Abstract
Tartary buckwheat (Fagopyrum tataricum Gaertn.) contains high concentration of flavonoids, which are mainly represented by rutin, anthocyanins, and proanthocyanidins. WD40 transcription factors (TFs) play significant roles in the transcriptional regulation of the anthocyanin biosynthetic pathway. In this study, a WD40-repeat protein gene (designated as FtWD40) was identified and characterized from tartary buckwheat. The bioinformatics analyses showed that the putative FtWD40 shared a high level of similarity with MtWD40-1, which is a positive regulator in anthocyanin biosynthesis of Medicago truncatula. The yeast one-hybrid assay indicated that FtWD40 had transcriptional activation activities. During florescence, FtWD40 was highly expressed in flowers compared to other organs. Furthermore, its overexpression in tobacco resulted in a remarkable deepening of petal pigmentation in flowers due to a significant increase in anthocyanins accumulation. Meanwhile, the expression of dihydroflavonol-4-reductase (DFR) and anthocyanin synthase (ANS) was upregulated 1.95- and 1.56-fold, respectively. In contrast, the expression level was lower for flavonol synthase (FLS) in the transgenic lines. To the best of our knowledge, this is the first functional characterization of a WD40 transcription factor (FtWD40) from tartary buckwheat that controls the anthocyanin pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tartary buckwheat (Fagopyrum tataricum Gaertn), an important nutrient-rich cereal crop with pharmacological values, is mostly grown in Asian countries (Park and others 2011). Tartary buckwheat has been recognized as a health-promoting food, because of its abundant nutrients and high levels of anti-oxidants, including rutin and anthocyanins (Li and others 2015). Anthocyanins, as a kind of flavonoids, have received extensive attention owing to their prospective health benefits, such as anti-mutagenic, anti-microbial, anti-inflammatory, anti-oxidant, and anti-hypertensive properties (Lai and others 2016; Lim and others 2016).
The anthocyanin biosynthetic pathway has been studied in multiple species (Grotewold 2006; Koes and others 2005). Anthocyanin biosynthesis is always in response to multiple stress factors such as light, salt, and temperature (Das and others 2012). Furthermore, the literature has shown that the response is largely attributed to the regulation of anthocyanin-related gene expression through transcription factors involving MYB, bHLH, and WD40 (Grotewold 2006; Lepiniec and others 2006; Lin-Wang and others 2010). The three transcription factors generally activate anthocyanin biosynthesis as a MBW complex, in which the WD40 protein usually stabilizes the complex through binding to the bHLH, and interacts with MYB in the complex (Hichri and others 2011). In addition, WD40 also can independently bind to its DNA targets related to anthocyanin biosynthesis (Dong and others 2014). TRANSPARENT TESTA GLABRA1 (TTG1), which belongs to the WD40-repeat protein family in Arabidopsis thaliana, has been identified to effectively promote anthocyanin accumulation by increasing the expression of ANS and DFR (Mehrtens and others 2005). Ectopic expression of the grapevine gene WDR1 enhances anthocyanin accumulation in shoots and rosette leaves, compared to wild-type plants in Arabidopsis (Matus and others 2010). In other cases, WD40 interacts with bHLHs or MYBs, and regulates the expression of anthocyanin-related genes as a crucial part of the MBW. In Arabidopsis, TTG1 interacts with both MYB TF TT2 and bHLH TF TT8 (Baudry and others 2004). However, in some other species, the WD40 protein has been shown to only interact with bHLH, but not with MYB (Dubos and others 2008; Grotewold and others 2000). Yeast two-hybrid (Y2H) and Bimolecular Fluorescence Complementation (BiFC) assays demonstrated that the apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation (An and others 2012). So, it is necessary to clone more WD40 genes, and characterize their function in the anthocyanins biosynthetic pathway of plants.
Recent studies of TFs in tartary buckwheat have mainly focused on MYBs and bHLHs. In our previous studies, FtMYB1/2 were found to control proanthocyanidin biosynthesis (Bai and others 2014). FtMYB3 inhibited anthocyanin synthesis in transgenic tobacco plants by decreasing the expression of chalcone isomerase (CHI), dihydroflavonol 4-reductase (DFR), and anthocyanidin synthase (ANS). In addition, FtbHLH1 was involved in anthocyanin biosynthesis of tartary buckwheat (unpublished). However, WD40 transcript factor is unknown in tartary buckwheat so far. To further study the mechanism of the single transcription factor or MBW complex in anthocyanin biosynthesis of tartary buckwheat, we turned our attention to the WD40 transcription factor family. In this study, a WD40-type gene, FtWD40, was firstly isolated and characterized from tartary buckwheat. We evaluated the expression profiles of the gene in tartary buckwheat. The results of stable transformation experiments in Nicotiana tabacum strongly indicated that FtWD40 plays a significant role in the anthocyanin biosynthesis pathway in tartary buckwheat. Possible mechanisms of how FtWD40 exerts biological effect in tobacco are discussed as well as its possible importance for plant metabolic engineering.
Materials and Methods
Plant Materials and Growth Conditions
About 500 seeds of tartary buckwheat seeds (“Xiqiao No.2”) were sown in the experimental field of Sichuan Agriculture University, Ya’an Sichuan, China, and the germination rate was above 90%. The tender roots, stems, leaves, and flowers were harvested separately during the florescence stage at 6 pm, and each sample was derived from 3 plants. All samples were frozen in liquid nitrogen immediately and kept at −80 °C for further use. Three independent biological replicates were measured for each sample. To detect the transcription level of the FtWD40 gene under different abiotic stress treatments, tartary buckwheat seeds were germinated in a growth chamber at 25 °C and approximately 60% humidity for 48 h in the dark and then grown in 1/2 Hoagland’s solution liquid medium with a 16 h light/8 h dark cycle at 25/22 °C (day/night) for two weeks. Wild-type and T1 transgenic tobacco (Nicotiana tabacum T12) were germinated in a growth chamber at 25 °C and approximately 60% humidity (16 h light/8 h dark).
Abiotic Stress Treatments
To examine whether FtWD40 expression is mediated by any external stresses, FtWD40 transcription levels in tartary buckwheat were examined after exposure to abiotic stresses that are known to promote anthocyanin accumulation (Shin and others 2016). Two-week-old tartary buckwheat seedlings were treated with the following conditions: 150 mM NaCl, 30% PEG-6000, 100 μM abscisic acid (ABA), 1 mM salicylic acid (SA), UV-B light (302 nm, 0.1 mW/cm2), and 4 °C (Su and others 2014; Zhou and others 2015), separately. For all the treatments, the seedlings (3 plants) were collected at 0, 6, 12, 24, and 48 h (0 h was non-treated control) and immediately frozen in liquid nitrogen for total RNA extraction. Three independent biological replicates were measured for each sample.
Cloning of FtWD40
The RNA was extracted from the various plant organs using an RNAout kit (Tiandz, China), and cDNA was synthesized with a RevertAid First Strand cDNA Synthesis kit (MBI, USA). The cDNA of flowers was used as a PCR template to amplify the gene of FtWD40 from tartary buckwheat. A pair of degenerate primers, WD40-S and WD40-A, was designed according to the conserved region of known WD40 genes from other plants, such as Arabidopsis thaliana and maize. To get the full-length cDNA sequence of FtWD40, rapid-amplification of cDNA ends (RACE) was performed using a Smart Race cDNA Amplification kit (Takara, Japan). Primers were used to obtain both ends of the FtWD40 gene as follow: F5-GSP, F5-NGSP1, and F5-NGSP2 for the 5′primer, and F3-GSP, F3-NGSP1, and F3-NGSP2 for the 3′primer. The Open Reading Frame (ORF) sequence of FtWD40 was obtained with primers WD40-F and WD40-R. The PCR products were subcloned into the pMD@19-T simple vector (Takara, Japan) and sequenced. All primer sequences are listed in Table S1.
Molecular Characterization of FtWD40
The sequence analysis of the FtWD40 gene was conducted using BLASTx and BLASTn programs in the National Center of Biotechnology Information (NCBI) database. The nucleotide and amino acid sequences were aligned using the Clustal X software. Phylogenetic analysis was performed using MEGA (Version 5).
Transactivation Assay
The transcription activation assay was performed in the yeast strain AH109 with LacZ and His reporter genes. The ORF sequence of the FtWD40 was ligated into the yeast expression vector pBridge (Biovector Inc., USA) after double digestion with EcoRI and BamHI. The pBridge empty plasmids and AH109 cells was used as a negative controls, and pBridge-GmMYBJ6 (Yang and others 2009) served as a positive control. The resulting plasmids were transformed into yeast AH109 cells using a lithium acetate-mediated method (Gietz and others 1992). The yeast transformants were verified by colony PCR and plated on the SD/-His-Trp medium for 3 d at 30 °C. Transcription activation was evaluated according to the growth status of yeast cells and 5-bromo-4-chloro-3-indolyl-β-D-galacto-pyranoside (X-gal). The primers (WDf, WDr) used in vector construction are listed in Table S3.
Stable Transformation in Tobacco
The plasmids used in the stable expression assay were constructed by ligating the ORF sequence of FtWD40 with pCAMBIA1301 using KpnI and BamHI. The recombinant plasmids and the pCambia1301 vector (as an internal control) were transformed into Nicotiana tabacum (T12) by utilizing the Agrobacterium-mediated leaf disk transformation method (Horsch and others 1985). Transgenic plants were cultured on Murashige and Skoog (MS) medium and screened in 1/2 MS medium containing hygromycin (50 mg L−1, w/v). Positive lines were selected and verified by PCR. The primers used in vector construction are listed in TableS3.
Analysis and Determination of Total Anthocyanin Content in Plant Organs
All fresh samples, including transgenic tobaccos and tartary buckwheat, were homogenized in liquid nitrogen and analyzed according to the reported method (Rabino and Mancineli 1986). 1 mL of acidic methanol (1% HCl, v/v) was added to 200 mg of fresh plant material. The samples were incubated for 18 h at room temperature under moderate shaking. The extracts were diluted with an equal volume of water, and then an equal volume of chloroform was added to the extract. The samples were vortexed gently for a few seconds followed by centrifugation for 5 min at 15,000×g. The aqueous phase was used for the determination of absorbance at 530 nm and 657 nm. The anthocyanins were quantified using the following equation: QAnthocyanins=(A530 −0.25 × A657) × M− 1, where QAnthocyanins is the amount of anthocyanin, A530 and A657 are the absorptions at the indicated wavelengths, and M is the weight of the plant material used for extraction [g]. Three independent biological replicates were measured for each sample.
Quantitative Real-Time PCR (qRT-PCR) Analysis
The qRT-PCR analysis was performed to confirm the FtWD40 expression in tartary buckwheat under abiotic stresses and to analyze the expressions of anthocyanin-related genes in transgenic tobacco. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA) from samples. The RNA was then converted into first-strand cDNA using ReverTra Ace (Toyobo, Osaka, Japan) with a random hexamer. Transcription levels of FtWD40 and six anthocyanin-related genes, including phenylalanine ammonialyase (PAL, GenBank ID: AB289452), chalcone synthase (CHS, GenBank ID: AF311783), anthocyanidin synthase (ANS, GenBank ID: EB427369), flavonoid3′-hydroxylase (F3′H, GenBank ID: AB289449), dihydroflavonol 4-reductase (DFR, GenBank ID:EF421429), and flavonol synthase (FLS, GenBank ID: AB289451), were analyzed using qRT-PCR. The primers for qRT-PCR were designed by the Primer Premier 5 software based on obtained FtWD40 and other gene cDNA sequences (Table S2). The gene FtH3 (Histone 3, GenBank ID: HM628903) and β-actin (GenBank ID: AB158612) were used as the reference genes in tartary buckwheat and tobacco, respectively. qRT-PCR was performed with a SYBR Premix EX Taq Kit (TaKaRa, Japan) in a total reaction volume of 20 μL, which contained 10 μL of SYBR Green Mix, 0.5 μM of each primer, and 1 μL of cDNA. The reactions were carried out in a CFX96 Real-Time PCR thermocycler (Bio Rad, USA) according to the manufacturer’s protocol. The PCR protocol was as follows: 95 °C for 5 min, followed by 39 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. The data were evaluated using the 2−ΔΔCT method (Livak and Schmittgen 2001). Three independent biological replicates were measured for each sample.
Statistical Analyses
Three independent biological replicates were measured for each sample and the data presented as the mean ± standard error (SE). Where applicable, data were analyzed by Student’s t test in a two-tailed analysis. Values of P < 0.05 or <0.01 were considered to be statistically significant.
Results
Cloning of FtWD40 from F. tataricum
Our aim was to isolate WD40 transcription factors (TFs) that may be involved in flavonoid biosynthesis in F. tataricum. Using homology cloning and RACE technology, full-length cDNA sequences of FtWD40 (1097 bp, GenBank ID: KX059426) were characterized from flowers of F. tataricum, which contained a 1035 bp ORF and encoded a protein of 344 amino acids. Sequence alignments with the NCBI database showed the FtWD40 protein was a member of the WD40 family.
Molecular Characterization of FtWD40
The amino acid sequence of FtWD40 was aligned with previously reported WD40 proteins. The majority of the homology was observed within the WD40 repeat motifs, normally a 40-amino acid tandem repeat characterized by Gly-His (GH) and Trp-Asp (WD) doublet residues (Van Nocker and Ludwig 2003). The four hypothetical WD40 repeat motifs were found in the predicted amino acid sequence of FtWD40 (Fig. 1).
Alignment of FtWD40 with related WD40 proteins by Clustal X: AtTTG1 (NM_122360) from Arabidopsis thaliana, VvWD (NP_001268006) from Vitis vinifera, PfWD (BAB58883) from Perilla frutescens, PhAN11 (AAC18914) from Petunia x hybrida, GhTTG1 (AAM95641) from Gossypium hirsutum, and MtWD40-1 (ABW08112) from Medicago truncatula. The WD40 conserved domains were indicated with black boxes
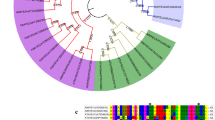
A phylogenetic tree was drawn to establish the biological functions and evolutionary relationships between FtWD40 and other known WD40s (Fig. 2). The dendrogram was classified into two clusters, the PALE ALEURONE COLOR1 (PAC1) and the MP1 clades. Mapping experiments have positioned Mp1 to the long arm of chromosome 5 close to Pac1. Analysis of the Mp1 sequences revealed a lack of a nuclear localization signal, suggesting that it might encode a cytosolic protein. Meanwhile, the Pac1 was involved in anthocyanin accumulation (Hernandez and others 2000). Phylogenetic analysis showed that FtWD40 is highly homologous to AtTTG1, PfWD40, PhAN11, and VvWDR1, which contribute to regulating anthocyanins.
Phylogenetic relationships between FtWD40 and WD40s from Fagopyrum tataricum and other plant species. The GenBank accession numbers are as follows: PfWD40 (BAB58883) from Perilla frutescens, PhAN11 (AAC18914) from Petunia x hybrida, VvWDR1 (ABF66625) from Vitis vinifera, PsWD40-2 (AIU98520) from Paeonia suffruticosa, AtTTG1 (NP_197840) from Arabidopsis thaliana, PgWD40 (ADV40946) from Punica granatum, MtWD40-1 (ABW08112) from Medicago truncatula, LjTTG1 (BAH28880) from Lotus japonicus, ZmPAC1 (AAM76742) from Zea mays, ZmMP1 (AAR01949) from Zea mays, IpWD40 (BAE94397) from Ipomoea purpurea, AtAN11 (AAC18912) from Arabidopsis thaliana, MiWD40 (CAE76645) from Matthiola incana, AtWD1 (NP_172751) from Arabidopsis thaliana, PpWD40-1 (ADN52336) from Pyrus pyrifolia, RcLWD1 (XP_002512788) from Ricinus communis, VvWDR2 (NP_001268006) from Vitis vinifera, PsWD40-1 (AIU98519) from Paeonia suffruticosa. FtWD40 were highlighted in red. (Color figure online)
Expression Pattern of FtWD40 in Various Tissues
To verify whether there is a relationship between FtWD40 expression and anthocyanin accumulation in different tissues, the expression pattern of FtWD40 was analyzed in four organs of tartary buckwheat using qRT-PCR. Tissues, including roots, stems, leaves, and flowers, were harvested separately from the whole mature plants. During florescence, the expression level of FtWD40 was the highest in flowers, moderate in roots, and lowest in leaves and stems. The level of FtWD40 transcription in flowers was 1.2-, 2.1-, and 1.9-fold higher than that in roots, stems, and leaves, respectively (Fig. 3a). In addition, the anthocyanin content analysis showed a similar profile and the expression of FtWD40 in various tissues (Fig. 3b).
a Relative expression level of FtWD40; b Total anthocyanin accumulation in different tissues. FtH3 was used as an internal control. The expression data were normalized to leaf (set at 1) using the comparative threshold cycle method. Error bars represent the standard deviation of triplicate runs for qRT-PCR. The values shown with different lowercase letters within different organs are significantly different (Tukey test: P < 0.05)
Transcriptional Analysis of the FtWD40 Gene Under Different Stresses
The result of qRT-PCR showed substantial differences of FtWD40 transcript abundance in response to multiple stresses except SA treatment (Fig. 4). Under 4 °C and NaCl stresses, the transcription of FtWD40 continuously increased after 12 h and reached the maximum level at 24 to 48 h. For UV-B and PEG-6000 treatment, FtWD40 expression showed no obvious change in 12h, but a remarkable increase was detected from 24 h. For ABA treatment, the highest activation was detected after 12 h of treatment. SA stress had no obvious effect on the transcription of FtWD40. On the whole, NaCl, ABA, and low temperature had more pronounced effects on FtWD40 expression than other treatments.
Expression analysis of FtWD40 gene in tartary buckwheat seedlings under ABA, SA, 4 °C, UV-B, NaCl, and drought treatments. The mRNA expression patterns were examined with a qRT-PCR assay. The 2− ΔΔCT method was used to determine the relative expression, and the expression levels of genes in 0 h (no treated) were set to “1”. FtH3 was used as a housekeeping gene. Each value represents the mean of three replicates, and error bars indicate standard deviations (± SD). *P < 0.05 and **P < 0.01 indicate significant differences between the control and stressed tartary buckwheat
Transcription Activation of FtWD40
The transcriptional activity of FtWD40 was analyzed using a yeast assay system. The ORF of FtWD40 was cloned into the pBridge vector and then transformed into yeast AH109. Yeast cells containing pBridge-FtWD40 and pBridge-GmMYBJ6 grew well in SD/-Trp/-His medium, whereas cells containing pBridge did not grow (Fig. 5a). Meanwhile, there was no color change in the negative control, and the positive control and pBridge-FtWD40 had a significant blue reaction in the presence of X-gal (Fig. 5b).
Transactivation analysis of FtWD40 in yeast. a Before the galactosidase filter lift assay. b After the galactosidase filter lift assay. WD: pBridge-FtWD40; PC (Positive Control): pBridge-GmMYBJ6; NC1 (Negative Control): pBridge pasmid ; NC2 (Negative Control): AH109. Fusion proteins of pBridge-FtWD40, pBridge-GmMYBJ6 and pBridge were expressed in yeast strain AH109. The transformants were streaked on the SD/Trp−/His− medium. The plates were incubated for 3 d and subjected to X-gal assay
Identification of Transgenic Tobacco Lines
The ORF of FtWD40 was ectopically expressed in tobacco using the binary vector pCAMBIA1301-FtWD40 (Fig. 6a). Seven independent transgenic tobacco lines that overexpressed the FtWD40 gene were obtained from Hyg-resistance selection and were cultured under the same conditions (Supplemental Fig. 1). PCR analysis confirmed the presence of the transformed FtWD40 gene in all transgenic lines, and the absence of endogenous FtWD40 in wild-type (WT) and control check (CK, plant was transformed with empty pCAMBIA1301 vector) tobacco plants (Fig. 6b). The qRT-PCR analysis showed that the expression level of FtWD40 was significantly higher in the transgenic plants, especially #2, #5, and #7, than in CK (Fig. 6c). Therefore, the three lines were used for further experiments.
Molecular analyses of the FtWD40-overexpressing tobacco plants. a Schematic diagram of the T-DNA region of the binary plasmid pCAMBIA1301-FtWD40. LB left border; 35S T, CaMV 35S terminator; hpt II, hygromycin phosphotransferase II gene; 35S, cauliflower mosaic virus (CaMV) 35S promoter; NOS T, nopaline synthase terminator; FtWD40, tartary buckwheat WD40 transcription factor gene; gusA, β-glucuronidase gene; RB, right border. b PCR analysis of transgenic plants. Lane M DL2000 DNA marker; Lane W water as negative control; Lane P plasmid pCAMBIA1301-FtWD40 as positive control; Lane CK wild-type; Lanes #1-#7 transgenic plants. c Expression analysis of the FtWD40 gene in transgenic tobacco plants and CK. The tobacco Ntaction gene was used as an internal control. Data are presented as mean ± SD (n = 3). * and ** indicate a significant difference from that of WT at P < 0.05 and <0.01, respectively, by Student’s t test
Ectopic Expression of FtWD40 Enhances Anthocyanin Accumulation in Transgenic Tobacco
Interestingly, there is no substantial difference in morphology between the transgenic plants and wild-type. However, transgenic tobacco lines harboring FtWD40 were affected in flower color, with a remarkable deepening of petal pigmentation compared with the CK tobacco (Fig. 7a). This change in the corolla color of transgenic tobacco was already visible prior to anthesis. To confirm that the deeper flower color was attributed from an increased pigment levels synthesized from the anthocyanin pathway, the total anthocyanin was determined. In the transgenic tobacco lines #2, #5, and #7, anthocyanin contents were improved to 3.38-, 2.85-, and 3.25-fold, respectively, as compared to the CK (Fig. 7b). Notably, the expression of FtWD40 in transgenic tobacco correlated with the pigmentation enhancement observed in the petals.
a Floral phenotypes of transgenic tobacco plants expressing tartary buckwheat FtWD40. b Quantification of total anthocyanins in transgenic tobacco seedling leaves. The CK plant was transformed with empty pCambia1301 vector. #-n (n represents the line number) transgenic lines were transformed with pCambia1301-FtWD40 vector. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). ** indicate a significant difference from that of CK at p < 0.01, by Student’s t-test
Expression of Genes Involved in Anthocyanin Biosynthesis in Transgenic Tobacco
To determine whether the gene expression could potentially account for the remarkable increase of pigmentation in flowers, the expression levels of anthocyanin-related genes were evaluated using qRT-PCR with NtActin as an internal control. It showed that these genes can be grouped into three categories in terms of their responsiveness to FtWD40 (Fig. 8 ). The first category, phenylalanine ammonialyase (PAL), chalcone synthase (CHS) and flavonoid 3′-hydroxylase (F3′H), showed an inconspicuous correlation to the FtWD40 expression. The expression of the three genes was similar to the CK. The effect of the constitutive FtWD40 expression was more pronounced for dihydroflavonol 4-reductase (DFR) and anthocyanidin synthase (ANS). A distinct increase (approximately 95%) in the transcript of DFR was observed and, to a lesser extent, a slighter increase (approximately 56%) in the latter gene. The single member of the third category is the flavonol synthase (FLS), whose expression was markedly repressed. The data indicated that only two genes (DFR and ANS) from the lower end of the flavonoid pathway, more directly related to the biosynthesis of anthocyanins, were dramatically FtWD40 responsive, and the alteration of DFR was most drastic.
Analysis of anthocyanin-related gene expression levels in FtWD40 transgenic tobacco seedling leaves. The transcripts of six anthocyanin biosynthetic genes were detected by qRT-PCR from transgenic lines and compared with control lines. The 2−ΔΔCT method was used to determine the relative expression, and the expression levels of genes in CK were set to “1”. β-actin was used as a housekeeping gene. Each value represents the mean of three replicates, and error bars indicate standard deviations (±SD). The CK plant was transformed with empty pCambia1301 vector. #-n (n represents the line number) transgenic lines were transformed with pCambia1301-FtWD40 vector. *P < 0.05 and **P < 0.01 indicate significant differences between control and transgenic tobacco
Discussion
In this study, we isolated FtWD40 encoding a WD-repeat protein that is possibly involved in the regulation of anthocyanin biosynthesis. The amino acid sequence of FtWD40 showed a high identity with other proteins from dicotyledonous plants, such as PhAN11, GhTTG1, AtTTG1, and PFWD (Table S4). Through analyzing the FtWD40 amino acid sequence, we observed the presence of four predicted WD repeats that are remarkably well conserved among plants, which suggested that FtWD40 is a highly conserved protein in the process of evolution. In addition, we detected a relationship between anthocyanin accumulation and FtWD40 expression. FtWD40 was ubiquitously expressed in all tissues examined, and the expression level was significantly correlated with the anthocyanin content in various organs. These results suggested that FtWD40 might be involved in the metabolism of anthocyanins in tartary buckwheat as a transcriptional activator. The majority of the WD40 genes were responsive to abiotic stresses such as salinity, drought, and low temperature. BnSWD1, which is a novel WD40 repeat-containing protein, was upregulated after treatment with abscisic acid, salicylic acid, and methyl jasmonate (Lee and others 2010). Similarly, FtWD40 was highly sensitive to abiotic stresses, suggesting that it had a protective role via regulation of anthocyanin biosynthesis (Dong and others 2014). Indeed, tartary buckwheat benefits from the anthocyanin accumulation, which protects seedlings from UV radiation, temperature change, and drought (Tsurunaga and others 2013). The phylogenetic tree placed FtWD40 into dicotyledon subgroups involved in anthocyanin biosynthesis. So, we propose that FtWD40 is a positive regulator of anthocyanin biosynthesis in tartary buckwheat.
Three major transcription factor families, including MYB, bHLH, and WD40, generally activate anthocyanin biosynthesis as a part of the MBW complex (Espley and others 2007; Lin-Wang and others 2014). It is worth noting that WD40 is an important part of the WD40/MYB/bHLH complex and interacts with bHLHs or MYBs to regulate anthocyanin or proanthocyanidin biosynthesis (Gonzalez and others 2008; Qi and others 2011; Ramsay and Glover 2005; Schaart and others 2013). In Arabidopsis, functional analysis of TTG1/WDR, AtMYB123/MYB, and AtbHLH42/bHLH indicated that these three proteins synergistically control anthocyanin accumulation (Mehrtens and others 2005). In petunia, the transcription factors AN2 and JAF13 or AN1 are thought to positively regulate the anthocyanin biosynthetic pathway (Albert and others 2011). Nevertheless, our results showed that overexpression of the FtWD40 gene can improve anthocyanin accumulation in N. tabacum without co-transferring other factors, which is consistent with PFWD from Perilla frutescens (Sompornpailin and others 2002). In addition, the overexpression of IbWD40 increased the expression of the anthocyanin biosynthesis-related gene (Dong and others 2014). Thus, overexpression of FtWD40 alone could also improve the synthesis of plant anthocyanins.
Furthermore, transcription factors control the synthesis of anthocyanin by regulating the expression of downstream target genes. The expression of early and late biosynthetic genes (EBGs and LBGs, respectively) appears to be regulated separately by different TFs (Dubos and others 2010; Nesi and others 2001). We examined the effect of FtWD40 on anthocyanin-related genes in transgenic tobacco. Similarly, our result indicated that there was little or no enhancement of the earlier genes. However, FtWD40 not only upregulated the expression of NtDFR and NtANS but also reduced the NtFLS. It is generally known that DFR and FLS play an important role in anthocyanin and flavonol biosynthesis; DFR and FLS compete for the same substrate, dihydroflavonol (Davies and others 2003). This being so, FtWD40 increased DFR expression and suppressed FLS expression, which might switch biosynthesis from dihydroflavonol to anthocyanin. When we ectopically expressed FtWD40 in tobacco, the seedlings of transgenic lines accumulated more anthocyanins than the control plant (Fig. 6b). Simultaneously, the color of the flowers obviously deepened. Our results are similar to those of previous studies, in which overexpression of the proanthocyanidins (PAs) structural gene ANRs resulted in a visible decrease in flower color in tobacco (Han and others 2012). However, PAP1(MYB) overexpression can affect the transcription of all genes involved in the phenylpropanoid pathway (Tohge and others 2005). Further research is necessary to reveal the differences among different plant species.
Taken together, whether at the transcriptional level or metabolic level, evidence from our transgenic tobacco experiments showed that transcription factor FtWD40 could effectively increase anthocyanin accumulation in a heterogonous host. Because of the health-promoting properties and the economic market for anthocyanins in human health, there is a growing interest in the development of functional foods rich in anthocyanins (Zhang and others 2012). Furthermore, recent research has suggested that transcription factors, which have the potential ability to regulate multiple structural genes, are more effective compared to single structural genes for plant metabolic engineering (Aharoni and Galili 2011). In this respect, owing to their potential to regulate anthocyanin accumulation, our data may provide new clues and potential opportunities for increasing the commercial value of tartary buckwheat by metabolic engineering of anthocyanins.
References
Aharoni A, Galili G (2011) Metabolic engineering of the plant primary secondary metabolism interface. Curr Opin. Biotech 22:239–244
Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM (2011) Members of an R2R3 - MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J 65:771–784
An XH, Tian Y, Chen KQ, Wang XF, Hao YJ (2012) The apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J Plant Physiol 169:710–717
Bai YC, Li CL, Zhang JW, Li SJ, Luo XP, Yao HP, Chen H, Zhao HX, Park SU, Wu Q (2014) Characterization of two tartary buckwheat R2R3-MYB transcription factors and their regulation of proanthocyanidin biosynthesis. Physiol Plantarum 152:431–440
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Das PK, Shin DH, Choi S-B, Park Y-I (2012) Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells 34:501–507
Davies KM, Schwinn KE, Deroles SC, Manson DG, Lewis DH, Bloor SJ, Bradley JM (2003) Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 131:259–268
Dong W, Niu L, Gu J, Gao F (2014) Isolation of a WD40-repeat gene regulating anthocyanin biosynthesis in storage roots of purple-fleshed sweet potato. Acta Physiol Plant 36:1123–1132
Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L (2008) MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J 55:940–953
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20:1425
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J 53:814–827
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780
Grotewold E, Sainz MB, Tagliani L, Hernandez JM, Bowen B, Chandler VL (2000) Identification of the residues in the Myb domain of maize C1 that specify the interaction with the bHLH cofactor R. Proc.Natl Acad Sci 97:13579–13584
Han Y, Vimolmangkang S, Soria-Guerra RE, Korban SS (2012) Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J Exp Bot 65:2437–2447
Hernandez J, Pizzirusso M, Grotewold E (2000) The maize Mp1 gene encodes a WD-repeat protein similar to An11 and TTG. Maize Genet Coop Newsl 74:24.
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Horsch RB, Fry J, Hoffmann NL, Eichholtz DA, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Lai B, Du LN, Liu R, Hu B, Su WB, Qin YH, Zhao JT, Wang HC, Hu GB (2016) Two LcbHLH transcription factors interacting with LcMYB1 in regulating late structural genes of anthocyanin biosynthesis in Nicotiana and Litchi chinensis during anthocyanin accumulation. Front Plant Sci 7:166
Lee S, Lee J, Paek KH, Kwon SY, Cho HS, Kim SJ, Park JM (2010) A novel WD40 protein, BnSWD1, is involved in salt stress in Brassica napus. Plant Biotechnol Rep 4:165–172
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Li SJ, Bai YC, Li CL, Yao HP, Chen H, Zhao HX, Wu Q (2015) Anthocyanins accumulate in tartary buckwheat (Fagopyrum tataricum) sprout in response to cold stress. Acta Physiol Plantarum 37: 159–167
Lim SH, Song JH, Kim DH, Kim JK, Lee JY, Kim YM, Ha SH (2016) Activation of anthocyanin biosynthesis by expression of the radish R2R3-MYB transcription factor gene RsMYB1. Plant Cell Rep 35:641–653
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC (2010) An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol 10:50
Lin-Wang K, McGhie TK, Wang M, Liu Y, Warren B, Storey R, Espley RV, Allan AC (2014) Engineering the anthocyanin regulatory complex of strawberry (Fragaria vesca). Front Plant Sci 5:651
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆CT method. Methods 25:402–408
Matus J, Poupin M, Cañón P, Bordeu E, Alcalde J, Arce-Johnson P (2010) Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol Biol Rep 72:607–620
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138:1083–1096
Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13:2099–2114
Park NI, Li X, Suzuki T, Kim SJ, Woo SH, Park CH, Park SU (2011) Differential expression of anthocyanin biosynthetic genes and anthocyanin accumulation in tartary buckwheat cultivars ‘Hokkai T8’and ‘Hokkai T10’. J Agr Food Chem 59:2356–2361
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D (2011) The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23:1795–1814
Rabino I, Mancinelli AL (1986) Light, temperature, and anthocyanin production. Plant Physiol 81:922–924
Ramsay NA, Glover BJ (2005) MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Schaart JG, Dubos C, Romero De La Fuente I, van Houwelingen AM, de Vos RC, Jonker HH, Xu W, Routaboul JM, Lepiniec L, Bovy AG (2013) Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol 197:454–467
Shin DH, Choi MG, Kang CS, Park CS, Choi SB, Park YI (2016) A wheat R2R3-MYB protein PURPLE PLANT1 (TaPL1) functions as a positive regulator of anthocyanin biosynthesis. Biochem Bioph Res Co 469:686–691
Sompornpailin K, Makita Y, Yamazaki M, Saito K (2002) A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol Biol 50:485–495
Su LT, Li JW, Liu DQ, Zhai Y, Zhang HJ, Li XW, Zhang QL, Wang Y, Wang QY (2014) A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 538:46–55
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima Ji, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Tsurunaga Y, Takahashi T, Katsube T, Kudo A, Kuramitsu O, Ishiwata M, Matsumoto S (2013) Effects of UV-B irradiation on the levels of anthocyanin, rutin and radical scavenging activity of buckwheat sprouts. Food Chem 141:552–556
Van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4:50
Yang W, Wu Y, Tang Y (2009) Expressing and functional analysis of GmMYBJ6 from soybean. Yi Chuan 31:645–653
Zhang ZL, Zhou ML, Tang Y, Li FL, Tang YX, Shao JR, Xue WT, Wu YM (2012) Bioactive compounds in functional buckwheat food. Food Res Int 49:389–395
Zhou M, Wang C, Qi L, Yang X, Sun Z, Tang Y, Tang Y, Shao J, Wu Y (2015) Ectopic expression of Fagopyrum tataricum FtMYB12 improves cold tolerance in Arabidopsis thaliana. J Plant Growth Regul 34:362–371
Acknowledgements
This work was supported by the Science and Technology Department of Sichuan Province, PR China (2015HH0047).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yao, P., Zhao, H., Luo, X. et al. Fagopyrum tataricum FtWD40 Functions as a Positive Regulator of Anthocyanin Biosynthesis in Transgenic Tobacco. J Plant Growth Regul 36, 755–765 (2017). https://doi.org/10.1007/s00344-017-9678-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-017-9678-6