Abstract
Sudden cardiac death (SCD) is the most frequent cause of sudden unexplained death in forensic practice. The most common cause of SCD is coronary artery disease related to coronary atherosclerosis. Previous study suggested the possible application of connexin 43 (Cx43) and zonula occludens-1 (ZO1) immunostaining in the early diagnosis of myocardial ischemia. However, there appears to be insufficient data with regard to their mRNA levels. The present study investigated the cardiac mRNA levels of Cx43 and ZO1, using forensic autopsy materials consisting of 41 control cases without any disease or structural abnormality of the heart (group 1), 32 deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis (group 2), and 29 traumatic deaths with coronary atherosclerosis (group 3). Ten candidate reference genes were evaluated in the left ventricles of 10 forensic autopsy cases. EEF1A1, PPIA, TPT1, and RPL13A were identified as the most stable reference genes. Using these validated reference genes, mRNA levels of Cx43 and ZO1 were examined in the bilateral ventricles and atria of the heart. Relative mRNA quantification demonstrated decreased calibrated normalized relative quantity (CNRQ) values of Cx43 and ZO1 in bilateral ventricles of group 2. When using one conventional reference gene (GAPDH or ACTB) for normalization, nearly no difference was detected among the three groups. These findings indicate that ventricular gap junction remodeling may be a key contributor to rhythm disturbances. Analysis of cardiac Cx43 and ZO1 using real-time PCR is useful in diagnosis of SCD, and validation of reference genes is crucial.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sudden cardiac death (SCD) is one of the most important causes of death in forensic practice which can be caused by various heart diseases. The most common cause of SCD is coronary artery disease related to coronary atherosclerosis [1, 2]. It is not difficult to detect typical myocardial lesions using conventional methods. However, it is still challenging to diagnose an acute myocardial infarction/ischemia with only little time of survival, since positive pathological findings cannot be observed in the early stage of infarction/ischemia [3]. Previous studies have suggested that postmortem biochemistry, immunochemistry, and molecular biology are useful for investigating SCD [3–8].
The most frequent cause of SCD is ventricular tachyarrhythmia, followed by bradyarrhythmia [9]. SCD cases without structural cardiac alterations seen in forensic practice are thought to be caused by arrhythmic syndromes. Postmortem genetic testing is recommended in such cases, though it has not yet been included into a routine work of the conventional autopsy [1, 10].
Connexin 43 (Cx43) is a gap junction (GJ) protein which is widely and highly expressed in mammalian tissues that mediate cell-to-cell coupling. In the mammalian heart, Cx43 is the primary connexin expressed in both the atrium and ventricle and is thought to be responsible for propagation of the action potential [11]. Alteration of Cx43 in quantity, phosphorylation, and distribution can cause myocardial electrically conductive disorder and finally result in arrhythmias [12].
Zonula occludens-1 (ZO1) is a membrane-associated guanylate kinase that can stabilize Cx43 at GJ plaque through cytoskeletal anchoring. Disruption of ZO1 leads to altered stabilization of the GJ plaque equilibrium between the two adjacent cells leading to plaque internalization [13].
In the field of forensic pathology, the possible application of Cx43 and ZO1 immunostaining in the early diagnosis of myocardial ischemia has been reported [5]. However, there appears to be insufficient postmortem human data with regard to their mRNA levels in the diagnosis of acute infarction/ischemia.
In the present study, we examined the mRNA levels of Cx43 and ZO1 in the myocardium and discussed the availability of these molecules for the markers of SCD.
Materials and methods
Sample collection
A total of 102 forensic autopsy cases were selected from autopsy documents. The cause of death was carefully diagnosed on the basis of autopsy examination, including macromorphological, histological, and toxicological analyses. Cases were divided into three groups as follows: 41 deaths due to traumatic brain injury without any disease or structural abnormality of the heart (group 1), 32 deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis (group 2), and 29 traumatic deaths with coronary atherosclerosis (group 3). A thorough pathological analysis was performed as a part of our routine work, and cases with any preexisting disease or structural abnormality were excluded. The demographics of study subjects are described in Table 1. The postmortem interval was defined as the time from the estimated time of death to autopsy, and survival time was the period from the onset of fatal insult to death; these were estimated based on autopsy findings and circumstantial evidence recorded in autopsy documents. This work was approved by our institutional ethics committee.
Among individuals in group B, 90.6 % (29/32) had triple-vessel disease and 9.4 % (3/32) had double-vessel disease. Significant narrowing of coronary lumen (>75 % luminal stenosis) was seen in all coronary arteries: left anterior descending coronary artery (71.9 %, 23/32), left circumflex coronary artery (34.4 %, 11/32), and right coronary artery (15.6 %, 5/32). Acute thrombosis was detected in three deaths (9.4 %). On the basis of postmortem investigations, the cause of death was attributed to CAD-induced cardiac arrhythmia in the absence of macroscopic and microscopic abnormalities other than coronary artery atherosclerosis as well as mild interstitial fibrosis. Death was witnessed in 30 cases (93.8 %). All deaths occurred instantaneously or within 6 h of collapse.
Among individuals in group C, 79.3 % (23/29) had triple-vessel disease and 20.7 % (6/29) had double-vessel disease. Significant narrowing of coronary lumen (>75 % luminal stenosis) was seen in all coronary arteries: left anterior descending coronary artery (62.1 %, 18/29), left circumflex coronary artery (27.6 %, 8/29), and right coronary artery (6.9 %, 2/29).
Extraction of total RNA and cDNA synthesis
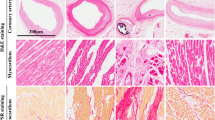
Tissue specimens were taken from consistent sites in the bilateral ventricles and atria of the heart at autopsy [14], then immediately submerged in 1 ml of RNA stabilization solution (RNAlaterTM, Ambion, Austin). Total RNA was isolated from 100 mg of sample using RNAiso Plus (Takara Bio, Inc., Shiga, Japan) according to the manufacturer’s instructions. After extraction, the RNA concentration was estimated by spectrophotometric analysis using a NanoDrop 1000 (Thermo Scientific, Wilmington, USA). The cDNA copies of total RNA were obtained using a High Capacity RNA-to-cDNA kit (Applied Biosystems Japan, Ltd.), then were adjusted to a concentration equivalent to 5 ng/μL of total RNA using nuclease-free water.
Evaluation of the quality and integrity of RNA samples
RNA purity was determined by using 260/280 absorbance (A260/A280) ratios. The RNA integrity number (RIN) was determined using a RNA 6000 Nano Labchip kit in an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, USA) following the manufacturer’s protocol [15, 16]. In the present study, myocardia from the left ventricles were used to determine the RIN.
Reference gene selection
Ten candidate reference genes were evaluated in the left ventricles of 10 forensic autopsy cases (5 from group 1 and 5 from group 2): tumor protein, translationally controlled 1 (TPT1), signal recognition particle 14 kDa (SRP14), eukaryotic translation elongation factor 1 alpha 1 (EEF1A1), peptidylprolyl isomerase A (PPIA), ribosomal protein L13a (RPL13A), tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ), TATA box binding protein (TBP), hydroxymethylbilane synthase (HMBS), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and beta-actin (ACTB). TPT1, SRP14, EEF1A1, PPIA, RPL13A, YWHAZ, TBP, and HMBS were chosen from the relevant literature and have been validated in human myocardium [17–19]. GAPDH and ACTB were conventional reference genes. Details are shown in Table 2.
Quantitative real-time reverse transcriptase polymerase chain reaction
The PCR primers and probes (TaqMan Gene Expression Assay) were purchased from Applied Biosystems, Inc. (Carlsbad, CA, USA). Details are shown in Table 2. Quantitative real-time reverse transcriptase polymerase chain reaction (RT-qPCR) reactions were run in 96-well reaction plates with an Applied Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). RT-qPCR was performed with 10 μL cDNA (corresponding to the cDNA reverse transcribed from approximately 50 ng RNA) in 20-μL reaction mix containing 10 μL TaqMan Gene Expression Master Mix (2×) and the abovementioned TaqMan Gene Expression Assays (lyophilized powder). Thermal cycling conditions included 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min, followed by 40 cycles of amplification at 95 °C for 15 s, and 60 °C for 1 min. The threshold cycle (Ct) was calculated by the instrument software automatically (threshold value at 0.2). Raw fluorescent data (normalized reporter values, Rn values) were also exported.
Amplification efficiency calculation
Amplification efficiencies were calculated from raw fluorescent data (Rn values), using a completely objective and noise-resistant algorithm, the Real-time PCR Miner program [20] (http://ewindup.info/miner/).
Data analysis
Normalization against validated reference genes
Raw Ct values and calculated amplification efficiencies of these 10 reference genes were imported into the qBaseplus software [21]. In the qBaseplus software, geNorm module was used to identify the most stable reference genes and determine the minimum number of reference genes [22].
After determining the minimum number of reference genes, raw Ct values and amplification efficiencies of targets and validated reference genes were imported into the qBaseplus software. Using a calibrator case (acute death due to blunt brain injury, 31-year-old male; 15-h postmortem), calibrated normalized relative quantity (CNRQ) values were exported from the qBaseplus software and statistically investigated.
Normalization against conventional reference gene (GAPDH or ACTB)
The expression levels of mRNA transcripts are described as the ratios of the targets normalized to the endogenous reference (GAPDH or ACTB), using the 2−△△Ct method, as the ratios for fold change relative to the abovementioned calibrator. The 2−△△Ct method assumes that the amplification efficiency of the reaction is ideal (100 %) and constant for each sample.
Statistic
All the RT-qPCR experiments were performed in triplicate, and results are reported as the mean ± SD. Correlation analyses between pairs of parameters were performed using linear regression (Pearson correlation analysis). One-way analysis of variance (ANOVA) with repeated measures, followed by post hoc Tukey tests, was used for comparisons of multiple groups. Statistical analyses were performed using GraphPad Prism version 5.0 (GraphPad Software, San Diego, USA). Values of p < 0.05 were considered as statistically significant.
Results
RNA concentration, purity and integrity
RNA concentrations ranged from 14.6 to 325.8 ng/μL (mean 190.2 ng/μL). There were no age or postmortem interval dependences on the Pearson correlation analysis (p > 0.05). RNA concentrations showed no significant differences among the three groups.
RNA purity, determined using 260/280 absorbance (A260/A280) ratios, ranged from 1.73 to 2.15 (mean 1.95). There were no age or postmortem interval dependences on the Pearson correlation analysis (p > 0.05). RNA purity showed no significant differences among the three groups.
RIN values showed substantial variations in each group. There were no age or postmortem interval dependences on the Pearson correlation (p > 0.05). RIN values were evidently lower in group 2 than others (Table 1).
Amplification efficiency
The amplification efficiencies of targets and reference genes ranged from 89.9 % (ACTB) to 108.2 % (TBP), showing small inter-individual variations (standard deviation, SD < 5 %) except for ZO1 (SD = 5.2 %). Details are shown in Table 2.
Reference genes validation
The geNorm module in the qBaseplus software ranked the 10 reference genes. The most stable one was EEF1A1, followed by PPIA, TPT1, and RPL13A. The least stable one was GAPDH (Fig. 1). Pairwise variation (V) was calculated based on normalization factor values (NFn and NFn + 1) after the inclusion of the least stable reference gene and indicated if the extra reference gene added to the stability of the normalization factor. The V-value was the lowest when the sixth most stable gene (SRP14) was added (Fig. 2). However, when the fourth most stable gene (RPL13A) was added, V3/4 showed a V-value of 0.145, below the threshold of 0.15. Therefore, four reference genes EEF1A1, PPIA, TPT1, and RPL13A, were selected for normalization.
Data analysis
Normalization against validated reference genes
Raw Ct values and amplification efficiencies of targets and four validated reference genes, EEF1A1, PPIA, TPT1, and RPL13A, were imported into the qBaseplus software. CNRQ values were exported and statistically investigated.
There were no gender-related differences or age dependence in CNRQ values of target genes on the Pearson correlation (p > 0.05).
CNRQ values of Cx43 and ZO1 were significantly lower in the bilateral ventricles, but not in the atria of the heart in group 2 (Figs. 3 and 4).
Cx43 mRNA expression levels after normalization against four validated reference genes. Bilateral ventricular CNRQ values of Cx43 were significantly lower in group 2 than groups 1 and 3. No difference was detected among the three groups in the bilateral atria. CNRQ calibrated normalized relative quantity, LV left ventricle, RV right ventricle, Group 1 control cases without any disease or structural abnormality of the heart, Group 2 deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis, Group 3 traumatic deaths with coronary atherosclerosis
ZO1 mRNA expression levels after normalization against four validated reference genes. Bilateral ventricular CNRQ values of ZO1 were significantly lower in group 2 than groups 1 and 3. No difference was detected among the three groups in the bilateral atria. CNRQ calibrated normalized relative quantity, LV left ventricle, RV right ventricle, Group 1 control cases without any disease or structural abnormality of the heart, Group 2 deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis, Group 3 traumatic deaths with coronary atherosclerosis
Normalization against GAPDH or ACTB
When ACTB or GAPDH alone was used for normalization, no difference was detected among the three groups, except for an increased Cx43 mRNA level in the left atrial wall of group 1 when normalized against ACTB (Supplementary Materials).
Discussion
Data normalization is extremely important in relative RT-qPCR. Ideally, reference genes should show stable and ubiquitous expression patterns which cannot be affected by any internal or external stimulus [23]. However, expressions of several conventional reference genes (GAPDH and ACTB) were shown to vary due to several factors, including some biological processes, and tissue or cell types. One conventional endogenous reference gene may not meet the criteria of an ideal reference gene [24].
In the field of forensic pathology, relative RT-qPCR has been used to investigate the systemic pathophysiological changes involved in the death process which cannot be observed directly by morphology in postmortem human materials [15, 25–27]. Previous studies, using different normalization strategies, have demonstrated contradicting results, suggesting the crucial role of selecting reference genes [15, 23]. However, postmortem studies on quantitative data normalized against only a single reference gene transcript are still regularly submitted and published [28, 29].
In the present study, utilizing geNorm module in the qBaseplus software, 10 candidate reference genes were evaluated. The V-value was the lowest when the sixth most stable gene (SRP14) was added. Further addition of genes increased V-values, indicating a negative influence on the normalization process. Since a threshold V-value of 0.15 is recommended as a cutoff value by geNorm to determine the optimal number of reference genes [22], only four reference genes (EEF1A1, PPIA, TPT1, and RPL13A, V3/4 = 0.145) were accurate enough in the present study. Therefore, EEF1A1, PPIA, TPT1, and RPL13A were used as reference genes to save on cost and time. As expected, inconsistent results were observed by different normalization methods. When those four validated reference genes, EEF1A1, PPIA, TPT1, and RPL13A, were used for normalization, decreased CNRQ values of Cx43 and ZO1 were detected in bilateral ventricles of deaths due to acute ischemic heart disease related to coronary atherosclerosis (group 2). When using only one conventional reference gene (GAPDH or ACTB) for normalization, no difference was detected among the three groups, except for an increased Cx43 mRNA level in the left atrial wall of group 1 when normalized against ACTB. Expression stability values of these six reference genes calculated by geNorm showed GAPDH as the least stable one, followed by ACTB. Therefore, gene expression levels that normalized against the four validated reference genes were believed to be accurate and reliable; GAPDH and ACTB, two conventional reference genes, were not suitable for normalization of human postmortem brain tissues.
Another significant factor influencing the accuracy of gene expression analysis is the integrity of RNA [30]. Unfortunately, unlike animal experimentation, RNA degradation is inevitable for postmortem human tissues. In the present study, RIN values were statistically lower in the myocardia of the left ventricles of group 2, indicating that RNA quality was more seriously affected in the cases of acute myocardial infarction/ischemia.
Ischemic and hypertrophic heart diseases are associated with ventricular arrhythmias, which may lead to death. Abnormal cellular coupling is implicated as having a crucial role in this fatal process. Myocardial GJs are the subcellular structure responsible for low-resistance electrical coupling between cardiac muscle cells. The GJ protein Cx43 is the most ubiquitous connexin and is thought to be responsible for propagation of the action potential [31]. Focal disorganization of gap junctional distribution and down-regulation of the major gap junctional protein Cx43 are typical features of myocardial remodeling in the failing heart. Increasing evidence indicates that Cx43 can interact with ZO1, and ZO1 can promote the formation and growth of GJ plaques. The loss of ZO1 staining in the failing hearts coincided with a reduction of connexin 43 at the same intercalated disks [32, 33]. Protein and/or mRNA down-regulation of Cx43 and ZO1 has been reported in several heart failure models as well as in the failing human heart [33–35]. Using experimental models, some studies suggested that ischemic preconditioned hearts retain higher Cx43 levels compared to non-preconditioned hearts [36, 37]. However, when compared to control group (normal hearts), lower Cx43 levels were detected in the ischemic preconditioned group. Considering from these observations, the down-regulations of Cx43 and ZO1 mRNA levels may be used as markers of fatal ventricular arrhythmia.
In the present study, though hearts in both groups 2 and 3 were heavier than those in group 1, CNRQ values of Cx43 and ZO1 (mRNA levels) were lower only in the bilateral ventricles of group 2, indicating the ventricular arrhythmias may be essentially involved in the death process of SCD. Further investigation using animal experiments is needed to confirm our hypothesis.
In fact, in contrast to undisturbed zones, significantly disturbed Cx43 can be observed in ischemia-exposed myocardium as well as epicardial border zone [38, 39]. However, the tissue specimens were taken from consistent sites in the bilateral ventricles and atria of the heart at autopsy. After a thorough pathological analysis, cases without apparent myocardial necrosis were selected. Therefore, the affected and non-affected myocardium specimens taken from the same case have not been examined, which is the major limitation of this study. Further investigation is needed.
Collectively, the present study indicates that ventricular gap junction remodeling may be a key contributor to rhythm disturbances. Analysis of cardiac Cx43 and ZO1 using real-time PCR maybe helpful in diagnosis of SCD related to coronary atherosclerosis, and validation of reference genes is crucial.
References
Campuzano O, Allegue C, Partemi S, Iglesias A, Oliva A, Brugada R (2014) Negative autopsy and sudden cardiac death. Int J Legal Med 128(4):599–606
Chappex N, Schlaepfer J, Fellmann F, Bhuiyan ZA, Wilhelm M, Michaud K (2015) Sudden cardiac death among general population and sport related population in forensic experience. J Forensic Legal Med 35:62–68
Zhu BL, Ishikawa T, Michiue T, Li DR, Zhao D, Kamikodai Y, Tsuda K, Okazaki S, Maeda H (2006) Postmortem cardiac troponin T levels in the blood and pericardial fluid. Part 2: analysis for application in the diagnosis of sudden cardiac death with regard to pathology. Legal Med 8(2):94–101
Zhu BL, Ishikawa T, Michiue T, Li DR, Zhao D, Bessho Y, Kamikodai Y, Tsuda K, Okazaki S, Maeda H (2007) Postmortem cardiac troponin I and creatine kinase MB levels in the blood and pericardial fluid as markers of myocardial damage in medicolegal autopsy. Legal Med 9(5):241–250
Kawamoto O, Michiue T, Ishikawa T, Maeda H (2014) Immunohistochemistry of connexin43 and zonula occludens-1 in the myocardium as markers of early ischemia in autopsy material. Histol Histopathol 29(6):767–775
Mayer F, Propper S, Ritz-Timme S (2014) Dityrosine, a protein product of oxidative stress, as a possible marker of acute myocardial infarctions. Int J Legal Med 128(5):787–794
Ortmann C, Pfeiffer H, Brinkmann B (2000) A comparative study on the immunohistochemical detection of early myocardial damage. Int J Legal Med 113(4):215–220
Chen JH, Michiue T, Ishikawa T, Maeda H (2012) Pathophysiology of sudden cardiac death as demonstrated by molecular pathology of natriuretic peptides in the myocardium. Forensic Sci Int 223(1–3):342–348
Bayes de Luna A, Coumel P, Leclercq JF (1989) Ambulatory sudden cardiac death: mechanisms of production of fatal arrhythmia on the basis of data from 157 cases. Am Heart J 117(1):151–159
Cittadini F, De Giovanni N, Alcalde M, Partemi S, Carbone A, Campuzano O, Brugada R, Oliva A (2015) Genetic and toxicologic investigation of sudden cardiac death in a patient with arrhythmogenic right ventricular cardiomyopathy (ARVC) under cocaine and alcohol effects. Int J Legal Med 129(1):89–96
Kar R, Batra N, Riquelme MA, Jiang JX (2012) Biological role of connexin intercellular channels and hemichannels. Arch Biochem Biophys 524(1):2–15
Michela P, Velia V, Aldo P, Ada P (2015) Role of connexin 43 in cardiovascular diseases. Eur J Pharmacol 768:71–76
Gilleron J, Carette D, Fiorini C, Benkdane M, Segretain D, Pointis G (2009) Connexin 43 gap junction plaque endocytosis implies molecular remodelling of ZO-1 and c-Src partners. Commun Integr Biol 2(2):104–106
Chen JH, Michiue T, Ishikawa T, Maeda H (2012) Molecular pathology of natriuretic peptides in the myocardium with special regard to fatal intoxication, hypothermia, and hyperthermia. Int J Legal Med 126(5):747–756
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H (2013) Molecular pathology of brain edema after severe burns in forensic autopsy cases with special regard to the importance of reference gene selection. Int J Legal Med 127(5):881–889
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H (2012) Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med 126(6):943–952
Caselli C, D’Amico A, Caruso R, Cabiati M, Prescimone T, Cozzi L, Cannata A, Parodi O, Del Ry S, Giannessi D (2013) Impact of normalization strategy on cardiac expression of pro-inflammatory cytokines: evaluation of reference genes in different human myocardial regions after left ventricular assist device support. Cytokine 63(2):113–122
Pilbrow AP, Ellmers LJ, Black MA, Moravec CS, Sweet WE, Troughton RW, Richards AM, Frampton CM, Cameron VA (2008) Genomic selection of reference genes for real-time PCR in human myocardium. BMC Med Genet 1:64
Koppelkamm A, Vennemann B, Fracasso T, Lutz-Bonengel S, Schmidt U, Heinrich M (2010) Validation of adequate endogenous reference genes for the normalisation of qPCR gene expression data in human post mortem tissue. Int J Legal Med 124(5):371–380
Zhao S, Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol: J Comput Mol Cell Biol 12(8):1047–1064
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8(2):R19
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (7):research0034.1–research0034.11. doi:10.1186/gb-2002-3-7-research0034
Huth A, Vennemann B, Fracasso T, Lutz-Bonengel S, Vennemann M (2013) Apparent versus true gene expression changes of three hypoxia-related genes in autopsy derived tissue and the importance of normalisation. Int J Legal Med 127(2):335–344
de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A (2007) Evidence based selection of housekeeping genes. PLoS One 2(9):e898
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H (2013) Molecular pathology of pulmonary edema in forensic autopsy cases with special regard to fatal hyperthermia and hypothermia. Forensic Sci Int 228(1–3):137–141
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H (2012) Molecular pathology of pulmonary edema after injury in forensic autopsy cases. Int J Legal Med 126(6):875–882
Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H (2014) Molecular pathology of brain matrix metalloproteases, claudin5, and aquaporins in forensic autopsy cases with special regard to methamphetamine intoxication. Int J Legal Med 128(3):469–474
Patel N, Crider A, Pandya CD, Ahmed AO, Pillai A (2015) Altered mRNA levels of glucocorticoid receptor, mineralocorticoid receptor, and co-chaperones (FKBP5 and PTGES3) in the middle frontal gyrus of autism spectrum disorder subjects. Mol Neurobiol. doi:10.1007/s12035-015-9178-2
Narayanan KL, Chopra V, Rosas HD, Malarick K, Hersch S (2015) Rho kinase pathway alterations in the brain and leukocytes in Huntington’s disease. Mol Neurobiol. doi:10.1007/s12035-015-9147-9
Fleige S, Pfaffl MW (2006) RNA integrity and the effect on the real-time qRT-PCR performance. Mol Asp Med 27(2–3):126–139
Rhett JM, Jourdan J, Gourdie RG (2011) Connexin 43 connexon to gap junction transition is regulated by zonula occludens-1. Mol Biol Cell 22(9):1516–1528
Hunter AW, Barker RJ, Zhu C, Gourdie RG (2005) Zonula occludens-1 alters connexin43 gap junction size and organization by influencing channel accretion. Mol Biol Cell 16(12):5686–5698
Laing JG, Saffitz JE, Steinberg TH, Yamada KA (2007) Diminished zonula occludens-1 expression in the failing human heart. Cardiovasc Pathol: Off J Soc Cardiovasc Pathol 16(3):159–164
Ai X, Pogwizd SM (2005) Connexin 43 downregulation and dephosphorylation in nonischemic heart failure is associated with enhanced colocalized protein phosphatase type 2A. Circ Res 96(1):54–63
Dupont E, Matsushita T, Kaba RA, Vozzi C, Coppen SR, Khan N, Kaprielian R, Yacoub MH, Severs NJ (2001) Altered connexin expression in human congestive heart failure. J Mol Cell Cardiol 33(2):359–371
Brandenburger T, Huhn R, Galas A, Pannen BH, Keitel V, Barthel F, Bauer I, Heinen A (2014) Remote ischemic preconditioning preserves connexin 43 phosphorylation in the rat heart in vivo. J Transl Med 12:228
Chen Z, Luo H, Zhuang M, Cai L, Su C, Lei Y, Zou J (2011) Effects of ischemic preconditioning on ischemia/reperfusion-induced arrhythmias by upregulatation of connexin 43 expression. J Cardiothorac Surg 6:80
Peters NS, Coromilas J, Severs NJ, Wit AL (1997) Disturbed connexin43 gap junction distribution correlates with the location of reentrant circuits in the epicardial border zone of healing canine infarcts that cause ventricular tachycardia. Circulation 95(4):988–996
Vetterlein F, Muhlfeld C, Cetegen C, Volkmann R, Schrader C, Hellige G (2006) Redistribution of connexin43 in regional acute ischemic myocardium: influence of ischemic preconditioning. Am J Physiol Heart Circ Physiol 291(2):H813–819
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 81401556), Guangdong Natural Science Foundation (No. 2014A030310504 and 2014A030310293), and the Special Foundation of President of School of Public Health and Tropical Medicine of Southern Medical University (No. GW201402).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
This work was approved by our institutional ethics committee.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Ye Xue, Rui Zhao and Si-Hao Du contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Cx43 mRNA expression levels after normalization against GAPDH. No difference was detected among three groups. CNRQ, calibrated normalized relative quantity; LV, left ventricle; RV, right ventricle; Group 1, control cases without any disease or structural abnormality of the heart; Group 2, deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis; Group 3, traumatic deaths with coronary atherosclerosis. (PPT 534 kb)
Supplementary Material 2
ZO1 mRNA expression levels after normalization against GAPDH. No difference was detected among three groups. CNRQ, calibrated normalized relative quantity; LV, left ventricle; RV, right ventricle; Group 1, control cases without any disease or structural abnormality of the heart; Group 2, deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis; Group 3, traumatic deaths with coronary atherosclerosis. (PPT 519 kb)
Supplementary Material 3
Cx43 mRNA expression levels after normalization against ACTB. No difference was detected among three groups, except for an increased Cx43 mRNA level in the left atrial wall of Group 1 than in Group 2. CNRQ, calibrated normalized relative quantity; LV, left ventricle; RV, right ventricle; Group 1, control cases without any disease or structural abnormality of the heart; Group 2, deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis; Group 3, traumatic deaths with coronary atherosclerosis. (PPT 492 kb)
Supplementary Material 4
ZO1 mRNA expression levels after normalization against ACTB. No difference was detected among three groups. CNRQ, calibrated normalized relative quantity; LV, left ventricle; RV, right ventricle; Group 1, control cases without any disease or structural abnormality of the heart; Group 2, deaths due to acute ischemic heart disease related to coronary atherosclerosis without apparent myocardial necrosis; Group 3, traumatic deaths with coronary atherosclerosis. (PPT 551 kb)
Rights and permissions
About this article
Cite this article
Xue, Y., Zhao, R., Du, SH. et al. Decreased mRNA levels of cardiac Cx43 and ZO1 in sudden cardiac death related to coronary atherosclerosis: a pilot study. Int J Legal Med 130, 915–922 (2016). https://doi.org/10.1007/s00414-016-1353-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-016-1353-0