Abstract
The R2R3-MYB transcription factors play an important role in regulating secondary metabolism biosynthesis and abiotic stress in plants. In this article, we report the identification of the transcription factor gene, FtMYB31, from the popular Eurasian crop tartary buckwheat (Fagopyrum tataricum) that enhances accumulation of the nutritionally beneficial compound rutin in transgenic tobacco leaves. The FtMYB31 complete cDNA coding sequence was isolated from the leaves of tartary buckwheat, and multiple protein sequence alignments and conserved domain analysis showed it contained a typical R2R3 MYB domain. Subcellular location experiments showed the FtMYB31 protein is localized in nucleus. The phylogenetic tree clustered FtMYB31 with VvMYBPA1 from Vitis vinifera, and AtMYB123 from Arabidopsis thaliana, belonging to the Subgroup 5 cluster. Comparison by qRT-PCR of FtMYB31 transcripts and those from rutin synthesis-related genes showed a relationship between FtMYB31, Ft4CL, and FtUFGT transcripts and rutin content in different tissues of F. tataricum, with correlation coefficients of − 0.68, 0.69, and 0.47, respectively. Transgenic experiments indicated that FtMYB31 upregulated CHS, F3H, and FLS genes in transgenic tobacco and enhanced the accumulation of rutin and total flavonols. These results suggest that FtMYB31 encodes an R2R3-MYB transcription factor that positively regulates flavonol biosynthesis in tartary buckwheat and tobacco and is a possible target for genetically modifying tartary buckwheat to enhance the content of beneficial compounds such as rutin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Buckwheat (Polygonaceae) is a pseudocereal with two major species of agricultural significance: common buckwheat (Fagopyrum esculentum Moench, sweet buckwheat) and tartary buckwheat (Fagopyrum tataricum (L.) Gaertn., bitter buckwheat) (Ohnishi 1998; Logacheva et al. 2011). Tartary buckwheat is a very healthy food enriched in bioactive compounds, such as rutin, a type of flavonol, and has great potential for development and also has applications as a medicinal plant (Kalinová et al. 2018). The rutin content in tartary buckwheat ranges from approximately 1% to 5% of the dry weight in flowers, leaves, and seeds (Kreft et al. 2002, 2006; Jiang et al. 2007). Rutin is beneficial for several aspects of growth and physiology of tartary buckwheat, including the enhancement of defense systems against UV radiation and abiotic stress, desiccation, and salinity (Suzuki et al. 2005; Sun et al. 2011, Gao et al. 2016). In humans, rutin has been reported to have antioxidant and anti-inflammatory activities and anticancer properties. It can also reduce the fragility of blood vessels and the risk of diabetes (Ahmed et al. 2014; Gou et al. 2014).
The flavonol biosynthesis pathways have been well documented in plants such as Arabidopsis thaliana and Solanum lycopersicum (Koes et al. 2005; Bovy et al. 2007). The precursor phenylalanine can be metabolized to different types of flavonol compounds via different biosynthesis pathways. Most enzymes involved in this pathway have been well studied. Phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate: CoA ligase (4CL) are involved in the general phenylpropanoid pathway; and chalcone synthase (CHS), chalcone isomerase (CHI), and flavanone-3-hydroxylase (F3H) are early biosynthetic genes (EBGs). Dihydro flavanol 4-reductase (DFR) is involved in the proanthocyanidin (PA) and anthocyanin pathways; UDP-glucose: flavonoid 3- glucosyltransferase (UFGT) is involved in the anthocyanin and flavonol pathway (Saito et al. 2013). Rutin is a glycosylated flavonol, derived from dihydroquercetin by sequential reactions catalyzed by flavonol synthase (FLS), glucosyltransferase (GT), and rhamnosyltransferase (RT) (see Supplementary Figure S1). Besides structural genes, it is well known that transcription factors (TFs), such as MYBs, WD40, and bHLH, play an essential role in regulating the overall activity of flavonol biosynthesis (Stracke et al. 2007, 2010).
In the recent decades, R2R3-MYB genes have been extensively studied and members of the MYB family have been found to be involved in phenylpropanoid metabolism (Grotewold et al. 1994; Hichri et al. 2011), anthocyanin biosynthesis, and flavonol accumulation (Meng et al. 2014; Aharoni et al. 2010). In Arabidopsis, 126 R2R3 MYB TF-encoding genes have been classified into 24 subgroups (Dubos et al. 2010), and 71 putative R2R3-MYB TFs have been identified from the common buckwheat genome database (Yasui et al. 2016). Due to the increasing demands from consumers and industrial applications, the rutin content of crops requires further enhancement to achieve satisfactory yields. Tomato and tobacco plants with enhanced rutin content have been achieved via the heterologous expression of the AtMYB12 transcription factor from Arabidopsis. A subsequent study on callus from AtMYB12-expressing tobacco also showed higher rutin levels than control plants (Pandey et al. 2012). In addition, overexpression of AtMYB11, a homolog of AtMYB12, significantly increases the rutin content in tobacco and tomato leaves (Pandey et al. 2015, Li et al. 2015). FtMYB123L was identified as a homolog of AtMYB123/TT2 from a floral transcriptome analysis, and was inferred involved in the flavonoid synthesis (Zhou et al. 2013).
The aim of this study was to isolate and identify an R2R3 MYB transcription factor that regulates the flavonol biosynthesis pathway in tartary buckwheat. In addition, we analyzed the sequence characteristics and tissue specific gene expression of the identified R2R3 MYB gene. To understand its function and relationship to flavonol biosynthesis, the gene was studied in transgenic tobacco leaves, and the flavonoid content was found to be significantly higher than that of WT, and the ectopic expression FtMYB31 upregulated gene expression level and increased rutin content in tobacco.
Materials and Methods
Plant Materials and Growth Conditions
The seeds of the tartary buckwheat cultivar “Heifeng No.1” were donated by the Institute of Crop Genetic Resources (Shanxi Academy of Agricultural Sciences, Shanxi, China). Three seeds were germinated in each pot and grown in a greenhouse at Shanxi Agricultural University (northern China, 37°25′N, 112°29′E) under a light intensity of 200 μmol m−2 s−1, 24 ± 2 °C, and 16 h photoperiod. When the plants were full grown, the leaves, flowers, and grains were collected and stored at − 80 °C for further experiments. All T3 transgenic tobacco lines were planted in a greenhouse under the same growth condition as described above. The leaves of T3 transgenic lines were collected for further analyses. All experimental materials had three individual biological replicates.
Sequence Retrieval, Alignment, and Phylogenetic Analysis
A local protein blast database was constructed from the transcriptome data of leaves of F. tartaricum for further analysis using BioEdit 7.2.1 software (sequenced by Biomarker technologies, Beijing, China; unpublished). Using the conserved R2R3-MYB motif as the target sequence, a total of ten R2R3-MYB proteins in F. tartaricum, twenty from Arabidopsis, and one from Vitis vinifera, were obtained from Genbank (https://www.ncbi.nlm.nih.gov/) for sequence alignment and phylogenetic analyses. The 31 R2R3 MYB members were aligned using MUSCLE in MEGA 7.0 software with default parameters. The alignments achieved were used as an input to construct the phylogenetic tree with 10 000 bootstrap replicates with gaps deleted using MUSCLE 3.6 based on the neighbor-joining method. One R2R3-MYB gene named FtMYB31 clustered with VvMYBPA1, and this gene was selected to be cloned from leaves of F. tartaricum.
Subcellular Localization Analysis of FtMYB31
A 35S:FtMYB31-GFP construct was transiently expressed in tobacco leaves using the agrobacterium micro-injection methods (Xu et al. 2014). The transgenic cells were observed under a fluorescence microscope (OLYMPUS, FV10-ASW, USA) after 2 days’ cultivation.
RNA Extraction, cDNA Synthesis, and Gene Cloning
Total RNA was isolated from leaves, flowers, and grains of F. tartaricum and leaves of T3 transgenic tobacco lines using the MiniBEST Plant RNA Extraction Kit (Takara, Japan). The RNA quality was checked using a NanoDropTM2000 spectrophotometer (Thermo Fisher Scientific, USA) and electrophoresis. Using PrimeScriptTM RT reagent Kit (Takara, Japan) according to the manufacturer’s instructions, 1 μg total RNA was reverse transcribed to generate the first strand cDNA library. Gene specific forward and reverse primers were designed, and FtMYB31 cDNA was cloned using RT-PCR. The RT-PCR reaction system was conducted in a total volume of 50 µL containing 25 µL Premix ExTaq (Takara, Japan), 2 µL first-strand cDNA template, and 0.4 µM each primer (all primers are listed in Supplementary Table S1). The RT-PCR reaction program was set as follows: preheating at 94 °C for 5 min, 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 90 s, followed by a final extension at 72 °C for 10 min. The amplified products were tested by agarose gel electrophoresis, and the bands were recovered from the gel using the Gel DNA Purification Kit (Sangon Co., Shanghai of China). These DNA fragments were subcloned into the pMD19-T vector (Takara, Japan) and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The nucleotide sequence of FtMYB31 has been submitted to GenBank.
Gene Expression Analysis
For real-time-quantitative PCR analysis, expressions of all of genes in this study were measured using a PCR mix containing 1 μL of diluted cDNA (10 ng), 10 μL of 2 × SYBR Green PCR Master Mix (Takara, Japan), and 200 nM of each gene specific primer in a final volume of 20 μL. Assays with no template were also performed for each primer pair as a control. All the PCR amplifications were performed in 96-well optical reaction plates (Bio-rad, USA) under the following conditions: 20 secs at 95 °C, 3 secs at 95 °C, and 40 cycles of 30 secs at 60 °C. The specificity of amplicons was verified by melting curve analysis (60 °C to 95 °C) after 40 cycles. Three biological replicates were analyzed for each gene, and each sample run three technical replicates as well. Results are expressed as change in expression levels relative to the histone gene (tartary buckwheat) or ubiquitin gene (tobacco) using the 2−ΔΔct method (Livak and Schmittgen 2001).
Expression Vector Construction and Genetic Transformation
The complete coding sequence of FtMYB31 cDNA with BamH I and Sma I digestion site (see Figure. S2), under the control of the cauliflower mosaic virus 35S promoter in the binary vector pBI121, was transferred into Agrobacterium tumefaciens strain EHA105 by electroporation. For transient expression, healthy leaves of wild-type tobacco were chosen and injected with Agrobacterium tumefaciens EHA105, which had been transformed with either FtMYB31 or empty vector (EV). Further control leaves were injected with water.
For generating stable transgenic lines, tobacco plants were transformed using Agrobacterium-mediated transformation (Horsch et al. 1985). EV-infected tobacco plants were also generated as a control. The T0-transformed tobacco seeds were harvested, sterilized, and plated on solid half strength MS medium supplemented with 100 mg/L kanamycin to screen for positive transgenic lines, which were detected by RT-PCR. Finally, T3-positive transgenic tobacco lines were obtained for further analysis of rutin content and gene expression (Figure S3).
Determination of Total Flavonoid and Rutin Content
Assay of total flavonoid content was carried out by the method described by Liu and Zhu (2007). Briefly, 0.1 g dried transgenic tobacco leaves were extracted in methanol, then 1 mL diluted solution was added to 0.7 mL of 5% NaNO2 and 10 mL of 30% methanol and mixed for 5 min. Next, 0.7 mL of 10% AlCl3 was added to the mixture. After 5 min, 5 mL of 1 mol/L NaOH was added and the absorbance of the solution was measured at 500 nm with a spectrophotometer. The results were calculated as mg/g dry weight and compared with the rutin standard curve.
High Performance Liquid Chromatography (HPLC) was used to analyze the rutin content following Sun’s method (2011). Briefly, the fresh tissues (approximately 0.2 g) were ground into a fine powder and macerated in 1 mL of methyl alcohol. The samples were placed in an ultrasonic bath at 50 °C for 50 min for extraction, centrifuged at 12,000 rpm for 10 min to obtain the supernatant, then filtered through 0.45 μm PTFE filers into HPLC vials. An HPLC analysis was performed with a C18 column (150 mm × 4.6 mm, 5 μm) (Agilent, U.S.A). The mobile phase consisted of 46% methanol to 54% water, and the sample was run on an Ultimate 3000 HPLC System (Thermo scientific, USA). The retention time of rutin was 3.8 ± 0.1 min at 257 nm. The concentrations of rutin were determined using a standard curve (Figure S4).
Visualization of Flavonoid
The wild-type tobacco leaves that had been infiltrated with Agrobacterium were cut into pieces (0.5 cm × 0.5 cm) and stained for 5 to 15 min using saturated (0.25%, w/v) DPBA with 0.02% (v/v) Triton X-100. Stained leaves were then visualized with an epifluorescence microscope equipped (excitation 450–490 nm, Leica DM5500, German) as described in Murphy et al. (2000).
Results
Sequence Characteristics and Subcellular Localization of FtMYB31 Gene
The cloned FtMYB31 cDNA (GenBank accession number: KM588380) comprised 912 nucleotides encoding 303 amino acid residues. By comparing cDNA and genomic sequences the gene was shown to have no introns. The predicted molecular weight of the protein encoded by this gene was 34,013.16 Daltons, and the isoelectric point was 7.741. Based on the conserved protein domain analysis, the FtMYB31 protein contained two Myb DNA-binding domains (ranging from amino acid residues 14-61 and 67-112). When compared with homologous proteins from Arabidopsis thaliana, Vitis vinifera, and Fagopyrum tartaricum, the multisequence alignment and motif analyses indicated that the protein possessed typical R2 (WX19WX19W) and R3 (F/IX18W) motifs (Fig. 1b, c). The FtMYB31 protein showed the highest amino acid sequence similarity to VvMYBPA1 from Vitis vinifera, with a 60% identity by BLAST in the nonredundant protein database. The phylogenetic tree distinguished three groups of R2R3-MYB genes in F. tataricum and other species (Fig. 1a). FtMYB31 clustered with VvMYBPA1and AtMYB123. Because the Arabidopsis AtMYB123 genes was functionally identified as flavonoid biosynthesis transcriptional regulators, we inferred that FtMYB31 was an R2R3-MYB SG5 gene.
To examine whether FtMYB31 was localized in the nucleus, as would be expected of a typical R2R3-MYB TF protein, the fusion protein FtMYB31-GFP was transiently expressed in tobacco leaves. Fluorescence microscopy showed that the free GFP control was located in both the cytosol and nucleus, whereas FtMYB31-GFP protein was exclusively observed in the nucleus (Fig. 2). Therefore, the subcellular localization of FtMYB31 is consistent with the putative role of a TF protein.
Tissue-Specific Expression Analyses of FtMYB31 and Flavonol Biosynthesis-Related Genes
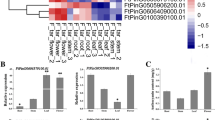
We performed qRT-PCR to investigate the transcript levels of flavonol biosynthesis-related genes and FtMYB31 in different tissues of F. tartaricum. FtCHS gene expression was detected in flowers and leaves, with a level in seeds only 1% of that found in leaves. This indicates that the upstream genes of flavonoid biosynthesis may not be highly expressed in mature seeds. Higher expression levels of the Ft4CL gene were noted in leaves compared with the flowers and seeds and FtF3H expression was 1.36-fold higher in flowers in comparison with leaves. Notably, FtFLS was much more highly expressed in flowers and seeds, 107.2-fold and 11.8-fold, respectively, compared to leaves and FtUFGT and FtMYB31 transcripts were up to 2.1-fold and 16.7-fold higher in seeds (Fig. 3a). The correlations between rutin content and gene expression level were visualized by heatmap analysis. The rutin content was higher in flowers (50.15 mg/g FW) than grains (18.07 mg/g FW) and leaves (24.17 mg/g FW). Based on the hierarchical clustering analysis, the expression pattern of FtMYB31 clustered with Ft4CL, FtUFGT, and rutin content, with a correlation coefficient of − 0.68, 0.69, and 0.47, respectively. FtCHS, FtFLS, and FtF3H clustered in another group, with a correlation coefficient of − 0.96, 0.24, and 0.29, respectively, when compared with FtMYB31 (Fig. 3b).
Relative gene expression levels of flavonol biosynthesis-related gene and FtMYB31 in different tissues (flowers, leaves, and seeds) from tartary buckwheat (a). Hierarchical Clustering and correlation analysis of rutin content and expression pattern of flavonol biosynthesis pathway-related genes (b). All data were normalized with log2. *Indicated significant different at p < 0.05 according to Ducan’s multiple range test
Overexpression of FtMYB31 in Tobacco Strongly Induced Flavonol Accumulation and Upregulated Expression of Flavonol Biosynthesis-Related Genes
To understand whether the FtMYB31 gene regulated flavonol biosynthesis in vivo, we constructed a transient expression vector and infiltrated it into wild-type tobacco leaves and the visualized the flavonol compounds produced with DPBA staining under a fluorescence microscope (Fig. 4a). The DPBA was injected by sterile syringe near the vein easy for observation. No fluorescence signal was detected in the control leaves of wild-type tobacco infiltrated with water and the empty vector after 24 h. In comparison, strong orange fluorescence was observed close to the injection site of tobacco leaves infiltrated with 35S: FtMYB31. Qualitative detection visualized the change of flavonoid levels by flavonoid staining. However, due to the detection sensitivity, it is difficult to visualize the lower flavonoid levels in wild type. Flavonoid accumulation assays confirmed the lower levels of flavonoid in WT. The total flavonol content in the infiltrated leaves were higher than the control at 24 h and 48 h after injection, up to 5.5 and 5.6 mg/g, compared with injected water (2.1 mg/g) and EV (2.3 mg/g), respectively, and there was no significant change after injecting 72 h (Fig. 4b).
Flavonol accumulation stimulated by transient expression (visualized after 24-h injection) of FtMYB31 in tobacco leaves in vivo. a Flavonol staining in leaves of control and FtMYB31-infiltrated tobacco lines. Leaves were photographed after 1 h. Bar = 500 μm (Note: excitation at 488 nm provides excellent visualization of DPBA-flavonoid complexes). CK indicates plants injected with H2O, EV indicates plants injected with empty vector. b Total contents of flavonoid in transgenic tobacco lines after injecting for 24 h, 48 h, and 72 h
We then transformed tobacco with Agrobacteria containing 35S: FtMYB31 and transgenic lines were obtained. Several independent FtMYB31 transgenic tobacco lines were grown to the T3 generation (Fig. 5a), positive transformants were identified via PCR amplifications using gene-specific primers and two independent transgenic lines were chosen for determination of FtMYB31 expression level (Fig. 5b, c). The result showed a sustainable expression in OE 31-1 and OE 31-2 lines. Then, the content of total flavonol, quercetin, and rutin were measurement using both spectrophotometric and HPLC assays. The OE31-1 and OE31-2 transgenic lines showed a higher total flavone content than wild-type (WT) or empty vector (EV) plants, with approximately 7.23 and 7.86 mg/g FW, respectively. The quercetin content was 1.11 and 1.10 mg/g FW, respectively, and the rutin content was 2.56 and 2.78 mg/g FW, which is up to 78.1% and 74.5% higher, respectively, than in the wild type (Fig. 5s).
FtMYB31 transgenic tobacco lines causing accumulation of the flavonols content and enhancement of the rutin-related gene expression level. Transgenic FtMYB31 tobacco lines grow in the pot (a). Positive transgenic lines were screened by agarose gel (b). FtMYB31 gene expression in transgenic lines were identified by semiquantitative RT-PCR (c). Flavonoid contents (d) and gene expression patterns (e) in T3 transgenic FtMYB31 tobacco lines compared to the wild type
The expression of flavonol biosynthesis-genes and FtMYB31 in the two independent transgenic lines was measured by qRT-PCR (Fig. 5e). The result showed that NtCHS, NtF3H, and NtFLS were more highly expressed in OE 31-1 and OE 31-2 leaves compared to WT and EV transformed plants. In OE31-1 and OE31-2 transgenic tobaccos, NtCHS expressions were 1.14- and 1.22-fold higher than those in WT plants, NtF3H expressions were 1.96- and 2.47-fold higher in the same transgenic tobaccos, and NtFLS expressions were 2.49- and 2.93-fold greater than that found in the WT plants.
Discussion
Flavonoids are important secondary metabolites produced in tartary buckwheat, which have benefits for human health. However, the underlying molecular mechanisms regulating flavonol biosynthesis in different tissues of tartary buckwheat are still not clearly known. Here, we identified and cloned an R2R3-MYB homologous gene, FtMYB31, from tartary buckwheat by protein homology. Stracke et al. (2001) classified 126 R2R3-MYBs into 25 subgroups using protein sequence and motif analysis in Arabidopsis. The fifth subgroup factor MYB123 (Stracke et al. 2007) and the seventh subgroup factors PFG1/MYB12 (Mehrtens et al. 2005), PFG2/MYB11, and PFG3/MYB111 (Stracke et al. 2010) which function in flavonoid biosynthesis have been reported. In the present study, sequence analysis showed that FtMYB31 has a typical R2R3-conserved domain. Based on the phylogenic tree, it clustered with members of subgroup five of the R2R3-MYBs in Arabidopsis, which function as regulators of flavonol biosynthesis. Other studies have also found that R2R3- MYB TFs with similar functions cluster together in the same subgroup (Zhao et al. 2014; González et al. 2016) and extensive phylogenetic analysis of the R2R3-MYB proteins has been performed in model plants such as Arabidopsis, rice, and wheat (Chen et al. 2006; Katiyar et al. 2012). In the phylogenetic analysis, Arabidopsis MYB123/TT2, barley MYB31, and grapefruit MYBA1 clustered one clade. Zhou (2013) identified the TT2 gene from floral transcriptomic and suggested that TT2 controlled flavonoid metabolism in buckwheat, and VvMYBAP1 shown to control the flavonoid metabolism in grapefruit (Bogs et al. 2007). These genes are highly homologous, which supports the theory that FtMYB31 has a similar function to the genes in this clade.
We tested whether R2R3-MYB could regulate the accumulation of flavanols, especially rutin content in tartary buckwheat. We found that FtMYB31 gene expression level was correlated with the expression levels of CHS, 4CL, F3H, FLS, and also the rutin content in different tissues of tartary buckwheat. Previously, we have shown that UV-C, salicylic acid, and MeJA may enhance rutin biosynthesis and in parallel activate CHS, 4CL, F3H, and FLS genes in all tissues of tartary buckwheat (Sun et al. 2011, Hou et al. 2015). These results implied that the FtMYB31 gene may be involved in regulating rutin biosynthesis in tartary buckwheat. Transient and stable expressions in transgenic plants indicated that FtMYB31 regulated these genes related to the rutin synthesis pathway in tartary buckwheat. To date, nearly 23 MYB transcription factors from tartary buckwheat have been listed in the NCBI database. Eight of these TFs respond to multiple abiotic stressors and phytohormones; they have higher expression levels under NaCl, PEG, cold, or UV-B treatment and also play important roles in ABA, SA, and MeJA crosstalk involving multiple stress signaling pathways (Gao et al. 2016). A further two MYBTFs, FtMYB1 and FtMYB2, have been identified as R2R3-MYBs and are thought to regulate proanthocyanidin biosynthesis (Bai et al. 2014). Zhang et al. (2018) discovered that jasmonate-responsive MYB factors could inhibit the jasmonate signaling pathway, resulting in repression of rutin biosynthesis. These results enhance our understanding of the regulatory mechanism of rutin. In our study, the upstream genes of the flavonoid synthesis pathway, such as Ft4CL and FtCHS, showed higher expression level in leaves than the other organs. The central and downstream genes, such as FtF3H and FtFLS, showed higher expression levels in flowers, whereas the last gene leading to rutin biosynthesis, FtUFGT, showed the highest expression level in mature seeds. Our results demonstrate that FtMYB31 is specifically expressed in seeds of tartary buckwheat, and it can regulate flavonoid synthesis in tartary buckwheat and tabacoo.
Our results showed that FtMYB31 could positively regulate the flavonol synthase (FLS) gene, increasing flavonol biosynthesis in tartary buckwheat, and various transgenic tobacco lines expressing the FtMYB31 gene developed in this study accumulated significantly enhanced levels of total flavonols and rutin in leaf tissue. Previously, Pandey et al. (2012) reported that the expression of AtMYB12 in tobacco leads to a several-fold higher accumulation of flavonols in transgenic lines, compared to WT plants. Other reports have shown that overexpressing FtMYB15 and FtWD40 in transgenic tobacco increased the anthocyanin content and expression of corresponding biosynthetic genes (Luo et al. 2018). For example, the overexpression of FtWD40 increased anthocyanin content in tartary buckwheat, but not flavonol synthase (FLS) in the transgenic lines (Yao et al. 2017). The increase in flavonol content in the transgenic lines developed in this study suggests that FtMYB31 TF activates the flavonol biosynthesis branch of the pathway. Transcription factors have been demonstrated as efficient tools for metabolic engineering of this pathway, but often exhibit unexpected effects in heterologous systems relative to that in the homologous system. In the present study, FtMYB31 has been successfully expressed in tobacco in a heterologous system and its expression can enhance flavonoid biosynthesis. Although homologous expression systems are still quite challenging for tartary buckwheat, our work lays a foundation for genetically modifying it for the future improvement of nutrition values.
References
Aharoni A, De Vos CH, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP (2010) The strawberry famyb1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28(3):319–332
Ahmed A, Khalid N, Ahmad A, Abbasi NA, Latif MSZ, Randhawa MA (2014) Phytochemicals and bio-functional properties of buckwheat: a review. J Agr Sci 152(3):349–369
Bai YC, Li CL, Zhang JW, Li SJ, Luo XP, Yao HP, Chen H, Zhao HX, Park SU, Wu Q (2014) Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiol Plantarum 152(3):431–440
Bogs J, Jaffé FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143(3):1347–1361
Bovy A, Schijlen E, Hall RD (2007) Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): the potential for metabolomics. Metabolomics 3(3):399–412
Chen YH, Yang XY, He K, Liu MH, Li JG, Gao ZF, Lin ZQ, Zhang YF, Wang XX, Qiu XM, Shen YP, Li Z, Deng XH, Luo JC, Deng XW, Chen ZL, Gu HY, Qu LJ (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60(1):107–124
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15(10):573–581
Gao F, Yao HP, Zhao HX, Zhou J, Luo XP, Huang YJ, Li CL, Chen H, Wu Q (2016) Tartary buckwheat FtMYB10 encodes an R2R3-MYB transcription factor that acts as a novel negative regulator of salt and drought response in transgenic Arabidopsis. Plant Physiol Bioch 109:387–396
González M, Carrasco B, Salazar E (2016) Genome-wide identification and characterization of R2R3 MYB family in Rosaceae. Genomics Data 9:50–57
Gou XB, Jing-Man XU, Wang JX, Jiang Y, Han SY (2014) The protective effect of extract of rutin from buckwheat on renal injury of type 2 diabetes rats. China J Mod Med 24(7):7–10
Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P, gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76(3):543–553
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62(8):2465–2483
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple method for transferring genes into plants. Science 227:1229–1231
Hou SY, Sun ZX, Linghu B, Wang YG, Huang KS, Xu DM, Han YH (2015) Regeneration of buckwheat plantlets from hypocotyl and the influence of exogenous hormones on rutin content and rutin biosynthetic gene expression in vitro. Plant Cell Tiss Org 120(3):1159–1167
Jiang P, Burczynski F, Campbell C, Pierce G, Austria JA, Briggs CJ (2007) Rutin and flavonoid contents in three buckwheat species Fagopyrum esculentum, F. tataricum, and F. homotropicum, and their protective effects against lipid peroxidation. Food Res Int 40(3):356–364
Kalinová JP, Vrchotová N, Tříska J (2018) Contribution to the study of rutin stability in the achenes of tartary buckwheat (Fagopyrum tataricum). Food Chem 258:314–320
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13(1):1–19
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10(5):236–242
Kreft S, Strukelj B, Gaberscik A, Kreft I (2002) Rutin in buckwheat herbs grown at different UV-B radiation levels: comparison of two UV spectrophotometric and an HPLC method. J Exp Bot 53(375):1801–1804
Kreft I, Fabjan N, Yasumoto K (2006) Rutin content in buckwheat (Fagopyrum esculentum, Moench) food materials and products. Food Chem 98(3):508–512
Li Y, Chen M, Wang SL, Ning J, Ding XH, Chu ZH (2015) AtMYB11 regulates caffeoylquinic acid and flavonol synthesis in tomato and tobacco. Plant Cell Tiss Org 122(2):309–319
Liu B, Zhu Y (2007) Extraction of flavonoids from flavonoid-rich parts in tartary buckwheat and identification of the main flavonoids. J Food Eng 78(2):584–587
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 25:402–408
Logacheva MD, Kasianov AS, Vinogradov DV, Samigullin TH, Gelfand MS, Makeev VJ, Penin AA (2011) De novo sequencing and characterization of floral transcriptome in two species of buckwheat (Fagopyrum). BMC Genomics 12(1):1–30
Luo XP, Zhao HX, Yao PF, Li QQ, Huang YJ, Li CL, Chen H, Wu Q (2018) An R2R3-MYB transcription factor FtMYB15 involved in the synthesis of anthocyanin and proanthocyanidins from tartary buckwheat. J Plant Growth Regul 37(1):76–84
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138(2):1083–1096
Meng X, Yin B, Feng HL, Zhang S, Liang XQ, Meng QW (2014) Overexpression of R2R3-myb gene leads to accumulation of anthocyanin and enhanced resistance to chilling and oxidative stress. Biol Plantarum 58(1):121–130
Murphy A, Peer WA, Taiz L (2000) Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 211(3):315–324
Ohnishi O (1998) Search for the wild ancestor of buckwheat III. The wild ancestor of cultivated common buckwheat, and of tatary buckwheat. Econ Bot 52(2):123–133
Pandey A, Misra P, Chandrashekar K, Trivedi PK (2012) Development of AtMYB12 expressing transgenic tobacco callus culture for production of rutin with biopesticidal potential. Plant Cell Rep 31(10):1867–1876
Pandey A, Misra P, Trivedi PK (2015) Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep 34(9):1515–1528
Saito K, Yonekura-sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in arabidopsis: structural and genetic diversity. Plant Physiol Bioch 72:21–34
Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol 4(5):447–456
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50(4):660–677
Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Femie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188(4):985–1000
Sun ZX, Hou SY, Yang WD (2011) Effect of exogenous L-phentermine and UV-C on the accumulation of rutin compounds and the expression of rutin biosynthesis genes in Fagopyrum tartaricum. Sci Agric Sin 44(23):4772–4780
Suzuki T, Honda Y, Mukasa Y (2005) Effects of UV-B radiation, cold and desiccation stress on rutin concentration and rutin glucosidase activity in tartary buckwheat (Fagopyrum tataricum) leaves. Plant Sci 168(5):1303–1307
Xu Q, Yin XR, Zeng JK, Ge H, Song M, Xu CJ, Li X, Ferquson IB, Chen KS (2014) Activator-and repressor-type MYB transcription factors are involved in chilling injury induced flesh lignification in loquat via their interactions with the phenylpropanoid pathway. J Exp Bot 65:4349–4359
Yao PF, Zhao HX, Luo XP, Gao F, Li CL, Yao HP, Chen H, Park SU, Wu Q (2017) Fagopyrum tataricum FtWD40, functions as a positive regulator of anthocyanin biosynthesis in transgenic tobacco. J Plant Growth Regul 36(3):1–11
Yasui Y, Hirakawa H, Ueno M, Matsui K, Katsube-Tanaka T, Yang SJ, Aii J, Sato S, Mori M (2016) Assembly of the draft genome of buckwheat and its applications in identifying agronomically useful genes. DNA Res 23(3):215–224
Zhang K, Logacheva MD, Meng Y, Meng Y, Hu JP, Wan DP, Li L, Janovska D, Wang ZY, Georglew M, Yu Z, Yang FY, Yan M, Zhou ML (2018) Jasmonate-Responsive MYB factors spatially repress rutin biosynthesis in Fagopyrum Tataricum. J Exp Bot 69(8):1955–1966
Zhao P, Li Q, Li J, Wang L, Ren Z (2014) Genome-wide identification and characterization of R2R3MYB family in Solanum lycopersicum. Mol Genet Genomics 289(6):1183–1207
Zhou ML, Tang Y, Zhang KX, Li FL, Yang PY, Tang YX, Wu YM, Shao JR (2013) Identification of TT2 gene from floral transcriptome in Fagopyrum tataricum. Food Res Int 54(1):1331–1333
Acknowledgements
We would like to thank Professor Donald Grierson, University of Nottingham, UK, for discussions and help with the manuscript. This work was supported by the National Key R&D program of China (2017YFE0117600), Shanxi youth science and technology research fund (201801D221296), NSFC (No. 31301385), Research Project Supported by Shanxi Scholarship Council of China (2017-069), China Agriculture Research System (CARS-07-A1), and Shanxi key innovative platform for germplasm enhancement and molecular breeding in major crops (201605D151002).
Author information
Authors and Affiliations
Contributions
ZS, SH, and YH designed the experiments, analyzed the transcriptome data, and wrote the manuscript. BL carried out RNA extraction, cDNA synthesis, and gene cloning. RL and LW carried out expression vector construction and genetic transformation. RL and YH planted and collected plant materials. MZ, LL, and HL supervised the research and modified the manuscript. ZS and BL contributed equally. All the authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Z., Linghu, B., Hou, S. et al. Tartary Buckwheat FtMYB31 Gene Encoding an R2R3-MYB Transcription Factor Enhances Flavonoid Accumulation in Tobacco. J Plant Growth Regul 39, 564–574 (2020). https://doi.org/10.1007/s00344-019-10000-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-019-10000-7