, ranging from cognitive and visual impairments to motor disabilities (Dobson and Giovannoni, 2019). Of these, motor impairment is the most dominant and restricting. Gait and postural control changes are present already in recently diagnosed minimally impaired patients and worsen over time (Martin et al., 2006; Burschka et al., 2012). Reduction in mobility together with fatigue increase risk of fall and injury, endangering the patient and reducing self-confidence (Giannì et al., 2014). More than 40% of people with multiple sclerosis report walking difficulties, which are identified as the most challenging disease aspect. However, the majority of people with multiple sclerosis do not voluntarily seek medical advice regarding walking impairments (Heesen et al., 2008; Asch, 2011; LaRocca, 2011).
Despite motor deficit being a major concern of people with multiple sclerosis, detailed motor function evaluation is rarely performed in practice. Consequently, the literature relating motor deficits to brain imaging in multiple sclerosis is limited, often focusing on single neuroimaging modality (Sbardella et al., 2015; Jakimovski et al., 2018; Steenwijk et al., 2014; Tona et al., 2014). Moreover, the studies generally feature a small sample size, low degree of impairment, and high number of statistical tests, weakening the findings reproducibility. These limitations call for methods that not only detect group differences in neuroimaging features, but also provide continuous quantitative prediction of the level of symptoms, and ultimately may prospectively characterize and predict health development in a clinical setting. Therefore, a methodological shift from pure hypothesis testing to predictive modeling (in statistical, or even forecasting sense) is needed, along with the increase of the sample sizes; machine learning is thus a natural candidate.
Machine learning has the potential to allow integration of multiparametric datasets with identification of feature sets that promise diagnostic or prognostic relevance for individual patients. Note that apart from promising improvement of the diagnostic and prognostic capabilities by providing accurate classification and prediction models, machine learning can also help getting insight into the disease character and mechanisms in case that the estimated models are reasonably interpretable. Thus, it has become more widespread in clinical research recently (Woo et al., 2017), but its implementation is not always straightforward. Particularly for limited sample sizes, care is needed to prevent model overfitting and sustain generalization to a wider population.
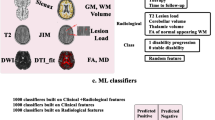
Progress has been made towards the application of machine learning methods in multiple sclerosis (Stafford et al., 2020). The most common task is the classification of subjects into patients and controls, sometimes also considering the degree of impairment and the phenotype of the disease (Bendfeldt et al., 2012; Kocevar et al., 2016; Zurita et al., 2018; Marzullo et al., 2019; Eitel et al., 2019; Tozlu et al., 2021). Most of the classification approaches use the support vector machines (SVM) (Vapnik and Chapelle, 2000), a broadly used algorithm in neuroscience for its ability to deal with the common high data dimensionality (especially in cases, when the number of features exceeds the number of instances), and for its computational effectiveness (learning time is considerably shorter than in neural networks). Although theoretically, no prior dimensionality reduction is needed, it is usually desirable. Previous studies commonly employed Fisher Score (FS) (He et al., 2006), which quantifies the power of variables to distinguish between the studied classes. However, the consequences of working only with a pre-selected set of features optimized for predictive power is often poorly discussed and, if not done carefully (i.e., blind to the test dataset), may lead to overfitting.
In this work, we build imaging-based models that provide efficient yet interpretable biomarkers for motor impairment in multiple sclerosis. We first identify imaging biomarkers across imaging modalities using machine learning for the simple task of distinguishing people with multiple sclerosis from controls. We subsequently construct predictive models for more subtle differences in motor performance among people with multiple sclerosis.
Materials and methods
The study design
Participants with multiple sclerosis were recruited from multiple sclerosis centers across the Czech Republic. The inclusion criteria were: positive diagnosis of multiple sclerosis (Polman et al., 2011); spastic paraparesis as a prominent clinical feature; stable clinical status for at least three months preceding the study (determined by neurologist); physical ability to undergo clinical testing – consistent with the Expanded Disability Status Scale (EDSS) score \(\le\) 7.5. Participants with disturbed mobility for reasons not related to multiple sclerosis (e.g. fractures, pregnancy, stroke) were excluded. All phenotypes of multiple sclerosis were accepted: RR – Relapsing-remitting; PP – Primary Progressive; and SP – Secondary Progressive. We analyzed 64 participants with multiple sclerosis and 65 healthy controls statistically matched for age and sex (Table 1). Participants were informed about the experimental setup and provided written informed consent in accordance with the Declaration of Helsinki. The study design was approved by the Ethics Committee of the Faculty Hospital Královské Vinohrady.
Clinical assessment
Participants with multiple sclerosis underwent a set of standardised assessments with the Berg Balance Scale (BBS) (Podsiadlo and Richardson, 1991; Berg et al., 2009); Timed Up and Go (TUG) test, The Multiple Sclerosis Impact Scale (MSIS); and Multiple Sclerosis Walking Scale (MSWS) (Hobart et al., 2001, 2003; Fischer et al., 1999).
MRI data acquisition
Imaging was performed with a 3T magnetic resonance scanner (Siemens Trio Tim, Erlangen, Germany) using a 12-channel phased-array head coil. The protocol consisted of T1-weighted and T2-weighted anatomical scans, DTI and resting-state fMRI. The parameters were: DWI: TR = 9,100 ms; TE = 96 ms; FOV = 260 x 211.25 mm; 64 contiguous axial slices; 2 mm thickness; b = 0 and 1100 s/mm; 64 gradient directions; voxel size = 2.03 x 2.03 x 2mm; Resting-State fMRI: BOLD single-shot echo-planar images TR = 2500 ms, TE = 30 ms, flip angle = 70\(^{\circ }\), 64 \(\times\) 64 matrix, FOV = 192 mm\(^{2}\), 44 contiguous axial slices, 3 mm thick, 240 volumes, acquisition time = 10 min. T1 volumetric imaging: TR = 2300 ms, TE = 4.63 ms, flip angle = 10\(^{\circ }\), matrix = 256 \(\times\) 256, FOV = 256 mm\(^{2}\), 156 contiguous sagittal slices, 1 mm thick.
Image processing
DTI was preprocessed using the FSL tools (FMRIB Software Library v5.0, http://www.fmrib.ox.ac.uk/fslwiki, FMRIB, Oxford, UK) and MRtrix3 v3.0_rc3 (Tournier et al., 2019). The data were denoised (dwidenoise) (Veraart et al., 2016b, a), Gibbs ringing artefacts were corrected (mrdegibbs) (Kellner et al., 2016). The volumes with low quality (visually checked) were discarded. Subsequently, eddy-current induced distortions and movement displacement was corrected by the eddy tool. Finally, we generated FA maps (Basser et al., 1994; Westin et al., 2002) and applied TBSS (Smith et al., 2006). We parcellated the resulting skeletonized images using the white matter ICBM-DTI-81 atlas (Mori et al., 2005) containing 48 regions and computed the mean FA for each region, which resulted in 48 FA features per subject.
The fMRI data were preprocessed using a combination of the SPM12 software package (Wellcome Department of Cognitive Neurology, London, UK); CONN toolbox (McGovern Institute for Brain Research, MIT, USA) running under MATLAB (The Mathworks) and FSL routines. Due to near-gaussianity of fMRI data (Hartman et al., 2011; Hlinka et al., 2011), we quantified FC for each pair of AAL atlas regions (Tzourio-Mazoyer et al., 2002) by the Pearson correlation coefficient (116 regions provide 6,670 FC features per subject).
Segmentation of the T1 images into white matter, gray matter, and cerebrospinal fluid was a part of the CONN pipeline employed for fMRI processing. We computed the GM region volumes using the AAL atlas (116 GMV features per subject). Additionally, the T1 scans were processed using Voxel-Based Morphometry in CAT12 (Keller and Roberts, 2008), using default settings involving bias-field and noise correction, segmentation into gray and white matter, and normalization to MNI space using DARTEL algorithm to a 1.5 mm isotropic adult template (Yassa et al., 2009). Total intracranial volume was estimated to be used as a covariate in statistical analyses.
Statistical analysis
Classification
For the classification task, we developed two strategies:
-
Fisher-score as a dimensionality reduction step, classification by support vector machines with linear kernel (FS-SVM)
-
Principal component analysis as a dimensionality reduction step, classification by logistic regression (PCA-LR)
The first approach uses the SVM classifier that can, in principle, deal with high-dimensional data, although it may be prone to overfitting, especially when it is combined with informed feature selection procedure such as the commonly used Fisher-score (He et al., 2006). In this approach, the features are selected into the classifier based on their individual classification power on the data in the training set. The box constraint (parameter C) for SVM was fixed to 1.
The latter approach uses a classical method of linear regression that can only deal with a small number of input features. To obtain these, we use PCA of the original features, a standard dimension reduction approach that avoids overfitting by being blind to the patient/control labels. Unlike in the Fisher score method, the transformation of the data is based on their explained variance in general and is not influenced by class labels.
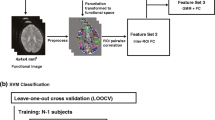
For each strategy, we constructed four classifiers: based on FA, FC, GMV, and their combination, using leave-one-out cross-validation (see Fig. 1). All neuroimaging features were transformed to z-scores prior to the dimensionality reduction step and model fitting.
A general scheme of the classification pipeline: In every iteration, one subject is left out. Either FS or PCA is computed on the training dataset, and the desired transformation (n terms of the number of components or features) is then applied. The classifier is trained using N-1 subjects and subsequently tested on the remaining subject. After N rounds, the classification accuracy is evaluated. Full lines on the image illustrate the data flow, whereas the dashed lines stand for data transformations
To further assess how the classification accuracy depends on the number of features, we evaluated each classifier for a range of thresholds. For the FS-SVM combination, we included 1%, 10%, 25%, 50%, 75%, and 100% of the features with the top Fisher Score. For the low-dimensional PCA-LR, we included one to ten PCA components. We consider sensitivity, specificity, and overall accuracy as measures classification quality and compare the classifiers using the McNemar test. All analyses were conducted using Matlab2018b, The MathWorks, Natick, 2018.
Prediction of the motor impairment
We applied an analogous approach for the prediction of motor impairment. In particular, we replaced SVM by support vector regression (SVR) and LR by linear regression (LinR). For dimensionality reduction, we sorted the features according to the Spearman correlation with the predicted scale instead of using FS. The use of PCA based on the variability explained remained unchanged. In each iteration, we performed a dimensionality reduction step on N-1 patients, constructed a model and predicted the clinical scale score for the remaining patient. Model quality was assessed by the correlation between the prediction and the original scale. Prior to the regression analysis, Box-Cox normalization was applied to the clinical scales. To avoid outlier and nongaussianity effects, Spearman correlation was employed for the final model fit assessment.
Results
Classification
All neuroimaging modalities allowed successful classification (albeit not necessarily at the same degree of complexity), with the best modality-specific accuracy ranging from 75 to 96 percent (Fig. 2, Table 3, Table 4).
Classification accuracy of Support Vector Machines classifiers (A) and Logistic Regression classifiers (B): overall accuracy (full line), sensitivity (dashed line with dots) and specificity (dashed line) for different thresholds. Classifiers based on fractional anisotropy (FA), functional connectivity (FC), and gray matter volume (GMV) correspond to orange, green and gray line respectively, the classifier based on the combination of features corresponds to the blue color
The Spearman correlation of the Support Vector Regression models prediction with the clinical scales: Models based on fractional anisotropy, functional connectivity, and gray matter volume correspond to orange, green and gray line respectively, models based on the combination of all features corresponds to the blue color. EDSS – Expanded Disability Status Scale; BBS – Berg Balance Scale; TUG – Timed Up and Go Test; MSIS – Multiple Sclerosis Impact Scale; MSWS – Twelve Item Multiple Sclerosis Walking Scale
The Spearman correlation of the Linear regression models prediction with the clinical scales: Models based on fractional anisotropy, functional connectivity, and gray matter volume correspond to orange, green and gray line respectively, models based on the combination of all features corresponds to the blue color. EDSS – Expanded Disability Status Scale; BBS – Berg Balance Scale; TUG – Timed Up and Go Test; MSIS – Multiple Sclerosis Impact Scale; MSWS – Twelve Item Multiple Sclerosis Walking Scale
The SVM classifier based on FA performed the best; the baseline classifier with a single FA feature reached 85% accuracy, the accuracy for the full feature set was 96%. The accuracy generally grew with feature count, although did not show significant performance differences beyond using 25% of the features (Fig. 2A, Fig. 5).
The SVM classifiers based on FC or GMV achieved about 70 percent accuracy and were not generally improving with feature count.
We also assessed the possibility to further improve the classification by combining the features across modalities. The baseline classifier based on the combination of features from all modalities (containing 69 features) performed better than the corresponding classifier based on FA (90% vs 85% respectively), however, with a rising amount of features, the accuracy declined to 81% (full dataset).
In the low-dimensional PCA-LR approach, models using only the first PCA component of FA or GMV reached an accuracy of 78% and 69% respectively (Fig. 2B). The first component of FC provided insignificant classification; nevertheless, the third alone reached an accuracy of 70%. The lack of classification power of the first two components in fMRI suggests that, unlike the previous two modalities, the main source of variance does not relate strongly to the patients-controls differences. Notably, the three components with substantial classification power (the first FA, the first GMV, and the third FC component) were significantly correlated (Spearman correlations: \(R(FA, GMV)=0.6\), \(R(FA, FC)=0.47\), \(R(GMV, FC)=0.46)\), therefore the combined model was not more effective.
Moreover, the model consisting of the first, second, and the third PCA component of FA reached an accuracy of 90%, not significantly worse than the full FA data SVM classifier (\(p=0.121\)). Similarly, the classifiers based on the GMV or FC did not perform significantly worse than their counterparts in FS-SVM models (except one and two-dimensional FC models). Nevertheless, their accuracy was inferior to that of FA.
Prediction of the motor impairment
In contrast to the classification for the prediction of the motor impairment, the results were not dominated by the FA. The best results in the SVR prediction were reached when using the FC (Fig. 3, 5). Especially for the MSWS scale, the prediction was consistently significant, reaching up to \(R=0.79\). The FA and GMV prediction ability varied with the number of features included and did not exceed \(R=0.50\) for any combination of feature type, count and clinical scale. The results of PCA in combination with linear regression complement those reached by SVR (Fig. 4, Table 6). While FC components were only successful in predicting the MSWS scale, when using the GMV features, the low-dimensional model significantly predicted almost all scales (EDSS, BBS, TUG, and MSWS).
Discussion
Classification
In our study, the model using only the regional average of FA in combination with SVM reached among the highest accuracy yet reported in the literature – 96% (sensitivity = 98%; specificity = 94%). The performance improved with features added. These results justify SVM use for classification based on similar imaging datasets. However, the accuracy of low-dimensional PCA-LR is also relatively high - a simple three-dimensional model using the first three FA components reached 90%.
Interestingly, the combined models using FA, FC and GMV were not superior to individual modality models. For SVM this may be caused by the almost perfect accuracy for FA diminishing space for improvement. The lack of accuracy improvement in the low-dimensional scenario is due to high correlation between the well-performing components of each modality - the three modalities thus did not provide synergistic performance effect.
We observed similar performance of the approaches, contrasting with the commonly observed dominance of SVM in classification tasks. One might speculate concerning potential data overfitting in practice due to the class-informed choice of features in combination with a small data size - something we avoided by careful design of the pipeline. From this perspective, we would advocate the use of PCA as a class-independent data transformation method that is not prone to this type of bias. Moreover, it reduces the model dimensionality, providing more easily interpretable results.
Zurita et al. (2018) also used the SVM approach in combination with the FS feature selection, reaching 88.9% classification accuracy between patients (EDSS higher than 1.5) and healthy controls, combining structural and functional connectivity. However, this classifier used almost 6,000 features; whereas our PCA-based classification reached 89.9% accuracy using only three. Contrary to our observation, their classification based on the FA was less accurate than using FC. Dominance of our FA-based model may stem from using robust regional averages and TBSS skeleton rather than noisy high-dimensional voxel-wise analysis. Our complementary analysis in which we applied SVM to the FC matrices generated using the Harvard-Oxford Atlas slightly outperformed the AAL atlas results, reaching 80% accuracy using 75% of features; still not matching the values reported (Zurita et al., 2018); additional investigation with liberal preprocessing only decreased the performance.
Another study (Kocevar et al., 2016) used structural connectivity matrices, to derive graph-theoretical indices. SVM classification using combined features reached 92% precision comparing 12 subjects with clinically isolated syndrome to 24 healthy controls. While a small sample was analyzed, the results line up with other literature exploring the classification potential of structural connectivity graph using other classification methods (Marzullo et al., 2019; Charalambous et al., 2019).
Neuroimaging correlates of motor disability
We did not identify any study that applied machine learning analysis to explore the relationship of specialized motor impairment scales and brain imaging, although models have been proposed combining structural, functional and cognitive impairment (Schoonheim et al., 2010, 2015). Tommasin et al. (2018) reported that general disability (measured through EDSS) had a direct linear relationship with lesion load, the inverse of thalamic volume, and functional connectivity in bi-frontal region pairs. With a similar approach and atlas for FC quantification, we did not observe correlation with EDSS (or other clinical measures), after appropriate multiple comparison correction. However, the SVR consistently confirmed significant predictive power of FC, indicating a presence of a more complex, multivariate relationship. Jakimovski et al. (2018) studied the correlation between walking disability (represented by 25 Foot Walk and the expanded TUG) and selected MRI-derived measures and DTI-measures maps; the DTI measures were not associated with the scales. Using regression analysis, the best predictor for the expanded TUG proved to be the cortex volume, \(\text {R}^{2}\) = 0.176, comparable with our results in TUG (\(\text {R}^{2}\) = 0.153 with the first GMV component). Steenwijk et al. (2014) reported partial correlations between different measures of the corticospinal tract and disability. Although corticospinal tract FA was correlated neither with EDSS, nor with MSWS, there was a significant correlation for the cortical thickness of the cortical area connected to it.
Our results indicate that the first GMV principal component is significantly correlated with numerous motor impairment scales. The same holds for the first FA component. For FC, the first and the second components played a role in regression, with the most noticeable results in the MSWS scale. Overall, the results of PCA-LinR are more reliable than the SVR results. The prediction quality did not vary significantly with dimensionality, whereas the SVR prediction using the FA and GMV features varied with the number of features.
Classification of phenotypes
The study included numerous phenotypes of multiple sclerosis. Acknowledging that there are structural differences between the subtypes (Schoonheim et al., 2015; Bozzali et al., 2002; Rovaris et al., 2002; Filippi and Rocca, 2011; Filippi et al., 2019), we further investigated the SVM’s ability to separate the relapsing-remitting and the secondary progressive group. The highest accuracy of the FS-SVM classifier was using all the GMV features – 75.9% (sensitivity 64.0%; specificity 81.6%). Considerable successes have been reported by groups which based the classifier on the graph representation of structural connectivity and combined the information with either SVM or graph convolutional neural networks (Kocevar et al., 2016; Marzullo et al., 2019), albeit the insufficient sample size calls for further research.
Limitations
The application of machine learning methods in neuroscience is generally limited by sample size. We analyzed 64 multiple sclerosis patients and 65 healthy controls, which is a considerable size in this area of research. Nevertheless, the group of multiple sclerosis patients was not homogeneous in terms of the phenotype, potentially decreasing the classification power and accuracy of the regression models. Importantly, we took transparent precautions not to overfit the models, in particular, feature selection was carried out in a manner blind to the labels in the testing set. On the other hand, the degree of patients’ impairment was on average higher than in other studies, which makes this project unique, but also warrants caution when interpreting the comparison with other works in the classification task.
However, beyond the numerous advantages, there are limitations associated with machine learning application. For SVM, the biggest concern may be associated with the dimensionality of the FC. The number of support vectors rose drastically with the number of features added to the model; nevertheless, the accuracy remained unchanged and inferior to the FA. The same applies to the PCA, where there were substantial differences between the variability explained by the first component among the three modalities. While the first component of GMV explained 71.7%, for FC, it was only 8.6%. Thus, while we can generally recommend using logistic regression combined with prior dimension reduction by PCA, selecting the first component is not necessarily optimal.
Finally, in this work, we focused on the comparison of two specific analytical pipelines, for which we presented the results. However, as is the case in most data-oriented work, it is possible that better performance could be achieved using different algorithms. For example, as suggested by on of the reviewers, methods with regularization might be applied, reducing the need for dimensionality reduction, although also making it more challenging to control for the number of features included. To explore this direction, we performed logistic regression with the LASSO regularization known for the sparsity of its solutions; the observed performance is comparable to that logistic regression run on the PCA variables (Table 7).
Conclusions
In the current study, we confirmed the widespread decrease of white matter integrity in patients with multiple sclerosis and demonstrated that localized white matter changes are related to motor symptoms. Using state-of-the-art machine learning methods, we showed that the white matter changes are specific and sensitive enough to provide 96% accuracy in recognizing patients from healthy controls. We further proposed an approach to construct low-dimensional classifiers with competitive performance, paving the way to robust and interpretable clinical tools.
We employed multimodal machine learning models, showing that the utilization of synergy between the modalities remains a challenge in the current settings, both due to potential redundancy between the features and problems with increasing the dimension of the feature set.
Finally, we fitted predictive models for the motor disability in multiple sclerosis, suggesting that while the disease itself is most apparent in decreased white matter integrity, the functional motor changes may indeed be more reflecting the accumulated cortical atrophy and changes in functional connectivity.
Data availability
The raw imaging data were measured in the Institute for Clinical and Experimental Medicine and are not publicly available due to restrictions imposed by the administering institution. Raw anonymized data can be shared based on a formal data sharing agreement based on a submission of a project outline, that may - depending on the nature of the data requested - require project amendment approval from the local ethics committee. Informal requests for information shall be sent to the corresponding author. Part of the preprocessed fMRI data is available for download (Bučková et al. 2021).
References
Asch, P. V. (2011). Impact of mobility impairment in multiple sclerosis 2 - patients’ perspectives. European Neurological Review, 6(2), 115–120.
Basser, P. J., Mattiello, J., & LeBihan, D. (1994). MR diffusion tensor spectroscopy and imaging. Biophysical Journal, 66(1), 259–267.
Bendfeldt, K., Klöppel, S., Nichols, T. E., Smieskova, R., Kuster, P., Traud, S., Mueller-Lenke, N., Naegelin, Y., Kappos, L., Radue, E.-W., & Borgwardt, S. J. (2012). Multivariate pattern classification of gray matter pathology in multiple sclerosis. NeuroImage, 60(1), 400–408.
Berg, K., Wood-Dauphine, S., Williams, J. I., & Gayton, D. (2009). Measuring balance in the elderly: preliminary development of an instrument. Physiotherapy Canada, 41(6), 304–311.
Bozzali, M., Cercignani, M., Sormani, M. P., Comi, G., & Filippi, M. (2002). Quantification of brain gray matter damage in different MS phenotypes by use of diffusion tensor MR imaging. American Journal of Neuroradiology, 23(6), 985–988.
Bučková, B., Kopal, J., Řasová, K., Tintěra, J., & Hlinka, J. (2021). Open access: The effect of neurorehabilitation on multiple sclerosis–unlocking the resting-state fMRI data. Frontiers in Neuroscience, 15, 615.
Burschka, J. M., Keune, P. M., Menge, U., Oy, U.H.-V., Oschmann, P., & Hoos, O. (2012). An exploration of impaired walking dynamics and fatigue in multiple sclerosis. BMC Neurology, 12(1), 161.
Charalambous, T., Tur, C., Prados, F., Kanber, B., Chard, D. T., Ourselin, S., Clayden, J. D., Wheeler-Kingshott, C. A. M. G., Thompson, A. J., & Toosy, A. T. (2019). Structural network disruption markers explain disability in multiple sclerosis. Journal of Neurology, Neurosurgery & Psychiatry, 90(2), 219–226.
Dobson, R., & Giovannoni, G. (2019). Multiple sclerosis-a review. European Journal of Neurology, 26(1), 27–40.
Filippi, M., Brück, W., Chard, D., Fazekas, F., Geurts, J. J. G., Enzinger, C., Hametner, S., Kuhlmann, T., Preziosa, P., Rovira, A., Schmierer, K., Stadelmann, C., & Rocca, M. A. (2019). Association between pathological and MRI findings in multiple sclerosis. The Lancet Neurology, 18(2), 198–210.
Filippi, M., & Rocca, M. A. (2011). MR imaging of multiple sclerosis. Radiology, 259(3), 659–681.
Fischer, J. S., Rudick, R. A., Cutter, G. R., & Reingold, S. C. (1999). The multiple sclerosis functional composite measure (MSFC): an integrated approach to MS clinical outcome assessment. Multiple Sclerosis Journal, 5(4), 244–250.
Giannì, C., Prosperini, L., Jonsdottir, J., & Cattaneo, D. (2014). A systematic review of factors associated with accidental falls in people with multiple sclerosis: a meta-analytic approach. Clinical Rehabilitation, 28(7), 704–716.
Heesen, C., Böhm, J., Reich, C., Kasper, J., Goebel, M., & Gold, S. (2008). Patient perception of bodily functions in multiple sclerosis: gait and visual function are the most valuable. Multiple Sclerosis Journal, 14(7), 988–991.
Hlinka, J., Paluš, M., Vejmelka, M., Mantini, D., & Corbetta, M. (2011). Functional connectivity in resting-state fMRI: Is linear correlation sufficient? NeuroImage, 54(3), 2218–2225.
Hobart, J., Lamping, D., Fitzpatrick, R., Riazi, A., & Thompson, A. (2001). The multiple sclerosis impact scale (MSIS-29)a new patient-based outcome measure. Brain, 124(5), 962–973.
Hobart, J., Riazi, A., Lamping, D., Fitzpatrick, R., & Thompson, A. (2003). Measuring the impact of MS on walking ability: The 12-item MS walking scale (MSWS-12). Neurology, 60(1), 31–36.
Jakimovski, D., Weinstock-Guttman, B., Hagemeier, J., Vaughn, C. B., Kavak, K. S., Gandhi, S., Bennett, S. E., Fuchs, T. A., Bergsland, N., Dwyer, M. G., Benedict, R. H. B., & Zivadinov, R. (2018). Walking disability measures in multiple sclerosis patients: Correlations with MRI-derived global and microstructural damage. Journal of the Neurological Sciences, 393, 128–134.
Keller, S. S., & Roberts, N. (2008). Voxel-based morphometry of temporal lobe epilepsy: An introduction and review of the literature. Epilepsia, 49(5), 741–757.
Kellner, E., Dhital, B., Kiselev, V. G., & Reisert, M. (2016). Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine, 76(5), 1574–1581.
LaRocca, N. G. (2011). Impact of walking impairment in multiple sclerosis. The Patient: Patient-Centered Outcomes Research, 4(3), 189–201.
Leray, E., Moreau, T., Fromont, A., & Edan, G. (2016). Epidemiology of multiple sclerosis. Revue Neurologique, 172(1), 3–13.
Martin, C. L., Phillips, B. A., Kilpatrick, T. J., Butzkueven, H., Tubridy, N., McDonald, E., & Galea, M. P. (2006). Gait and balance impairment in early multiple sclerosis in the absence of clinical disability. Multiple Sclerosis Journal, 12(5), 620–628.
Mori, S., Wakana, S., Zijl, P. C. M. V., & Nagae-Poetscher, L. M. (2005). MRI Atlas of Human White Matter. Elsevier.
Podsiadlo, D., & Richardson, S. (1991). The timed “up & go’’: A test of basic functional mobility for frail elderly persons. Journal of the American Geriatrics Society, 39(2), 142–148.
Polman, C. H., Reingold, S. C., Banwell, B., Clanet, M., Cohen, J. A., Filippi, M., Fujihara, K., Havrdova, E., Hutchinson, M., Kappos, L., Lublin, F. D., Montalban, X., O’Connor, P., Sandberg-Wollheim, M., Thompson, A. J., Waubant, E., Weinshenker, B., & Wolinsky, J. S. (2011). Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Annals of Neurology, 69(2), 292–302.
Rovaris, M., Bozzali, M., Iannucci, G., Ghezzi, A., Caputo, D., Montanari, E., Bertolotto, A., Bergamaschi, R., Capra, R., Mancardi, G. L., Martinelli, V., Comi, G., & Filippi, M. (2002). Assessment of normal-appearing white and gray matter in patients with primary progressive multiple sclerosis: A diffusion-tensor magnetic resonance imaging study. Archives of Neurology, 59(9), 1406–1412.
Sbardella, E., Tona, F., Petsas, N., Upadhyay, N., Piattella, M., Filippini, N., Prosperini, L., Pozzilli, C., & Pantano, P. (2015). Functional connectivity changes and their relationship with clinical disability and white matter integrity in patients with relapsing-remitting multiple sclerosis. Multiple Sclerosis Journal, 21(13), 1681–1692.
Smith, S. M., Jenkinson, M., Johansen-Berg, H., Rueckert, D., Nichols, T. E., Mackay, C. E., Watkins, K. E., Ciccarelli, O., Cader, M. Z., Matthews, P. M., & Behrens, T. E. J. (2006). Tract-based spatial statistics: Voxelwise analysis of multi-subject diffusion data. NeuroImage, 31(4), 1487–1505.
Stafford, I. S., Kellermann, M., Mossotto, E., Beattie, R. M., MacArthur, B. D., & Ennis, S. (2020). A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. Npj Digital Medicine, 3(1), 1–11.
Steenwijk, M. D., Daams, M., Pouwels, P. J. W., Balk, L. J., Tewarie, P. K., Killestein, J., Uitdehaag, B. M. J., Geurts, J. J. G., Barkhof, F., & Vrenken, H. (2014). What explains gray matter atrophy in long-standing multiple sclerosis? Radiology, 272(3), 832–842.
Tommasin, S., De Giglio, L., Ruggieri, S., Petsas, N., Giannì, C., Pozzilli, C., & Pantano, P. (2018). Relation between functional connectivity and disability in multiple sclerosis: a non-linear model. Journal of Neurology, 265(12), 2881–2892.
Tona, F., Petsas, N., Sbardella, E., Prosperini, L., Carmellini, M., Pozzilli, C., & Pantano, P. (2014). Multiple sclerosis: Altered thalamic resting-state functional connectivity and its effect on cognitive function. Radiology, 271(3), 814–821.
Tournier, J.-D., Smith, R., Raffelt, D., Tabbara, R., Dhollander, T., Pietsch, M., Christiaens, D., Jeurissen, B., Yeh, C.-H., & Connelly, A. (2019). Mrtrix3: A fast, flexible and open software framework for medical image processing and visualisation. Neuroimage, 202, 116137.
Tzourio-Mazoyer, N., Landeau, B., Papathanassiou, D., Crivello, F., Etard, O., Delcroix, N., Mazoyer, B., & Joliot, M. (2002). Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. NeuroImage, 15(1), 273–289.
Vapnik, V., & Chapelle, O. (2000). Bounds on error expectation for support vector machines. Neural Computation, 12(9), 2013–2036.
Veraart, J., Fieremans, E., & Novikov, D. S. (2016). Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Medicine, 76(5), 1582–1593.
Veraart, J., Novikov, D. S., Christiaens, D., Ades-aron, B., Sijbers, J., & Fieremans, E. (2016). Denoising of diffusion MRI using random matrix theory. NeuroImage, 142, 394–406.
Wallin, M. T., Culpepper, W. J., Nichols, E., Bhutta, Z. A., Gebrehiwot, T. T., Hay, S. I., Khalil, I. A., Krohn, K. J., Liang, X., Naghavi, M., Mokdad, A. H., Nixon, M. R., Reiner, R. C., Sartorius, B., Smith, M., Topor-Madry, R., Werdecker, A., Vos, T., Feigin, V. L., & Murray, C. J. L. (2019). Global, regional, and national burden of multiple sclerosis 1990–2016: a systematic analysis for the global burden of disease study 2016. The Lancet Neurology, 18(3), 269–285.
Woo, C.-W., Chang, L. J., Lindquist, M. A., & Wager, T. D. (2017). Building better biomarkers: brain models in translational neuroimaging. Nature Neuroscience, 20(3), 365–377.
Yassa, M. A., & Stark, C. E. L. (2009). A quantitative evaluation of cross-participant registration techniques for MRI studies of the medial temporal lobe. NeuroImage, 44(2), 319–327.
Eitel, F., Soehler, E., Bellmann-Strobl, J., Brandt, A. U., Ruprecht, K., Giess, R. M., Kuchling, J., Asseyer, S., Weygandt, M., Haynes, J.-D., et al. (2019). Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional mri using layer-wise relevance propagation. NeuroImage: Clinical, 24, 102003.
Hartman, D., Hlinka, J., Paluš, M., Mantini, D., & Corbetta, M. (2011). The role of nonlinearity in computing graph-theoretical properties of resting-state functional magnetic resonance imaging brain networks. Chaos: An Interdisciplinary Journal of Nonlinear Science, 21(1), 013119.
He, X., Cai, D., & Niyogi, P. (2006). Laplacian score for feature selection. In Y. Weiss, B. Schölkopf, J. C. Platt (Eds.), Advances in Neural Information Processing Systems (Vol. 18, pp. 507–514). MIT Press.
Kocevar, G., Stamile, C., Hannoun, S., Cotton, F., Vukusic, S., Durand-Dubief, F., & Sappey-Marinier, D. (2016). Graph theory-based brain connectivity for automatic classification of multiple sclerosis clinical courses. Frontiers in Neuroscience, 10(478), 1–11.
Marzullo, A., Kocevar, G., Stamile, C., Durand-Dubief, F., Terracina, G., Calimeri, F., & Sappey-Marinier, D. (2019). Classification of multiple sclerosis clinical profiles via graph convolutional neural networks. Frontiers in Neuroscience, 13, 1–15.
Schoonheim, M. M., Meijer, K. A., & Geurts, J. J. G. (2015). Network collapse and cognitive impairment in multiple sclerosis. Frontiers in Neurology, 6, 1–5.
Schoonheim, M. M., Geurts, J. J. G., Barkhof, F., & Radiology and nuclear medicine, Pathology, Anatomy and neurosciences, NCA - Multiple Sclerosis and Other Neuroinflammatory Diseases, and NCA - Neurodegeneration. (2010). The limits of functional reorganization in multiple sclerosis. Neurology, 74(16), 1246–1247.
Tozlu, C., Jamison, K., Gauthier, S. A., & Kuceyeski, A. (2021). Dynamic functional connectivity better predicts disability than structural and static functional connectivity in people with multiple sclerosis. Frontiers in neuroscience, 15, 1–12.
Westin, C. F., Maier, S. E., Mamata, H., Nabavi, A., Jolesz, F. A., & Kikinis, R. (2002). Processing and visualization for diffusion tensor MRI. Medical Image Analysis, 6(2), 93–108.
Zurita, M., Montalba, C., Labbé, T., Cruz, J. P., Dalboni da Rocha, J., Tejos, C., Ciampi, E., Cárcamo, C., Sitaram, R., & Uribe, S. (2018). Characterization of relapsing-remitting multiple sclerosis patients using support vector machine classifications of functional and diffusion MRI data. NeuroImage: Clinical, 20, 724–730.
Funding
This work was supported by Ministry of Health of the Czech Republic – DRO (Kralovske Vinohrady University Hospital – FNKV, IN: 00064173, National Institute of Mental Health – NIMH, IN: 00023752, and Institute for Clinical and Experimental Medicine – IKEM, IN: 00023001), Charles University (112616/GAUK/2016, 260533/SVV/2022, Q35, Q37, Q41, Cooperatio - Neuroscience); the Czech Technical University Internal Grant Agency (SGS19/169/OHK3/3T/13, SGS22/062/OHK3/1T/13), the Czech Science Foundation project No. 13-23940S, Czech Health Research Council Project No. NU21-08-00432, the Czech Academy of Sciences Strategy AV21 Research Programmes “Hopes and Risks of the Digital Age” and “Breakthrough Technologies for the Future – Sensing, Digitisation, Artificial Intelligence and Quantum Technologies”, and by the long-term strategic development financing of the Institute of Computer Science (RVO:67985807) of the Czech Academy of Sciences.
Author information
Authors and Affiliations
Contributions
BRB: Methodology, Formal analysis, Writing - Original Draft, Visualization JM: Data Curation AS: Software, Data Curation JK: Software, JT: Investigation RAD: Conceptualization, KR: Conceptualization, Project administration, Funding acquisition, JH: Conceptualization, Project administration, Funding acquisition, Methodology, Supervision. All authors participated on editing the manuscript and approved its final version.
Corresponding author
Ethics declarations
Ethics approval
The study design was approved by the Ethics Committee of the Faculty Hospital Královské Vinohrady.
Consent to participate
All participants were informed about the experimental setup and provided written informed consent in accordance with the Declaration of Helsinki.
Conflicts of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Tables 2, 3, 4, Figs. 5 and Tables 5, 6 and 7.
P-values of McNemar test between all thresholds and classifiers: Support Vector Machines (left), Logistic Regression (right). Threshold values in Support Vector Machines stand for the percentage of all features, in Logistic regression they represent the number of PCA component added to the model. FA – Fractional Anisotropy; FC – Functional Connectivity; GMV – Grey Matter Volume; all – combination of all three modalities
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rehák Bučková, B., Mareš, J., Škoch, A. et al. Multimodal-neuroimaging machine-learning analysis of motor disability in multiple sclerosis. Brain Imaging and Behavior 17, 18–34 (2023). https://doi.org/10.1007/s11682-022-00737-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-022-00737-3