Abstract
In contrast to yeast, plant interphase nuclei often display incomplete alignment (cohesion) along sister chromatid arms. Sister chromatid cohesion mediated by the multi-subunit cohesin complex is essential for correct chromosome segregation during nuclear divisions and for DNA recombination repair. The cohesin complex consists of the conserved proteins SMC1, SMC3, SCC3, and an α-kleisin subunit. Viable homozygous mutants could be selected for the Arabidopsis thaliana α-kleisins SYN1, SYN2, and SYN4, which can partially compensate each other. For the kleisin SYN3 and for the single-copy genes SMC1, SMC3, and SCC3, only heterozygous mutants were obtained that displayed between 77% and 97% of the wild-type transcript level. Compared to wild-type nuclei, sister chromatid alignment was significantly decreased along arms in 4C nuclei of the homozygous syn1 and syn4 and even of the heterozygous smc1, smc3, scc3, and syn3 mutants. Knocking out SYN1 and SYN4 additionally impaired sister centromere cohesion. Homozygous mutants of SWITCH1 (required for meiotic sister chromatid alignment) displayed sterility and decreased sister arm alignment. For the cohesin loading complex subunit SCC2, only heterozygous mutants affecting sister centromere alignment were obtained. Defects of the α-kleisin SYN4, which impair sister chromatid alignment in 4C differentiated nuclei, do apparently not disturb alignment during prometaphase nor cause aneuploidy in meristematic cells. The syn2, 3, 4 scc3 and swi1 mutants display a high frequency of anaphases with bridges (~10% to >20% compared to 2.6% in wild type). Our results suggest that (a) already a slight reduction of the average transcript level in heterozygous cohesin mutants may cause perturbation of cohesion, at least in some leaf cells at distinct loci; (b) the decreased sister chromatid alignment in cohesin mutants can obviously not fully be compensated by other cohesion mechanisms such as DNA concatenation; (c) some cohesin genes, in addition to cohesion, might have further essential functions (e.g., for genome stability, apparently by facilitating correct recombination repair of double-strand breaks).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The multi-subunit complexes containing two molecules of the “structural maintenance of chromosome” (SMC) protein family are important structural components of chromosome organization and function, including sister chromatid cohesion, condensation, DNA repair, gene expression, and development (reviewed in Dorsett 2007; Hirano 2006; Nasmyth and Haering 2005; Onn et al. 2008; Uhlmann 2008). The collinear alignment of sister chromatids defined as cohesion (Maguire 1990; Miyazaki and Orr-Weaver 1994) is mediated by the cohesin complex and is essential for correct chromosome segregation during mitosis and meiosis as well as for DNA recombination repair and transcription. There is increasing evidence that cohesin pathways and/or targeting mechanisms may vary between phylogenetic branches (reviewed in Peric-Hupkes and van Steensel 2008).
The cohesin complex consists of the conserved proteins SMC1, SMC3, SCC3, and an α-kleisin subunit called SCC1 in budding yeast (reviewed in Nasmyth and Haering 2005; Onn et al. 2008). In yeast, two cohesin pools are present. The first pool is recruited during G1 via the SCC2/SCC4 loading complex (Ciosk et al. 2000) at the centromeres and along chromosome arms. These cohesins can move from their loading sites to regions of convergent transcriptional termination (Lengronne et al. 2004) and dislocate from centromeres (“centromere breathing”) due to tension during pre-anaphase. The second pool of cohesin, loaded after replication, is partly retained during “breathing”. When sister centromeres re-associate after transient separation, cohesins are reloaded independently of the SCC2/SCC4 complex (Ocampo-Hafalla et al. 2007). Compared to euchromatic chromosome arm regions, cohesin is enriched ~3-fold in a 20–50-kb domain flanking the centromeres (Blat and Kleckner 1999; Tanaka et al. 1999; Weber et al. 2004) and at pericentric heterochromatin of fission yeast (Bernard et al. 2001). The enrichment of cohesin around centromeres, despite the separation of sister centromeres prior to anaphase onset, was explained by intermolecular cohesion of centromere-flanking DNA (Yeh et al. 2008). Along chromosome arms, the cohesion sites of ~0.8–1.0 kb are separated by only ~11-kb intervals (Glynn et al. 2004; Laloraya et al. 2000). Thus, fluorescence in situ hybridization (FISH) signals do not allow to distinguish yeast sister chromatids (Guacci et al. 1994) because the space between cohesion sites defines the length of potential lateral chromatin loops, and 11-kb loops of an expected length of less than 300 nm are at the limit of microscopic resolution.
In contrast to yeast, allelic loci of sister chromatids in human fibroblast nuclei may occupy distant positions (Volpi et al. 2001) and appear as double signals after replication in human lymphoma nuclei (Selig et al. 1992) when probed by FISH. Similar observations were made for interphase nuclei of angiosperm species. Whereas sister chromatids are often not completely aligned along chromosome arms, sister centromeres stay mostly aligned (up to an endopolyploidy level of 16C in Arabidopsis thaliana (L.) Heynh.; Schubert et al. 2006). The high frequency of local sister chromatid separation (on average occurring in more than 30% of homologues), the absence of preferential alignment sites, and the variability of the alignment extension (<500 kb to >1.2 Mb) along sister chromatid arms, as inferred from microscopic images after chromosome painting in interphase nuclei (Schubert et al. 2008), suggest that sister chromatid cohesion in higher plants is highly dynamic (Berr et al. 2006; Schubert et al. 2006, 2007, 2008).
Evidence from yeast has shown that the dynamic behavior (loading, moving, and diminishing) of cohesins during the cell cycle is closely related to transcription (Bausch et al. 2007; Bernard et al. 2008; Gullerova and Proudfoot 2008; Lengronne et al. 2004). The distribution of cohesins on mammalian chromosome arms is linked to the regulatory zinc-finger protein CTCF, responsible for cohesin recruitment and transcriptional insulation (reviewed in Gause et al. 2008; Parelho et al. 2008; Stedman et al. 2008; Wendt et al. 2008). The SCC2/SCC4 cohesin loading complex is conserved from yeast to human (Seitan et al. 2006; Watrin et al. 2006). In contrast to yeast, where cohesins, after moving from the sites of original loading, are located mostly between genes, cohesins and Nipped-B (corresponding to the SCC2 subunit of the yeast SCC2/SCC4 loading complex) bind consistently to the same sites throughout the entire non-repetitive part of the Drosophila genome (Misulovin et al. 2008).
Cohesin dynamics is also linked to DNA repair (reviewed in Onn et al. 2008; Ström and Sjögren 2007). The local pairing of a damaged chromatid with its intact sister is required to facilitate postreplicative homologous recombination repair of double-strand breaks (DSBs). Investigations in yeast (Cortés-Ledesma and Aguilera 2006; Ström et al. 2004, 2007; Ünal et al. 2004) and human (Kim et al. 2002) have shown that cohesins specifically accumulate at DSB ends, mediating de novo cohesion at these sites. This cohesin recruitment is promoted by the SMC5/6 repair complex loaded to DSB positions (Potts et al. 2006; reviewed in Cortés-Ledesma et al. 2007; Murray and Carr 2008). In A. thaliana, X-irradiation enhances positional sister chromatid alignment when the AtSMC5/6 complex is intact (K. Watanabe, M. Pacher, S. Dukowic, V. Schubert, H. Puchta, I. Schubert, unpublished results).
SCC3, present from yeast to human (reviewed in Losada and Hirano 2005; Onn et al. 2008), is essential for sister chromatid cohesion during mitosis and meiosis in Caenorhabditis elegans Maupas (Pasierbek et al. 2003; Wang et al. 2003) and A. thaliana (Chelysheva et al. 2005).
Several A. thaliana genes express potential components of cohesin, condensin, and SMC5/6 complexes (reviewed in Schubert 2009). Immunolocalization at various subcellular compartments was taken to suggest multiple functions for SMC3 (Lam et al. 2005). In addition to the single-copy genes SMC1, SMC3, and SCC3, A. thaliana has four α-kleisin genes, the SCC1 homologues SYN1, SYN2, SYN3, and SYN4. Arabidopsis SMC1, SMC3, and SCC3 were identified in somatic and meiotic tissues (Chelysheva et al. 2005; Lam et al. 2005; Liu et al. 2002). SYN1 mediates cohesion during meiosis (Bai et al. 1999; Bhatt et al. 1999; Cai et al. 2003). SYN2 and SYN3, mainly expressed in meristematic tissues, seem to be mitotic α-kleisins (Dong et al. 2001). SYN3 is enriched in the nucleolus; therefore, its additional involvement in controlling rDNA structure and transcription or in rRNA processing has been suggested (Jiang et al. 2007). Homozygous “knock out” mutants of either SYN2 or SYN4 are viable, probably because of the redundancy of the α-kleisin genes, although SYN2 plays an additional role in DNA repair after ionizing radiation (da Costa-Nunes et al. 2006). Four α-kleisin genes, showing different functions in somatic cells and during meiosis, were also reported for C. elegans (Mito et al. 2003; Pasierbek et al. 2001) and Oryza sativa L. (Tao et al. 2007; Zhang et al. 2004, 2006). Yeast and vertebrates contain two α-kleisins, the mitotic SCC1 and its meiosis-specific variant REC8 (reviewed in Lee and Orr-Weaver 2001; Nasmyth 2001), whereas Drosophila has no obvious REC8 ortholog in addition to RAD21 (corresponds to SCC1; Heidmann et al. 2004; Vass et al. 2003).
Besides cohesin, the protein SWI1 (Switch) with a partial similarity to SMC proteins is involved in sister chromatid cohesion and chromosome organization during meiosis in A. thaliana (Mercier et al. 2001, 2003).
The difference in sister chromatid alignment between yeast and higher plants, the dynamics of sister chromatid alignment along plant chromosomes, and the presence of four different α-kleisin genes in higher plants inspired us to analyze the consequences of “knocking out” separately the genes presumably encoding homologues of cohesins, as well as of SCC2 and SWI1 of A. thaliana. T-DNA insertion mutants were studied regarding their habit, fertility, and mRNA expression. Sister chromatid alignment frequencies were evaluated in differentiated interphase nuclei of these mutants after FISH with probes specific for mid-arm and centromeric positions. Mutants with severe effects on sister chromatid alignment in differentiated nuclei were tested as to (a) sister chromatid alignment in pro- and metaphase chromosomes, (b) the frequency of hyperploidy in 2C nuclei, and (c) the occurrence of mitotic disturbances (frequency of anaphase bridges).
Materials and methods
Plant material and genotyping
The SALK T-DNA insertion lines in Columbia (Col-0) were obtained from the Salk Institute Genomic Analysis Laboratory (http://signal.salk.edu/cgi-bin/tdnaexpress; Alonso et al. 2003) and provided by the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/). GABI T-DNA mutants (in Col-0) were generated in the context of the GABI-Kat program and provided by Bernd Weisshaar (MPI for Plant Breeding Research, Cologne, Germany; http://www.gabi-kat.de/; Rosso et al. 2003).
Seeds were germinated on agar, followed by further cultivation in soil under short day condition (8 h light/16 h dark) at 21°C. Genomic DNA was isolated from rosette leaves and used for PCR-based genotyping to identify hemizygous and homozygous T-DNA insertion mutants. The PCR primers used for genotyping are listed on Electronic supplementary Table 1, and their positions are shown together with the corresponding gene structure (http://mips.gsf.de/, MAtbB v2.0) in Fig. 1. PCR using the gene-specific primer sets yielded DNA fragments of ~1 kb representing the wild-type alleles. The PCR fragments specific for the “knocked out” allele yielded PCR products of ~0.5 kb. The positions of T-DNA insertion were confirmed by sequencing the PCR-amplified T-DNA junction fragments (Electronic supplementary Table 2).
A. thaliana genes involved in sister chromatid cohesion and scheme of the cohesin complex. (a) Cohesin complex, SWI1, and SCC2 gene structures. Exons are shown as blue boxes. UTRs are visible in gray. T-DNA insertions (SALK and GABI lines) are indicated. The positions and directions of primers are shown as horizontal arrows. Arabic numbers indicate gene-specific primers used for genotyping; roman numbers denote primers applied for RT and Real-time PCR. (b) Model of the cohesin complex of yeast (Nasmyth and Haering 2005) consisting of SMC1, SMC3, SCC3, and the α-kleisin SCC1. The latter is represented by four homologues (SYN1-4) in A. thaliana
mRNA expression analyses
Total RNA was isolated from rosette leaves using the RNeasy plant mini kit (QIAGEN) according to manufacturer’s instructions. Reverse transcription was performed using a First Strand cDNA Synthesis Kit (Fermentas) and 1 µg of total RNA as starting material.
RT-PCR and real-time PCR primers used to amplify transcripts are shown in Fig. 1 and Electronic supplementary Table 3.
Real-time PCR with SYBR Green was used to quantify the abundance of transcripts within 1 µg RNA using an iCycler from BIORAD. Initial denaturation was for 5 min. Then, 40 cycles were run with 10-s denaturation at 95°C, 20-s annealing at 60°C, and 20-s elongation at 72°C. Actin2 served as standard.
For RT-PCR, the following program was used: initial denaturation 2 min, 40-s denaturation, 30-s annealing, 40-s elongation for 35 cycles, 5-min final elongation. Elongation factor 1α served as standard.
Preparation of nuclei, probe labeling, and fluorescent in situ hybridization
Nuclei were isolated and flow-sorted using a FACS Aria (BD Biosciences) according to their ploidy level from rosette leaves after formaldehyde fixation as described (Pecinka et al. 2004).
To investigate mitotic divisions in cotyledons, 3-day-old seedlings were squashed in a drop of 45% acetic acid after fixation overnight in ethanol/acetic acid (3:1). The 178-bp centromeric repeat probe (pAL) was generated by PCR with specific primers from genomic DNA (Kawabe and Nasuda 2005) and subsequently labeled with digoxigenin-dUTP. For painting of the chromosome 1 top arm, 17 pools of in total 87 BACs were labeled with biotin-dUTP as described (Pecinka et al. 2004). The BACs were obtained from the Arabidopsis Biological Resource Center (Columbus, OH, USA). DNA was labeled by nick translation with digoxigenin-dUTP, biotin-dUTP, or Cy3-dUTP according to Ward (2002). Hybridization, post-hybridization washes, and FISH signal detection were as described (Schubert et al. 2001). Biotin was detected by avidin conjugated with Texas Red (1:1,000; Vector Laboratories), goat-anti-avidin conjugated with biotin (1:200; Vector Laboratories), and again with avidin conjugated with Texas Red; digoxigenin by mouse-anti-digoxigenin (1:250; Roche) and goat-anti-mouse conjugated with Alexa-488 (1:200; Molecular Probes). Cy3 was observed directly. Nuclei and chromosomes were counterstained with DAPI (1 µg/ml) in Vectashield (Vector Laboratories).
Microscopic evaluation, image processing, and statistics
Analysis of FISH signals was performed with an epifluorescence microscope (Zeiss Axiophot) using a ×100/1.45 Zeiss α plan-fluar objective and a 3-chip Sony (DXC-950P) color camera. The microscope was integrated into a Digital Optical 3D Microscope system (Schwertner GbR, Germany) to check signal separation/distances along x-, y-, and z-axis. Images were captured separately for each fluorochrome using appropriate excitation and emission filters. The images were merged using Adobe Photoshop 6.0 software (Adobe Systems, San Jose, USA).
In 2C nuclei, more than ten centromeric or more than two arm-specific FISH signals were taken to indicate hyperploidy as a measure of mitotic mis-segregation due to prematurely separated sister chromatids. In prometaphase as well as in 4C nuclei, local separation of sister chromatids at the tested arm positions is indicated by three or four FISH signals (Fig. 4). In 8C nuclei, based on cohesion of centromeres of each homologue and on the appearance of no more than two homologous chromosome territories (V. Schubert, unpublished), the two homologues possess four chromatids each, and thus, three to eight signals indicate local sister chromatid separation. Evaluation followed the criteria described by Schubert et al. (2008).
The differences of sister arm alignment frequencies and of anaphase bridge frequencies observed for mutants in comparison to wild type were compared by the two-sided Fisher’s exact test. The differences of centromeric FISH signals per nucleus were tested with the Kruskal–Wallis test (SigmaStat 3.1) and multiply compared against Columbia wild type with Dunn’s method at P < 0.01 level. Additionally, the frequencies of more than ten centromeric FISH signals indicating unambiguously sister centromere separation were compared against Columbia wild type by the one-sided Fisher’s exact test.
To distinguish between anaphase bridge frequency of 3-day-old wild type versus homozygous and/or heterozygous mutant seedlings that descended from heterozygous parents and could not be genotyped, the frequencies for each individual were grouped according to a significance table based on Fisher's exact test. The exact 95% binomial confidence intervals for the corresponding bridge frequencies were calculated with a program described by Fagan (1996).
Results
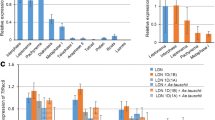
Fifteen A. thaliana T-DNA insertion mutants of cohesin, one of SWI1, and two of SCC2 genes were identified from SALK and GABI T-DNA insertion mutant collections. Presence and positions of T-DNA insertions were confirmed by genotyping via PCR using gene-specific and T-DNA specific primers (Fig. 1, Electronic supplementary Tables 1 and 2) and by sequencing the PCR products. Three insertions were found in introns, one in 5′ UTR and 14 in exons. Depending on the essentiality of the respective genes, homo- or only heterozygous lines were identified and characterized according to plant morphology, fertility, mRNA expression, the degree of sister chromatid alignment, and the occurrence of mitotic disturbances (Table 1, Figs. 2, 3, and 4). Homozygous mutants were analyzed by RT-PCR to confirm the absence of the corresponding transcripts. Real-time PCR using specific primers (Fig. 1, Electronic supplementary Table 3) applied to the heterozygous insertion mutants revealed only a slight decrease in expression of corresponding mRNAs (~77–97% of wild-type level; Fig. 2). Painting of chromosome 1 top arm with 87 labeled BACs was performed on flow-sorted 4C leaf nuclei to compare the compactness of sister arm territories and the frequency of whole sister arm alignment between wild type and cohesin mutants. In all T-DNA insertion lines, sister arm territory compactness (Fig. 4e) and sister arm separation frequency (~4%) were similar as in wild type nuclei (Schubert et al. 2006). The frequency of one or two FISH signals for single BACs per 4C nucleus was taken to indicate positional alignment, while three and four signals indicate separation of sister chromatids in one or both homologues at the corresponding position. Sister centromere separation is indicated by more than ten signals for the 178-bp centromeric repeat in 4-16C nuclei (Fig. 4) because wild-type nuclei up to a DNA content of 16C display very rarely more than ten signals (Table 1, Schubert et al. 2006).
T-DNA insertion mutants, except those for syn2, show decreased sister chromatid alignment in comparison to Columbia wild type, in some cases even at centromeres (marked by asterisks). The percentages were calculated per each BAC locus individually in relation to wild type and then averaged for each mutant line (for details, see Table 1). Transcript levels in relation to wild type are given for heterozygous mutants in parentheses
RT-PCR expression analysis of the α-kleisin and SWI1 genes in A. thaliana leaves of homozygous mutants compared to wild type. The elongation factor (EF1α) mRNA served as a control. a The α-kleisin mutants syn1 and 2 produce no transcript. The homozygous syn3 mutant SALK_119629 (T-DNA localized in the 5′ UTR) produces a wild-type-like transcript. A partially functional truncated mRNA is expressed in all three homozygous syn4 α-kleisin mutants. For the wild-type accession Columbia (Col), only one representative sample of primer combinations is shown. (b) The truncated SWI1 protein (probably over-expressed due to the 35 S promoter of the T-DNA in comparison to Col) does not prevent mutant sterility
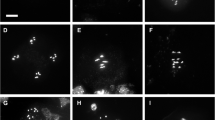
Representative examples of chromatin arrangement in A. thaliana mitotic and interphase nuclei of wild-type and cohesin T-DNA insertion lines. a Chromosomal location of FISH probes detecting centromeric 178-bp repeats (pAL), top arm territory (CT1top), and ~100 kb mid-arm segments (BACs). b Wild-type 4C nucleus with nine centromeric (pAL) signals (two of ten associated). c 4C nuclei of a homozygous syn1 mutant showing examples of positional sister chromatid alignment (left) and of sister chromatid separation of both homologues at position T1F9 (right). d 4C nuclei of a heterozygous syn3 mutant showing separation (left) and separation and alignment (right, arrow) at position T2P11 within the two homologous chromosome arm territories. e Extended sister centromere separation in 4C, 8C, and 16C syn4/syn4 nuclei. The 4C nucleus additionally shows paired chromosome 1 top arm territories, displaying a compactness similar to that of wild-type nuclei. The increase of number of centromeric signals is accompanied by extension of DAPI- intense heterochromatic chromocenters. f Pro-metaphase of a homozygous syn4 mutant (SALK_076116) with aligned sister chromatids at position F11P17 at both homologues (arrows), while 4C interphase nuclei revealed cohesion for only ~ 44% of homologues (Table 1). g Representative 2C nuclei of a homozygous syn4 mutant (SALK_020171) with ten (left) and four (right) centromeric FISH signals, indicating the absence of mitotic aneuploidy, in spite of decreased sister arm and centromere cohesion in differentiated 4C nuclei. The four signals of the right nucleus originate from fusion of the ten centromeres present. h Anaphases with one and two bridges of heterozygous ssc3 and syn3 mutants proving genomic instability, likely due to perturbed double-strand break repair in cells defective for genes encoding these proteins
SMC1 and SMC3 are essential for plant viability and sister chromatid alignment
Both T-DNA insertions in SMC1 are localized in the fifth intron; no homozygous mutants could be selected. Also, the progeny of the three smc3 mutants containing the T-DNA in exons 3, 8, and 11 only segregated into heterozygous and wild-type plants. This indicates the requirement of both genes for plant viability. Interestingly, heterozygous mutants for both genes displayed a significant decrease of positional sister chromatid alignment in spite of only a slight reduction of the transcript level (~78–90% of wild type). Although the average number of centromeric signals was higher in SALK_015308 than in wild-type nuclei, nuclei with more than ten signals, clearly indicating centromere separation, were not observed (Table 1, Fig. 2). Compared to wild type, no increase of anaphase bridge frequency was observed in smc1 and smc3 mutants (Table 1).
syn1 and syn4, but not syn2 mutants, show impaired sister arm and centromere alignment; syn2 and 4 mutants display genome instability
Both syn1 mutants with T-DNA insertions in exons 8 and 15, respectively, segregate sterile homozygous individuals, showing a smaller habit and significantly decreased positional mid-arm sister chromatid alignment frequencies. Although the average number of up to ten centromeric signals per nucleus was not significantly higher than in wild-type 4C nuclei, both mutants exhibited 4.5–9.2% of nuclei with up to 18 signals (Table 1), indicating occasional sister centromere separation. No highly significant increase in anaphase bridge frequencies was observed (Table 1).
The three homozygous syn4 mutant lines, all with inserts in the 11th exon, showed wild-type habit and fertility. RT-PCR expression analysis revealed the presence of a truncated product in all three lines (Fig. 3). However, the truncated transcript does not lead to a full rescue of the mutant because a significantly decreased positional sister chromatid alignment frequency was observed in 4C nuclei, while in prometaphase nuclei, sister chromatid alignment was similar as in wild type (Fig. 4f). Moreover, SYN4 seems to be involved in centromere cohesion because the average number of centromeric signals per nucleus was significantly higher for all three lines (up to 20 signals, see Table 1, Fig. 2) than in wild-type 4C nuclei (up to ten signals). The participation of SYN4 (tested for line SALK_076116) and likely also of SYN1 (tested for line SALK_137095) in centromere cohesion is supported by a further increase in centromeric FISH signal number in 8C and 16C nuclei (Fig. 4e), which is usually not observed in wild-type 8C and 16C nuclei. In all three syn4 mutants, anaphase bridge frequency was significantly (P < 0.001) increased (Table 1). One, two, or three bridges per cell could be observed, which most likely lead to cell lethality. More than ten centromeric FISH signals in 2C leaf nuclei as indication for mitotic aneuploidy due to precocious sister centromere separation were not found in lines SALK_076116 and SALK_020171 (Fig. 4g). At chromosome 1 mid-arm positions of BACs T7N9 and F11P17, no more than two FISH signals were found (tested for lines SALK_076116 and SALK_130085) again, indicating the absence of aneuploidy. Also, flow cytometric analysis showed no significant differences of the peak width (coefficient of variation) that could indicate aneuploidy in homozygous syn4 mutants (SALK_130085) compared to wild type.
Both homozygous syn2 mutants with insertions in exons 6 and 9, respectively, are fertile and of wild-type habit although no transcript was detectable by RT-PCR (Fig. 3). The syn2 mutations do not impair centromere cohesion because 4C nuclei of the SALK_015096 line only rarely showed more than ten centromeric signals, and no increase of signal numbers were found in 8C nuclei. At chromosomal positions of BAC T2P11 and T7N9, inserts of line SALK_015096 sister chromatid alignment was significantly decreased, while alignment frequency was even increased at position T1F9 and at all three positions tested in line SALK_044851, which also revealed bridges in 9.8% (P < 0.01) of anaphase nuclei (Table 1). Apparently, mutations of the syn2 gene can partially be compensated by other α-kleisins as to cohesion in 4C nuclei, but not as to its requirement for genome stability. The varying degrees of alignment along sister arms might indicate a locus-specific impact of SYN2 on cohesion.
SYN3 is essential for plant viability and genome stability and supports sister arm but apparently not centromere alignment
The homozygous syn3 mutant SALK_119629 is expressing a SYN3 transcript at about wild-type level. Apparently, the T-DNA insertion in the 5′ UTR occurs upstream the promoter region and thus does not impair the transcription of the gene (Fig. 3). Therefore, the plants show wild-type-like habit, fertility, and centromere cohesion. Nevertheless, positional sister chromatid alignment along arms was significantly decreased. Possibly, in spite of similar average expression, in some cells or cell types, expression might be reduced. It seems that already slightly reduced expression impairs sister chromatid cohesion.
The T-DNA insertion of line GABI_095A10 in the 6th exon has been confirmed by sequencing. Apparently, the gene is essential for plant viability because no homozygous progeny was obtained. Reduced expression of SYN3 mRNA (~77% of wild-type level) resulted in a significant decrease of positional sister chromatid alignment. The average number of centromeric FISH signals per nucleus was even lower than in wild type, and sister centromere cohesion seems not to be impaired in this mutant because 4C nuclei showed only very rarely more than ten centromeric FISH signals (Table 1, Fig. 2). In 21.9% (P < 0.001) of anaphases, bridges were found (Table 1, Fig. 4h), indicating a severe disturbance of genome stability.
SCC3 is essential for plant viability, genome stability, and sister arm alignment
No homozygous individuals were obtained from the SALK_ 021769 line, suggesting the need of SCC3 for plant viability. Although the reduction of SCC3 mRNA is mild (83.5% of wild-type level), positional sister chromatid alignment is significantly impaired in the heterozygous mutants with the T-DNA in the sixth exon (Table 1, Fig. 2). The average number of centromeric signals per nucleus was significantly higher than in wild type, but 4C nuclei with more than ten signals occurred not more often than in wild type (Table 1). In 15.9% (P < 0.001) of anaphases, bridges occurred (Table 1, Fig. 4h).
SWI1 is essential for fertility and genome stability and is involved in sister arm alignment
Previously, immunostaining and SWI1-GFP experiments in A. thaliana detected SWI1 expression exclusively in early meiocytes (Mercier et al. 2001, 2003). However, at least some transcription must occur also in leaf tissue, and homozygous individuals of line GABI_206H06 over-express a truncated SWI1 transcript (Fig. 3b), possibly using a second transcription start point (Mercier et al. 2001). This transcript does not lead to a fully functional protein because plants are sterile. Positional sister chromatid alignment frequencies are significantly decreased, whereas centromere cohesion is not impaired (Table 1, Fig. 2), indicating a hitherto not recognized impact of SWI1 (or its transcript) on sister arm cohesion in somatic nuclei. It remains unclear whether absence of the full length transcript or over-expression of the short transcript mediates the reduced positional sister arm cohesion in mutant 4C nuclei. A function of SWI1 for genome stability is indicated by 21.0% (P < 0.001) anaphases with bridges in heterozygous mutants and even 43.8% (P < 0.001) in one homozygous mutant (Table 1).
SCC2 is essential for plant viability, for genome stability, and probably for centromere cohesion
From both mutant lines with T-DNA inserted in exons 8 and 13, respectively, only heterozygous individuals could be obtained, indicating that the SCC2 subunit of the cohesin loading complex is needed to ensure plant viability. Sister chromatid alignment was significantly decreased at the position of BAC T7N9 but not at the position of BAC F11P17. The average number of centromeric signals was significantly increased in line SALK_058767 and ~10% of nuclei showed up to 13 signals. The relatively weak effect on sister arm cohesion could be due to the very low reduction (~95–97% of the wild-type transcript level) of SCC2 mRNA in the heterozygous mutants and possibly to the fact described for yeast (reviewed in Onn et al. 2008) that not all loaded cohesins contribute to cohesion along chromosome arms. In contrast to the chromosome arms, centromeres need a stronger, more dense cohesion, possibly mediated by preferential cohesin loading to centromeres or by additional factor(s) mediating centromere-specific cohesion, which is not warranted in the tested scc2 mutants. In 22.9% (P < 0.001) of anaphases of line SALK_151609, bridges were observed (Table 1). Recently, Sebastian et al. (2009) showed for the same insertion mutants that SCC2 is essential for seed development and that SCC2 depletion via RNAi causes a high degree of sterility and meiotic defects such as failure of homologue pairing, chromosome fragmentation, and segregation errors.
Discussion
Although different pathways and proteins can mediate sister chromatid cohesion, these cannot fully compensate all functions of cohesins in Arabidopsis mutants
There is increasing evidence that sister chromatid cohesion along chromosomes during the cell cycle is not only mediated by cohesins. Additionally, condensins, the SMC5/6 repair complex and components of the cohesin loading, the replication, and the transcription machinery (depending on tissue, cell cycle stage, and environmental conditions) seem to be involved. Specialized chromatin domains such as centromeres, telomeres, as well as rDNA tracts use distinct mechanisms for sister chromatid cohesion (Canudas et al. 2007; reviewed in Losada 2007). DNA catenation, a by-product of semiconservative replication (Sundin and Varshavsky 1980), provides an alternative mechanism for sister chromatid alignment at potentially any locus (Diaz-Martinez et al. 2008). Also, cohesion dissolution may follow various pathways (reviewed in Diaz-Martinez et al. 2008; Onn et al. 2008). It has been documented that the percentage of loss of cohesion in cohesin mutants depends on the locus analyzed; complete loss of cohesion has been documented only at telomeres, whereas pericentromeres, rDNA loci, and loci along chromosome arms remain cohesed (reviewed in Diaz-Martinez et al. 2008). Clearly, different alignment frequencies at centromeres (high) and along chromosome arms (lower) suggest also for higher plants various cohesion mechanisms for specific chromatin domains (Schubert et al. 2006, 2007).
In addition to the SCC2/SCC4 cohesin loading complex, active in late G1, cohesion establishing factors interact with components of the replication machinery such as the proliferating cell nuclear antigen, the replication factor C (RFC; reviewed in Guacci 2007; Skibbens et al. 2007), and the origin recognition complex (reviewed in Diaz-Martinez et al. 2008).
The yeast protein CTF18, found at the replication fork (Lengronne et al. 2006), is involved in the establishment of sister chromatid cohesion (Hanna et al. 2001) and also in DSB repair (Ogiwara et al. 2007). Its human homologue also interacts with the RFC complex (Merkle et al. 2003). Knocking out A. thaliana CTF18 decreases positional sister chromatid cohesion significantly in 4C mutant nuclei (V. Schubert, N. Takahashi, L. De Veylder, unpublished results).
Mutations in condensin subunits induced cohesion defects in Drosophila (Dej et al. 2004) and budding yeast that varied along the chromosomes (Lam et al. 2006; Vas et al. 2007).
The activation of cohesin loading mechanisms after DSB induction leads to genome-wide de novo establishment of cohesion in yeast (Ström et al. 2007; Ünal et al. 2007). DSB induction caused increased positional sister chromatid alignment at mid-arm segments in Arabidopsis wild-type nuclei but not in syn1 and smc6 mutants showing that both the cohesin and the SMC5/6 complex may be involved to enforce sister chromatid cohesion for DSB repair by homologous recombination (K. Watanabe, M. Pacher, S. Dukowic, V. Schubert, H. Puchta, I. Schubert, unpublished results).
In the present paper, we document decreased local sister chromatid alignment for the homozygous A. thaliana syn1, syn3 and syn4 mutants, and even for the heterozygous cohesin subunit and the swi1 mutants, indicating the participation of these proteins in sister chromatid cohesion. Moreover, the results suggest that not-cohesin-mediated alignment processes can at least not fully compensate cohesin functions in higher plants. The importance of SMC1, SMC3, SCC3, and SYN3 for plant viability allows to speculate that the mutant lethality is due not only to reduced cohesion along chromosome arms and/or centromeres in somatic nuclei but also to other (additional) functions of the corresponding cohesin components such as transcription regulation by SCC1 in mammals (reviewed in Gause et al. 2008). Furthermore, SYN1 and SWI1 were both found to be essential for plant fertility as already documented (Chelysheva et al. 2005; Mercier et al. 2001, 2003). The frequent occurrence of anaphase bridges during mitosis suggests that at least the α-kleisins SYN2, 3, and 4 as well as the proteins SCC3, SWI1, and SCC2 are required for genome stability, presumably by facilitating (via sister chromatid cohesion) homologous recombination for double-strand break repair. This is in concordance with the observation that SCC2 depletion reduces homologue pairing and causes chromosome fragmentation in A. thaliana meiocytes (Sebastian et al. 2009). Differential positional alignment frequencies at specific chromosomal mid-arm positions between differentiated 4C root and leaf nuclei (Schubert et al. 2006) might be an indication of the participation of cohesion in transcription and development of higher plants.
Our results for scc2 mutants show an effect on centromere rather than on sister arm alignment. The lack of homozygous mutants suggests that also the function of this protein cannot easily be substituted by other pathways and/or proteins.
Decreased sister arm cohesion in heterozygous mutants for SMC1, SMC3, SYN3, and SCC3 or in homozygous swi1 over-expressing a truncated protein (or decreased sister centromere cohesion in heterozygous mutants for SCC2) indicates that only a slight perturbation of the transcript level or a deviating protein structure, compared to the wild type, may significantly impair cohesion.
The four Arabidopsis α-kleisins may form cohesin complexes of potentially different function and may partially compensate each other
In higher plants, cohesins comprising four different α-kleisins have evolved. This raises the question whether they serve different functions. OsRAD21-4, one of the four α-kleisins of O. sativa, was reported to be meiosis specific (Zhang et al. 2006), and OsRAD21-3 was to be required for pollen mitosis (Tao et al. 2007).
Our data are in accordance with meiosis specificity of SYN1 found by Cai et al. (2003) because both homozygous syn1 mutants were sterile. In addition, SYN1 as well as SYN4 seem to be involved in centromere cohesion.
The α-kleisin subunit SCC1 of yeast cohesin is engaged in damage-induced cohesion (Heidinger-Pauli et al. 2008); similar in A. thaliana, SYN2 seems to be involved in DNA repair (da Costa-Nunes et al. 2006).
The syn1, syn3, and syn4 mutants showed decreased cohesion along chromosome arms. The role of SYN2 in sister arm alignment remains obscure because, with one exception, in both homozygous mutants, positional arm cohesion was even higher than in wild type. Perhaps, SYN2 is involved in fine tuning cohesin density at distinct loci, possibly depending on locus-specific transcriptional or other activities. It seems that SYN1, SYN2, and SYN4 can at least partially substitute each other because the homozygous mutants are viable (only syn1 mutants are smaller than wild-type plants and sterile) although cohesion is reduced in syn1 and syn4 mutants, and anaphase bridges appear frequently in syn2 and syn4 mutants. SYN3 can at least partially substitute the other α-kleisins, but homozygous syn3 mutants are lethal, probably because its complete loss may cause a sub-functional level of cohesion in mitotic and meiotic cells or due to an additional function of SYN3 in rDNA processing (Jiang et al. 2007). To clarify unambiguously these relationships, double and triple mutants have to be analyzed. In summary, A. thaliana α-kleisins can partially compensate each other and may have evolved other functions in addition to cohesion. SYN1 is involved in DNA repair by active sister chromatid alignment after X-irradiation to ensure genome stability (see above and K. Watanabe, M. Pacher, S. Dukowic, V. Schubert, H. Puchta, I. Schubert, unpublished results). SYN2, 3, and 4 are also required to prevent dicentric chromosome rearrangements.
Single cohesin mutations do not affect chromosome territory structure
Chromosome arms in A. thaliana interphase nuclei are organized in distinct territories (Pecinka et al. 2004). The compactness of sister arm territories and the frequency of whole sister arm alignment was not impaired in 4C nuclei of the cohesin, swi1 and scc2 mutants, in spite of decreased cohesion along arms (and in syn1, syn4, and scc2 mutants, even at centromeres). These findings and the observation that chromosome territory (CT) structure within wild-type nuclei up to an endopolyploidy level of 64C is not disturbed, although positional sister chromatid separation may reach 100% (Schubert et al. 2006; V. Schubert, unpublished results), suggest that the remaining cohesion (or other factors than cohesins) are sufficient to maintain CT structure and prevent intermingling of heterologous CTs.
References
Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Bai X, Peirson BN, Dong F, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11:417–430
Bausch C, Noone S, Henry JM, Gaudenz K, Sanderson B, Seidel C, Gerton JL (2007) Transcription alters chromosomal locations of cohesin in Saccharomyces cerevisiae. Mol Cell Biol 27:8522–8532
Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 294:2539–2542
Bernard P, Schmidt CK, Vaur S, Dheur S, Drogat J, Genier S, Ekwall K, Uhlmann F, Javerzat J-P (2008) Cell-cycle regulation of cohesin stability along fission yeast chromosomes. EMBO J 27:111–121
Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I (2006) Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J 48:771–783
Bhatt AM, Lister C, Page T, Fransz P, Findlay K, Jones GH, Dickinson HG, Dean C (1999) The DIF1 gene of Arabidopsis is required for meiotic chromosome segregation and belongs to the REC8/RAD21 cohesin gene family. Plant J 19:463–472
Blat Y, Kleckner N (1999) Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249–259
Cai X, Dong F, Edelmann RE, Makaroff CA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J Cell Sci 116:2999–3007
Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S (2007) Protein requirements for sister telomere association in human cells. EMBO J 26:4867–4878
Chelysheva L, Diallo S, Vezon D, Gendrot G, Vrielynck N, Belcram K, Rocques N, Márquez-Lema A, Bhatt AM, Horlow C, Mercier R, Mézard C, Grelon M (2005) AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J Cell Sci 118:4621–4632
Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K (2000) Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell 5:243–254
Cortés-Ledesma F, Aguilera A (2006) Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep 7:919–926
Cortés-Ledesma F, de Piccoli G, Haber JE, Aragón L, Aguilera A (2007) SMC proteins, new players in the maintenance of genomic stability. Cell Cycle 6:914–918
da Costa-Nunes JA, Bhatt AM, O'Shea S, West CE, Bray CM, Grossniklaus U, Dickinson HG (2006) Characterization of the three Arabidopsis thaliana RAD21 cohesins reveals differential responses to ionizing radiation. J Exp Bot 57:971–983
Dej KJ, Ahn C, Orr-Weaver TL (2004) Mutations in the Drosophila condensin subunit dCAP-G: defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics 168:895–906
Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ (2008) Chromosome cohesion—rings, knots, orcs and fellowship. J Cell Sci 121:2107–2114
Dong F, Cai X, Makaroff CA (2001) Cloning and characterization of two Arabidopsis genes that belong to the RAD21/REC8 family of chromosome cohesin proteins. Gene 271:99–108
Dorsett D (2007) Roles of the sister chromatid cohesion apparatus in gene expression, development, and human syndromes. Chromosoma 116:1–13
Fagan T (1996) QUICKBASIC program for exact and mid-p confidence interval for a binomial proportion. Comput Biol Med 26:263–267
Gause M, Schaaf CA, Dorsett D (2008) Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays 30:715–718
Glynn EF, Megee PC, Yu HG, Mistrot C, Ünal E, Koshland DE, DeRisi JL, Gerton JL (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2:1325–1339
Guacci V (2007) Sister chromatid cohesion: the cohesin cleavage model does not ring true. Genes Cells 12:693–708
Guacci V, Hogan E, Koshland D (1994) Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol 125:517–530
Gullerova M, Proudfoot NJ (2008) Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132:983–995
Hanna JS, Kroll ES, Lundblad V, Spencer FA (2001) Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol Cell Biol 21:3144–3158
Heidinger-Pauli JM, Ünal E, Guacci V, Koshland D (2008) The kleisin subunit of cohesin dictates damage-induced cohesion. Mol Cell 31:47–56
Heidmann D, Horn S, Heidmann S, Schleiffer A, Nasmyth K, Lehner CF (2004) The Drosophila meiotic kleisin C(2) M functions before the meiotic divisions. Chromosoma 113:177–187
Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7:311–322
Jiang L, Xia M, Strittmatter LI, Makaroff CA (2007) The Arabidopsis cohesin protein SYN3 localizes to the nucleolus and is essential for gametogenesis. Plant J 50:1020–1034
Kawabe A, Nasuda S (2005) Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol Genet Genomics 272:593–602
Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K (2002) Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem 277:45149–45153
Laloraya S, Guacci V, Koshland D (2000) Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol 151:1047–1056
Lam WS, Yang X, Makaroff CA (2005) Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. J Cell Sci 118:3037–3048
Lam WW, Peterson EA, Yeung M, Lavoie BD (2006) Condensin is required for chromosome arm cohesion during mitosis. Genes Dev 20:2973–2984
Lee JY, Orr-Weaver TL (2001) The molecular basis of sister-chromatid cohesion. Annu Rev Cell Dev Biol 17:753–777
Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430:573–578
Lengronne A, McIntyre J, Katou Y, Kanoh Y, Hopfner KP, Shirahige K, Uhlmann F (2006) Establishment of sister chromatid cohesion at the S. cerevisiae replication fork. Mol Cell 23:787–799
Liu CM, McElver J, Tzafrir I, Joosen R, Wittich P, Patton D, Van Lammeren AA, Meinke D (2002) Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J 29:405–415
Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116:321–329
Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19:1269–1287
Maguire MP (1990) Sister chromatid cohesiveness: vital function, obscure mechanism. Biochem Cell Biol 68:1231–1242
Mercier R, Vezon D, Bullier E, Motamayor JC, Sellier A, Lefèvre F, Pelletier G, Horlow C (2001) SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev 15:1859–1871
Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, Vezon D, Pelletier G, Jones GH, Franklin FC (2003) The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130:3309–3318
Merkle CJ, Karnitz LM, Henry-Sánchez JT, Chen J (2003) Cloning and characterization of hCTF18, hCTF8, and hDCC1—human homologs of an Saccharomyces cerevisiae complex involved in sister chromatid cohesion establishment. J Biol Chem 278:30051–30056
Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, Fay JC, Eisen MB, Pirrotta V, Biggin MD, Dorsett D (2008) Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma 117:89–102
Mito Y, Sugimoto A, Yamamoto M (2003) Distinct developmental function of two Caenorhabditis elegans homologs of the cohesin subunit Scc1/Rad21. Mol Biol Cell 14:2399–2409
Miyazaki WY, Orr-Weaver TL (1994) Sister-chromatid cohesion in mitosis and meiosis. Annu Rev Genet 28:167–187
Murray JM, Carr AM (2008) SMC5/6: a link between DNA repair and undirectional replication? Nat Rev Mol Cell Biol 9:177–182
Nasmyth K (2001) Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet 35:673–745
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Ocampo-Hafalla MT, Katou Y, Shirahige K, Uhlmann F (2007) Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma 116:531–544
Ogiwara H, Ohuchi T, Ui A, Tada S, Enomoto T, Seki M (2007) Ctf18 is required for homologous recombination-mediated double-strand break repair. Nucleic Acids Res 35:4989–5000
Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE (2008) Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol 24:105–129
Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschiager M (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132:422–433
Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J (2001) A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev 15:1349–1360
Pasierbek P, Födermayr M, Jantsch V, Jantsch M, Schweizer D, Loidl J (2003) The Caenorhabditis elegans SCC-3 homologue is required for meiotic synapsis and for proper chromosome disjunction in mitosis and meiosis. Exp Cell Res 289:245–255
Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I (2004) Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269
Peric-Hupkes D, van Steensel B (2008) Linking cohesin to gene regulation. Cell 132:925–928
Potts PR, Porteus MH, Yu HT (2006) Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J 25:3377–3388
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Schubert V (2009) SMC proteins and their multiple functions in higher plants. Cytogenet Genome Res (in press)
Schubert I, Fransz PF, Fuchs J, de Jong JH (2001) Chromosome painting in plants. Methods Cell Sci 23:57–69
Schubert V, Klatte M, Pecinka A, Meister A, Jasencakova Z, Schubert I (2006) Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172:467–475
Schubert V, Kim YM, Berr A, Fuchs J, Meister A, Marschner S, Schubert I (2007) Random homologous pairing and incomplete sister chromatid alignment are common in angiosperm interphase nuclei. Mol Genet Genomics 278:167–176
Schubert V, Kim YM, Schubert I (2008) Arabidopsis sister chromatids often show complete alignment or separation along a 1.2-Mb euchromatic region but no cohesion “hot spots”. Chromosoma 117:261–266
Sebastian J, Ravi M, Andreuzza S, Panoli AP, Marimuthu MPA, Siddiqi I (2009) The plant adherin AtSCC2 is required for embryogenesis and sister-chromatid cohesion during meiosis in Arabidopsis. Plant J . doi:10.1111/j.1365-313X.2009.03845.x
Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, Smith J, Bateman A, Krpic S, Hostert A, Rollins RA, Erdjument-Bromage H, Tempst P, Benard CY, Hekimi S, Newbury SF, Strachan T (2006) Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol 4:1411–1425
Selig S, Okumura K, Ward DC, Cedar H (1992) Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J 11:1217–1225
Skibbens RV, Maradeo M, Eastman L (2007) Fork it over: the cohesion establishment factor Ctf7p and DNA replication. J Cell Sci 120:2471–2477
Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J 27:654–666
Ström L, Sjögren C (2007) Chromosome segregation and double-strand break repair—a complex connection. Curr Opin Cell Biol 19:344–349
Ström L, Lindroos HB, Shirahige K, Sjögren C (2004) Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell 16:1003–1015
Ström L, Karlsson C, Lindroos HB, Wedahl S, Katou Y, Shirahige K, Sjögren C (2007) Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 317:242–245
Sundin O, Varshavsky A (1980) Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell 21:103–114
Tanaka T, Cosma MP, Wirth K, Nasmyth K (1999) Identification of cohesin association sites at centromeres and along chromosome arms. Cell 98:847–858
Tao J, Zhang L, Chong K, Wang T (2007) OsRAD21–3, an orthologue of yeast RAD21, is required for pollen development in Oryza sativa. Plant J 51:919–930
Uhlmann F (2008) Molecular biology: cohesin branches out. Nature 451:777–778
Ünal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D (2004) DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell 16:991–1002
Ünal E, Heidinger-Pauli JM, Koshland D (2007) DNA double-strand breaks trigger genome-wide sister-chromatid cohesion through Eco1 (Ctf7). Science 317:245–248
Vas AC, Andrews CA, Kirkland Matesky K, Clarke DJ (2007) In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Mol Biol Cell 18:557–568
Vass S, Cotterill S, Valdeolmillos AM, Barbero JL, Lin E, Warren WD, Heck MM (2003) Depletion of Drad21/Scc1 in Drosophila cells leads to instability of the cohesin complex and disruption of mitotic progression. Curr Biol 13:208–218
Volpi EV, Sheer D, Uhlmann F (2001) Cohesion, but not too close. Curr Biol 11:R378
Wang F, Yoder J, Antoshechkin I, Han M (2003) Caenorhabditis elegans EVL-14/PDS-5 and SCC-3 are essential for sister chromatid cohesion in meiosis and mitosis. Mol Cell Biol 23:7698–7707
Ward PB (2002) FISH probes and labelling techniques. In: Beatty B, Mai S, Squire J (eds) Fish. Oxford University, Oxford, pp 5–28
Watrin E, Schleiffer A, Tanaka K, Eisenhaber F, Nasmyth K, Peters JM (2006) Human Scc4 is required for cohesin binding to chromatin, sister-chromatid cohesion, and mitotic progression. Curr Biol 16:863–874
Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC (2004) The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol 2:1340–1353
Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, Yahata K, Imamoto F, Aburatani H, Nakao M, Imamoto N, Maeshima K, Shirahige K, Peters JM (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451:796–801
Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol 18:81–90
Zhang LR, Tao JY, Wang T (2004) Molecular characterization of OsRAD21–1, a rice homologue of yeast RAD21 essential for mitotic chromosome cohesion. J Exp Bot 55:1149–1152
Zhang L, Tao J, Wang S, Chong K, Wang T (2006) The rice OsRad21–4, an orthologue of yeast Rec8 protein, is required for efficient meiosis. Plant Mol Biol 60:533–554
Acknowledgments
We thank Martina Kühne, Joachim Bruder, and Rita Schubert for technical assistance and Andreas Houben for critical reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Uhlmann
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Table S1
PCR primers used to identify the T-DNA insertion alleles. (DOC 14 kb)
Table S2
Sequences of the left border junctions of the T-DNA insertion lines. (DOC 13 kb)
Table S3
RT and real-time PCR primers used to amplify transcripts. (DOC 14 kb)
Rights and permissions
About this article
Cite this article
Schubert, V., Weißleder, A., Ali, H. et al. Cohesin gene defects may impair sister chromatid alignment and genome stability in Arabidopsis thaliana . Chromosoma 118, 591–605 (2009). https://doi.org/10.1007/s00412-009-0220-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0220-x