Abstract
In higher eukaryotes, the condensin complex is a multisubunit apparatus that plays a pivotal role in the coordinated condensation of chromatin during mitosis. The catalytic subunits, CAP-E and CAP-C, members of the SMC family of ATPases, form a heterodimer, the activity of which is controlled by the non-SMC subunits CAP-D2, CAP-G and CAP-H. Here, we report the characterization of a T-DNA insertion mutant of the Arabidopsis CAP-C gene. Analysis of the progeny of selfed heterozygotes revealed that the homozygous null genotype is embryo lethal, with arrest occurring at or before the globular stage of development. Patterning defects associated with altered planes of cytokinesis were found in both the embryo and the suspensor. Crosses of heterozygotes with wild type plants revealed both male and female gametophytic defects. Stretched chromatin was observed between segregating mitotic chromosomes in pollen produced by selfed heterozygotes. Additionally, some plants heterozygous for the T-DNA insertion exhibited loss of apical dominance and mild fasciation, indicating a semi-dominant effect of the mutation. These results reveal a critical role for AtCAP-C during cell division and, unlike our previous studies on the AtCAP-E genes, suggest that no redundant factors for AtCAP-C exist in the Arabidopsis genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Proper development of multicellular organisms depends on coordination between two complex processes, the regulation of the cell cycle and the commitment of cells to differentiation. In the last decade, cell cycle studies have revealed a multitude of different regulatory molecules which influence cell cycle progression by orchestrating both nuclear and cytoplasmic processes (Dewitte and Murray 2003; Wignall et al. 2003). A successful cell cycle requires that the genetic material be faithfully duplicated and accurately segregated to opposite spindle poles, and during these processes chromatin undergoes dramatic structural changes. Interphase chromatin is relatively decondensed, facilitating events of DNA metabolism, but spatial and topological constraints require significant compaction of chromatin for mitosis. Compaction is accomplished in part by the condensin complex, a multisubunit enzyme that acts together with topoisomerase II to condense chromatids and disentangle them during prophase of mitosis. The catalytic subunits of the complex are members of the evolutionarily conserved structural maintenance of chromosomes (SMC) family whose members participate in a variety of processes including condensation, sister chromatid cohesion, repair/recombination and dosage compensation (see Losada and Hirano 2005 for a recent review). The SMC2/4 members (also referred to as CAP-E and CAP-C, respectively) form a heterodimer that appears to be designed as a flexible clamp, the opening and closing of which may be associated with ATP dependent supercoiling of DNA and thus compaction (Hirano et al. 2001; Bazett-Jones et al. 2002). The heterodimer is guided by three non-SMC proteins, CAP-D2, CAP-G and CAP-H, one or more of which may be phosphorylated by a cyclin dependent kinase to synchronize their activity with other cell cycle controls (Kimura et al. 1998; Sutani et al. 1999).
In plants, several SMC genes have been cloned from Arabidopsis. Mengiste et al. (1999) reported the characterization of a RAD18-like SMC protein that plays a role in recombinational DNA repair. Three different SMC genes are represented by titan mutants, which derive their name from the dramatic enlargement of endosperm nuclei (Tzafrir et al. 2002). ttn 3, encoding an SMC2 protein, exhibits aberrant endosperm nuclei and giant mitotic figures, but there are no significant defects in the embryo proper nor any post-embryonic defects. Two other titan mutants (ttn7 and ttn8, encoding SMC3 and SMC1, respectively) differ from ttn3 in that in addition to endosperm defects, embryo development is compromised, and embryos arrest very early in development (Liu et al. 2002). The lethality of these mutants parallels that found for orthologous gene mutations in other organisms. The viability of ttn3 (SMC2/AtCAP-E1) is due to functional redundancy conferred by a second, very similar gene, AtCAP-E2. Mutant analysis of these AtCAP-E genes revealed both male and female gametophytic defects, and in the double null mutant embryo arrest occurs at or before the globular stage and is characterized by defects in the organization of both the embryo and the suspensor (Siddiqui et al. 2003). Furthermore, reduction of AtCAP-E levels by antisense technology gave rise to plants exhibiting meristem structure and function defects, slow growth and precocious differentiation of certain cell types. Thus, the AtCAP-E proteins collectively are essential for embryogenesis, and reduction in protein levels perturb the normal structure and function of meristems.
To further research on the condensin complex in Arabidopsis, we screened for mutants in the SMC4 (AtCAP-C) gene, which was known to exist through the efforts of genome sequencing, but for which no mutant had been described. Here, we report the characterization of such a mutant that displays phenotypes similar to those exhibited by the AtCAP-E mutants, including altered segregation of alleles during gametogenesis and embryo lethality of null mutants.
Materials and methods
Isolation of an AtCAP-C T-DNA insertion mutant and complementation of the mutant phenotype
A BLAST search in the SAIL (Syngenta Arabidopsis Insertion Library) database with the AtCAP-C (encoding for the putative Arabidopsis SMC4 ortholog) genomic sequence (Accession number: NM_124236) revealed a line (86-D2) with a T-DNA insertion near the middle of the gene. The seeds of this line were germinated on Murashige and Skoog (MS) plates containing BASTA (10 μg/ml, Cresent Chemicals) to select for plants carrying a T-DNA insertion. Genomic DNA was isolated from BASTA resistant plants and the integration site of the T-DNA was determined by sequencing of a PCR amplification product generated by employing a gene-specific primer (SMC4-forward 5′-AGCCGGTCTGGAGTTGTGCAAATGG-3′) and the T-DNA left border primer (LB3-5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The insertion was found to be near the end of the seventh intron in the AtCAP-C gene. A total of 32 BASTA resistant plants were transferred to soil, and genotyping was conducted with two sets of primers, one employing a gene specific primer and the LB3 T-DNA primer (see above) to confirm the presence of a T-DNA insertion. The second set of gene specific primers (SMC4-forward, described above, and SMC4-backward 5′-GAGCTGTTTCTGAGCATCTGTGAAGG-3′), was employed with amplification of a 1,542 bp fragment being indicative of the presence of a wild type copy of the gene.
For mutant rescue, the wild type AtCAP-C gene was first amplified by PCR, using three sets of primers: SMC4pro forward 5′-TTGAAGTCGACAATTCTAGCTTGATGAAAAACC-3′ and SMC4 22333 Bam 5′-CACTTAATACATATAGAGGATCCAAACAAAAATTAC-3′ to amplify a 3.1 kb fragment containing promoter sequences; SMC4 22343 Bam backward 5′-TTTGGATCCTCTATATGTATTAAGTGTTACATGTGTT-3′ and SMC4 25613 Bam Backward 5′-TAAAGAAGCAGTGGGATAAGCTAATTAAAAACCTC-3′ to amplify a 3.3 kb fragment containing the proximal promoter region and the first eleven exons of the gene; SMC4 25584 Forward 5′-CTTGGCTCGTCTTCAAAAGAATTTCTAGAGAGG-3′ and SMC4 30321 backward 5′-AATTTGGCGCGCCATAGTTAGTGTTAATCCCAGCAT-3′ to amplify a 4.7 kb fragment containing the remainder of the coding sequence (underlined nucleotides correspond to engineered restriction sites). The 4.7 kbp product was cut with XbaI and AscI and introduced into pHPT-link, a modified binary vector derived from pGVPT-HPT (Becker et al. 1992). Because of non unique restriction enzyme sites the first two PCR products were cloned into pBluescript (Stratagene) and a 6.4 kbp SalI/XbaI fragment was released and mobilized into the SalI/XbaI sites of the intermediate construct. This reconstituted the wild type gene with the exception of engineered SalI (at −3.8 kbp relative to the beginning of the first exon), BamHI (at −686), and AscI (110 bp after the stop codon) sites. The rescue construct was used to transform Agrobacterium GV3101 and kanamycin/gentamycin resistant cells were used to transform 86D2 plants by the method of Clough and Bent (1998). Transgenic plants were selected on MS plates containing 15 μg/ml hygromycin and 10 μg/ml BASTA.
RNA analysis
RNA was prepared from various tissues of Columbia wild type and 86D2 plants using the Trizol protocol (Invitrogen) or a hot phenol/LiCl protocol (Verwoerd et al. 1989). RT-PCR was conducted using approximately 1 μg of total RNA, using oligo dT as a primer according to the product literature supplied with Superscript III reverse transcriptase (Invitrogen). Ten microliters of each reaction was diluted to 50 μl, mixed well, and 2 μl aliquots were used as templates for PCR with two sets of primers, one designed to amplify AtCAP-C cDNA and the other to amplify actin cDNA sequences. The AtCAP-C primers were 5′-GTCCCCATGATGAAGGTTTTCTTG-3′ (exon 5) and 5′-CTTTATATCAGCTTTCCAAGAGGTAG-3′ (exon 12), which generate a 1.8 kb product. The actin primers were: 5′-TGGTGAGGATATTCAGCCACTTGTC-3′ and 5′-TGTGAGATCCCGACCCGCAAGATCA-3′, and generate a 642 bp product. Conditions for PCR were: 1 min at 94°C, followed by 35 cycles of 94°C for 40 s, 53°C for 30 s, and 72°C for 2 min. A final extension cycle of 72°C for 5 min was carried out. Aliquots of each reaction were subjected to electrophoresis and based on the relative abundance of the actin gene amplification products, loading of the AtCAP-C reactions was adjusted accordingly.
Cytological analyses
Inflorescences were allowed to develop until the opening of the first flower bud. The entire inflorescence was then fixed in 3:1 ethanol and acetic acid. Fixation was done for 24–36 h, with the fixative being removed and replaced after 3 h. After fixation the buds were rinsed three times in 70% ethanol, and then either used immediately or stored at 4°C in 70% ethanol.
For squash preparations, an individual bud was placed in 45% acetic acid for 2 min, then transferred to a drop of water on a microscope slide. The anthers were removed from the bud, and all other organs were discarded. The water was then wicked away from the anthers and one drop of Vectamount with DAPI (Vector Labs) was added. Anthers were then sliced in half, and the contents were squashed out of the anther halves with the flat end of a spear. Anther debris was removed and a coverslip was added. The edges of the coverslip were sealed with fingernail polish. Microspore nuclei were examined using a 100× lens of a Zeiss Axiophot fluorescence microscope. Images were captured with a Sony video camera and converted to digital images using Northern Eclipse software.
To examine embryos, hand pollinations were performed and at specific times after fertilization, siliques were harvested and fixed for 4 h in 3:1 ethanol and acetic acid. The fixative was replaced with 70% ethanol and several changes were made over the next 4 h. To decolorize, the tissue was placed in an 8:2:1 solution of chloral hydrate and glycerol and water for one to several days. Siliques were dissected in this solution and embryos imaged by DIC microscopy.
Results
Identification of an Arabidopsis SMC4 gene and analysis of expression
Independent BLAST searches of the Arabidopsis genome database with the SMC4 sequence from humans and Xenopus indicated the presence of a single SMC4 ortholog in the Arabidopsis genome on chromosome V (TAIR Accession: 2152525; Locus: At5g48600). Annotation of the gene by AGI revealed that it is approximately 7.2 Kb in length, having 28 exons. To conform to the nomenclature used for SMC4 genes in higher eukaryotes, this gene was named AtCAP-C (Arabidopsis thaliana Chromosome Associated Protein-C).
A T-DNA insertion mutant in the AtCAP-C gene was identified in the Syngenta T-DNA mutant collection as line 86D2 (McElver et al. 2001). Sequencing of the PCR fragment amplified using the T-DNA specific left border primer and the gene specific primer revealed that the T-DNA insertion lies near the end of the seventh intron (Fig. 1a).
AtCAP-C gene structure and expression. a Intron/exon map showing the 28 exons (open boxes). The position of the Syngenta 86D2 T-DNA insertion is shown. The dots below exons 5 and 12 are the approximate positions of primer binding sites used for genotyping. b RT-PCR analysis using RNA from wild type cauline leaves, inflorescences, roots, rosette leaves, seedlings, siliques and stems (lanes 1–7, respectively), actin gene specific primers (top) and AtCAP-C gene specific primers (bottom). c RT-PCR analysis of RNA from the 86D2 mutant. Lane designations are the same as in b. Top panel, actin signal; bottom panel, AtCAP-C heterozygote signal
To examine wild type mRNA levels, we conducted RT-PCR with RNA from cauline leaves, inflorescences, roots, rosette leaves, seedlings, siliques, and stems of Columbia plants. Total RNA was prepared and reverse transcribed with oligo dT, then aliquots were used for PCR amplification of both AtCAP-C and actin gene fragments. Expression of AtCAP-C is highest in seedlings and inflorescences, and significantly lower but detectable in other tissues examined (Fig. 1b). This expression profile is similar to that observed for AtCAP-E (Siddiqui et al. 2003) and may be generally correlated with the mitotic indices of the tissues examined. To gauge AtCAP-C mRNA levels in 86D2 heterozygotes, we conducted RT-PCR as described above. Figure 1c shows that, while actin mRNA levels were similar to that observed in wild type, the intensity of AtCAP-C bands is reduced in all 86D2 samples, as might be expected since only one copy of the gene has suffered a mutation (see below).
Analysis of the AtCAP-C mutant phenotype
In order to identify mutant plants homozygous for the insertion at the AtCAP-C locus, we screened seeds generated from a segregating population of the AtCAP-C mutant line; 32 BASTA resistant plants were genotyped by PCR using primers flanking the T-DNA insertion site. Selection on BASTA ensures that resistant plants have at least one copy of the T-DNA insert. No homozygous mutant plants were identified among the 32 BASTA resistant plants that were genotyped. Consistently, all BASTA resistant plants were heterozygous for the T-DNA insertion. DNA gel blot analysis of genomic DNA from AtCAP-C +/− plants showed that the T-DNA was only inserted at the AtCAP-C locus in a tandem array of at least two copies of the T-DNA (data not shown). The inability to obtain a homozygous mutant of AtCAP-C strongly suggests that disruption of both copies of AtCAP-C is lethal and that AtCAP-C −/− individuals are lost early in development.
Embryogenesis is compromised in AtCAP-C mutants
The developing siliques of selfed AtCAP-C +/− plants were examined for aberrant seed phenotypes. We observed a large number of defective unfertilized ovules (compared to the wild type) that appeared as empty spaces between the apparently normal sibling seeds (Fig. 2a, b). Amongst the fertilized ovules, both green (harboring normally developing embryos) as well as white (containing arrested embryos) seeds were observed (Fig. 2b). Although DNA gel blotting revealed only one insertion site for the T-DNA in this line, to ensure that this defect is due to an insertion in the AtCAP-C gene, we transformed the 86D2 line with a wild type gene and following selection of T2 plants, found that the defective seed phenotype was rescued by the transgene (Fig. 2c).
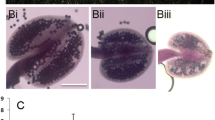
Silique morphology of wild type and mutant self crosses. a Mid mature silique of a wild type Columbia plant, illustrating normal morphology of developing seeds. b Silique of a selfed AtCAP-C +/− plant showing empty spaces and aborted seeds (arrows) segregating with normally developing siblings. c Silique of 86D2 plants (T2 generation) transformed with a wild type AtCAP-C gene showing rescue of the defective seed phenotype
Next, embryo development in both the normal and defective seeds was examined using differential interference contrast microscopy. While the wild type sibling seeds harbored embryos at about the heart stage of development (Fig. 3a), the mutant embryos were developmentally delayed and often arrested at what appears to be the globular stage (Fig. 3b). Occasionally, the embryo would proceed past this stage, but proper patterning was disrupted. Figure 3c shows such an embryo in which the cotyledons failed to separate. Aberrant development was associated with altered planes of cell division in both the embryo and the suspensor (Fig. 3b, d), a defect also observed for AtCAP-E1/2 mutant embryos (Siddiqui et al. 2003). Thus, as might be expected for the heterodimer partner of AtCAP-E, mutations in AtCAP-C results in patterning defects and embryonic lethality.
Cytological analysis of embryos and suspensors. Plants heterozygous for the T-DNA insertion were allowed to self and developing seeds were examined by Normarski optics to examine embryos and suspensors. a Normal sibling embryo at the heart stage of development. b Abnormal sibling arrested at approximately the globular stage of embryogenesis. c Abnormal sibling at approximately the heart stage that exhibits fused cotyledons. d Abnormal sibling arrested at the globular stage. Arrows point to altered planes of cytokinesis in suspensors and embryos
Genetic analysis of the AtCAP-C mutation
We screened seeds generated from self-crosses of AtCAP-C +/− plants to determine what percentage of seeds are resistant to BASTA. If all resulting genotypes are viable at the gametophyte stage we would expect the genotypic ratio in the sporophytes to be 1 AtCAP-C −/− (BASTA resistant): 2 AtCAP-C +/− (BASTA resistant): 1 AtCAP-C +/+ (BASTA sensitive). This genotypic ratio would lead to a 3:1 ratio of BASTA resistance: BASTA sensitivity if the AtCAP-C −/− plants are viable. Since we had evidence of embryo lethality in the null mutant plants we expected to see a ratio of less than 3:1. In the case of embryo lethality, one would expect the ratio of 2:1. Ratios lower than 2:1 indicate embryo lethality in homozygous mutants and failure in either female or male gametogenesis; ratios lower than 1:1 are indicative of inviability of both female and male gametophytes (Pagnussat et al. 2005). We observed a ratio of 1:0.18 (n=1,546; Chi squared tests revealed a P value of less than 0.005, data not shown), strongly suggesting gametogenic lesions in AtCAP-C − gametophytes. However, an alternative possibility is that the BASTA gene underwent silencing without T-DNA loss. To investigate this, we explanted 12 BASTA sensitive seedlings to soil, then genotyped each by PCR using a T-DNA left border primer and the gene specific primer, as well as with primers flanking the T-DNA insertion site. No fragment was amplified with the T-DNA specific and gene specific primers, however, a fragment of the expected size (for the wild type gene) was amplified with the gene specific primers flanking the T-DNA insertion site. This strongly suggested that the extreme skewing of the ratio against recovery of BASTA resistant plants was not due to the silencing or inactivation of the BASTA resistance gene on the T-DNA, but rather was due to a reduction in the recovery or viability of AtCAP-C +/− heterozygous seeds.
To examine male and female gametophytic affects separately, progeny of reciprocal genetic crosses between heterozygous AtCAP-C +/− individuals and wild type plants were performed and the resulting F1 seeds were scored for the segregation of the T-DNA locus conferring BASTA resistance to the progeny. Since AtCAP-C +/− plants would produce equal numbers of wild type and mutant gametes (C+, C−, respectively), 50% of the F1 progeny would be expected to be heterozygous for the T-DNA insertion (BASTA resistant) while the other 50% would be expected to be wild type (BASTA sensitive), if there were no sources of inviability.
When AtCAP-C +/− plants were used as the female parent, only 19.8% of the F1 progeny were BASTA resistant (n=164). This indicated that at most, 40% (19.8% observed versus 50% expected) of the mutant megaspores were able to combine with wild type pollen to produce viable, BASTA resistant embryos. Thus 60% of the mutant megaspores failed either in megasporogenesis or embryogenesis. If the skewing of the ratio against recovery of resistant plants were due exclusively to failure of embryogenesis in heterozygotes, without failure in gametogenesis, then the skewed ratios should have been the same in our reciprocal crosses. While we did see extensive skewing against recovery of BASTA resistant plants in both crosses, the effect was more dramatic when AtCAP-C − pollen was produced. When AtCAP-C +/− plants were used as the male parent, an even greater skewing against BASTA resistance was observed; only 4.8% of the F1 progeny were BASTA resistant (n=156). Thus there is clear evidence that AtCAP-C− microspores fail in microsporogenesis.
Pollen development in AtCAP-C +/− plants
The genetic data indicated that male gametogenesis is compromised in AtCAP-C mutant microspores. To look for the origin of lethality we examined pollen grain mitoses for aberrant segregation. In the Syngenta T-DNA collection, all mutants were generated in a background for the quartet mutation (Preuss et al. 1994). Therefore, we examined pollen from both 86D2 AtCAP-C heterozygotes and from control quartet plants (AtCAP-C +/+). Of 43 pollen grains from three different quartet plants, all appeared to undergo the pollen grain mitoses normally. We examined 46 pollen grain mitoses from two AtCAP-C heterozygotes in a quartet background and found 21 nuclei exhibiting obvious chromosome defects. The most obvious defect was failure of the chromosomes sets to properly separate at anaphase as indicated by the presence of chromatin bridges that connected the two chromatin bodies (Fig. 4). As the 86D2 parent plants were necessarily heterozygous, this frequency is near the 50% value expected if the microspores are compromised by a defective AtCAP-C allele, and supports a critical role for AtCAP-C in chromosome condensation and segregation during mitosis.
Cytological analysis of pollen grain mitoses. a Tetrad from a selfed 86D2 (AtCAP-C +/−) plant, illustrating two apparently normal mitoses (top), a chromatin bridge (lower right), and a fourth pollen grain out of the plane of focus (lower left). b–d Examples of other pollen grains with chromatin bridges in AtCAP-C − segregants. Bar=15 μm
Defects in AtCAP-C +/− heterozygotes
The chromosome bridges observed in the microspore mitosis can create chromosome fragments that likely compromise gametogenesis, and could cause abnormalities in heterozygous embryos and plants. We examined embryos from the AtCAP-C+/− plants crossed to wild type at 4–7 days after fertilization, but found very few aberrant embryos (data not shown).
However, many of the recovered AtCAP-C +/− heterozygotes did exhibit aspects of fasciation, most commonly manifested as supernumary shoots and phyllotaxy defects. To insure that these altered phenotypes were not due to any effect of the selection media but rather correlate with the interruption of the AtCAP-C gene, we planted unselected seeds to soil and screened the resulting plants for both phenotypic defects and the segregation of the AtCAP-C associated T-DNA. In all cases where phenotypic defects were observed, PCR reactions confirmed the existence of a T-DNA within the AtCAP-C gene (data not shown). Thus it is possible that some AtCAP-C+/− heterozygotes fail in embryogenesis, but at stages later than 4–7 days after fertilization.
We conclude that the AtCAP-C −/− genotype produces embryo lethality, and that gametogenesis is compromised to a variable extent in AtCAP-C − gametophytes. AtCAP-C − gametophytes that do survive might have some chromosome fragmentation due to mitotic abnormalities that occur in the gametophytic mitoses. These abnormalities may cause subsequent aberrations in heterozygous embryos and fasciation in the limited number of recovered mature heterozygous plants.
Discussion
Condensins are ubiquitous central components of the chromosome condensation machinery in eukaryotic cells (Losada and Hirano 2005). Interaction of these proteins with various partner proteins leads to the assembly of complexes that sponsor a variety of other cellular processes (Hagstrom and Mayer 2003).
We previously characterized the AtCAP-E genes of Arabidopsis (Siddiqui et al. 2003), and as might be expected for its heterodimer partner, many aspects of AtCAP-C expression and mutant phenotypes that are reported here are striking similar. Both AtCAP-C and AtCAP-E are expressed in all tissues, but expression levels are highest in young tissues where cells are continuing to cycle and have not yet reached the cell expansion phase when mitotic indices are much lower. Reducing the levels of either protein results in developmental defects, manifest as reduction in growth rate, loss of apical dominance, and phyllotaxy defects. Spores carrying null alleles exhibited a severe decline in viability and this we correlated with defects in chromosome segregation during cell division. In situations where spores participated in successful fertilization, developmental defects were observed in mutant embryos. These were manifest as slow growth, disorganization of embryos and/or suspensors, altered planes of cell division and ultimately, embryonic death. Based on the statistics of self crosses and reciprocal crosses with wild type, these defective mutants are likely AtCAP-C nulls. Lastly, heterozygous plants exhibit substantially normal development, but in some instances loss of apical dominance and phyllotaxy defects were observed.
In plants, perturbation of genes that coordinate different aspects of chromatin organization and cell cycling give rise to developmental defects similar to those we observed (Kaya et al. 2001; Hemerly et al. 2000; Hennig et al. 2003; Stevens et al. 2004). In the case of the fasciata mutants, which represent defects in chromatin assembly factors, structural and functional meristem defects could be correlated with altered expression of the WUSCHEL(WUS) and the SCARECROW(SCR) genes, which govern shoot and root meristem function, respectively (Kaya et al. 2001). Mutants in the AtCAP-E genes cause developmental anomalies (Siddiqui et al. 2003) and it has recently been shown that these mutants express RNAs that are normally transcriptionally silent (Takeda et al. 2004). Similarly, when the MSI1 protein is depleted by cosuppression, various developmental defects were observed and molecular analyses revealed that the temporal and spatial expression patterns of two homeotic genes, AGAMOUS and APETALA2, were altered (Hennig et al. 2003). Analyses of MSI1 in various systems suggests that the protein serves to guide heterochromatin formation and maintain some genes in repressed states. Lastly, mutations in genes involved in regulating other aspects of DNA metabolism and cell cycle progression also condition aberrant developmental phenotypes (Bundock and Hooykaas 2002; Takahashi et al. 2002; Stevens et al. 2004). Taken together, these studies provide strong evidence that changes in chromatin dynamics can alter gene expression, and serve to reinforce the importance of epigenetic regulation of genes in controlling development.
How altered expression of these developmental regulators underpins the observed phenotypic defects in the mutants remains a mystery. Defects in cell cycling, due to improper chromatin assembly or delays in replication or packaging, may simply activate checkpoint controls that lead to slower growth and reductions in organ size as is observed for some of the mutants. Fasciation and phyllotaxy defects are more difficult to decipher, but implicate differentiation programs and alterations in the machinery that specifies the proper planes of cytokinesis. We have demonstrated that the tunica/corpus organization of the SAM is perturbed in AtCAP-E antisense plants (Siddiqui et al. 2003) and that periclinal divisions in suspensors occur in both AtCAP-E and AtCAP-C compromised plants. It has been shown that the position of spindle poles influences the orientation of cytokinesis (O’Connell and Wang 2000; Kusch et al. 2003), and given the requirements for condensin in centromere organization and proper spindle assembly in animal cells (Hagstrom et al. 2002; Wignall et al. 2003) it seems likely that altered condensation of chromosomes leads to changes in the relative positioning of spindles to result in random patterning changes. In plants the spatial orientation of cell division depends on phragmoplast formation and a number of factors having roles in cytokinesis have been identified (Nacry et al. 2000). Phragmoplastin, a dynamin-like protein, can influence the orientation of cytokinesis as constitutive expression (Geisler-Lee et al. 2002) and localized expression (Wyrzykowska and Fleming 2003) can lead to alterations in cell division. There are at least 16 different dynamin-related proteins in Arabidopsis, and the four that have been characterized all interact with phragmoplastin (Hong et al. 2003). One of these (ADL1) is required for embryogenesis and seedling development (Kang et al. 2003). These dynamin-related proteins are thus prime targets to investigate as potential mediators of the cytokinetic defects in condensin deficient plants. The growing spectrum of annotated genes and technical advances in transcription profiling will permit the testing of this hypothesis and the identification of additional components of the pathways linking chromosome organization with alterations in cell fate and morphogenesis.
Abbreviations
- SMC:
-
Structural maintenance of chromosomes
- CAP:
-
Chromosome associated protein
- BASTA:
-
Glufosinate ammonium
- DAPI:
-
4′, 6-Diamidino-2-phenylindole
- MS:
-
Murashige and Skoog medium
References
Bazett-Jones DP, Kimura K, Hirano T (2002) Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol Cell 9:1183–1190
Becker D, Kemper E, Schell J, Masterson R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20:1195–1197
Bundock P, Hooykaas P (2002) Severe developmental defects, hypersensitivity to DNA-damaging agents, and lengthened telomeres in Arabidopsis MRE11 mutants. Plant Cell 14:2451–2462
Clough JS, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Dewitte W, Murray JAH (2003) The plant cell cycle. Annu Rev Plant Biol 54:235–264
Geisler-Lee CJ, Hong Z, Verma DPS (2002) Overexpression of the cell plate-associated dynamin-like GTPase, phragmoplastin, results in the accumulation of callose at the cell plate and arrest of plant growth. Plant Sci 163:33–42
Hagstrom KA, Mayer BJ (2003) Condensin and cohesin: more than chromosome compactor and glue. Nat Rev Genet 4:520–534
Hagstrom KA, Holmes VF, Cozzarelli NR, Meyer BJ (2002) C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes Dev 16:729–742
Hemerly A, Ferreira P, Van Montagu M, Engler G, Inze D (2000) Cell division events are essential for embryo patterning and morphogenesis: studies on dominant-negative cdc2aAt mutants of Arabidopsis. Plant J 23:123–130
Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130:2555–2565
Hirano M, Anderson DE, Erickson HP, Hirano T (2001) Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J 20:3238–3250
Hong ZL, Geisler-Lee CJ, Zhang ZM, Verma DPS (2003) Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol Biol 53:297–312
Kang BH, Busse JS, Bednarak SY (2003) Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 15:899–913
Kaya H, Shibahara K, Taoka K, Iwabuchi M, Stillman B, Araki T (2001) FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 104:131–142
Kimura K, Hirano M, Kobayashi R, Hirano T (1998) Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science 282:487–490
Kusch J, Liakopoulos D, Barral YS (2003) Spindle asymmetry: a compass for the cell. Trends Cell Biol 13:562–569
Liu C, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AAM, Meinke DW (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29:405–415
Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19:1269–1287
McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, Tossberg J, Nickle T, Levin JZ, Law M, Meinke D, Patton D (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159:1751–1763
Mengiste T, Revenkova E, Bechtold N, Paszkowski J (1999) An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J 18:4505–4512
Nacry P, Mayer U, Jurgens G (2000) Genetic dissection of cytokinesis. Plant Mol Biol 43:719–733
O’Connell CB, Wang YL (2000) Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell 11:1765–1774
Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie L-F, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132:603–614
Pruess D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264:1458–1460
Siddiqui NU, Stronghill PE, Dengler RE, Hasenkampf CA, Riggs CD (2003) Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130:3283–3295
Stevens R, Grelon M, Vezon D, Oh JS, Meyer P, Perennes C, Domenichini S, Bergounioux C (2004) A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16:99–113
Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13:2271–2283
Takahashi T, Matsuhara S, Abe M, Komeda Y (2002) Disruption of a DNA topoisomerase I gene affects morphogenesis in Arabidopsis. Plant Cell 14:2085–2093
Takeda S, Tadele Z, Hofmann I, Probst AV, Angelis KJ, Kaya H, Araki T, Mengiste T, Scheid OM, Shibahara K, Scheel D, Paszkowski J (2004) BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Genes Dev 18:782–793
Tzafrir I, McElver JA, Liu C, Yang LJ, Wu JQ, Martinez A, Patton DA, Meinke DW (2002) Diversity of TITAN functions in Arabidopsis seed development. Plant Physiol 128:38–51
Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17:2362
Wignall SM, Deehan R, Maresca TJ, Heald R (2003) The condensin complex is required for proper spindle assembly and chromosome segregation in Xenopus egg extracts. J Cell Biol 161:1041–1051
Wyrzykowska J, Fleming A (2003) Cell division pattern influences gene expression in the shoot apical meristem. Proc Natl Acad Sci USA 100:5561–5566
Acknowledgements
The authors wish to thank Dr. Nancy Dengler for advice and assistance with microscopy, Dr. Thomas Berleth for helpful discussions and Mr. David Douda for assistance with genotyping. We acknowledge the Arabidopsis Biological Resource Center for seeds of the quartet mutant, and Syngenta for providing the 86D2 lines. N.U.S. was supported by an Ontario Graduate Scholarship and a University of Toronto Research Fellowship. This research was supported by grants from the Natural Sciences and Engineering Research Council to C.A.H. and C.D.R.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Siddiqui, N.U., Rusyniak, S., Hasenkampf, C.A. et al. Disruption of the Arabidopsis SMC4 gene, AtCAP-C, compromises gametogenesis and embryogenesis. Planta 223, 990–997 (2006). https://doi.org/10.1007/s00425-006-0234-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0234-z