Abstract
Although the Galapagos are famous for their unique biodiversity, many groups of marine invertebrates from this isolated archipelago remain understudied or not investigated. One such group is the zoanthids (Order Zoantharia, =Zoanthidea, =Zoanthiniaria), anthozoans (Cnidaria) found in marine ecosystems worldwide. Zoanthid taxonomy has been in a state of disorganization and neglect due in large part to the morphological plasticity within species and questions about the accuracy of traditionally used morphological and ecological characteristics. However, recent studies utilizing molecular methodology combined with morphology have proven to be very useful in understanding zoanthid diversity. The results of a survey of zoanthids from the Galapagos and the east Pacific are reported in this study. Shallow water (<35 m) zoanthid specimens were identified using the molecular markers mitochondrial 16S ribosomal DNA (mt 16S rDNA), cytochrome oxidase subunit I (COI) gene, and the internal transcribed spacer of ribosomal DNA (ITS-rDNA). From the collected specimens seven putative zoanthid species-level clades from three known genera (Zoanthus, Palythoa, Parazoanthus) were identified at the molecular level. These identifications were further supported by morphological and ecological data. While almost all specimens belonged to known zoanthid genera, based on unique molecular and ecological data one group of specimens (designated unknown zoanthid sp. “03-103”) is potentially a novel undescribed genus. Additionally, the remaining three azooxanthellate Parazoanthus clades may also be undescribed new species, but due to the overall lack of zoanthid research and descriptions from neighboring areas (East Pacific, west coast of South America) further research is needed to clearly ascertain this. Additionally, notes on the four observed nominal azooxanthellate zoanthid species and a key to all eight nominal (seven from known genera, one from a potentially new genus) shallow water zoanthid species found thus far in the Galapagos Islands are provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Galapagos archipelago, straddling the equator off the western coast of South America and positioned in an unusual confluence of warm and cold currents, is widely recognized as a natural wonder for its remarkable endemic fauna and flora set in a primeval landscape. Although biological interest in the Galapagos has focused on the terrestrial environment (i.e., Darwin 1859), the less-explored marine ecosystem surrounding the islands is equally exceptional (Bustamante et al. 2000, 2002). Because of the high level of interest in these celebrated islands and the many scientific expeditions sent to the area, the marine fauna of Galapagos is better known than that of any other eastern Pacific oceanic island as well as the inshore fauna of coastal Ecuador and Colombia.
In general, the Galapagos can be described as a depauperate outpost of the Panamic Province. While diversity is high for groups with good dispersal potential, such as hydroids, bryozoans, cirripedians, and caridean shrimps, it is low for many other groups, such as molluscs, echinoderms, benthic polychaetes, hermatypic corals, and porcelain crabs. The natural barriers to colonization that have limited diversity have also contributed to high levels of endemism in many marine groups (Bustamante et al. 2000, 2002). These groups include polychaetes, brachyurans, caridean shrimps, barnacles, molluscs, echinoderms, bryozoans, sea anemones, gorgonians, hydroids, and ahermatypic corals (Bustamante et al. 2002).
Of the cnidarian fauna, groups that have received comprehensive study include the hydroids (Calder et al. 2003), sea anemones and cerianthids (Fautin et al. 2007), hermatypic corals (Glynn 2003), ahermatypic corals (Cairns 1991, 2001), and gorgonians in part (Williams and Breedy 2004; Breedy and Guzman 2005, 2007; Breedy et al. in press). However, most hexacorallian cnidarians remain largely unstudied. Zoanthids in particular were not researched by any of a series of marine expeditions to Galapagos in the nineteenth and twentieth centuries, and do not appear in any resulting publications, including the extensive series of the Allan Hancock Expeditions.
Although found in almost all marine ecosystems, zoanthids remain relatively unknown. While most families and genera are well established, the level of species diversity in this order remains unknown. Fautin (2007) lists 333 nominal species, but the true number of species may change due to the potential for cryptic (see Palythoa mutuki group in Reimer et al. 2006c) and/or undiscovered species (see Reimer et al. 2007a) or due to inadvertent re-description of varying morphotypes within species groups (see Zoanthus sansibaricus in Reimer et al. 2004). Identification of zoanthids to species level is often made difficult by their relatively plastic morphology (colony size, polyp shape, color, etc.) and relative lack of measurable morphological diagnostic characteristics. Zoanthids currently include five families, two primarily zooxanthellate (Zoanthidae, Sphenopidae) and three largely or completely azooxanthellate (Parazoanthidae, Epizoanthidae, Abyssoanthidae).
Recent investigations using molecular phylogenetic methods have begun to reorganize the classification of some zoanthid groups. Molecular methods have allowed a re-examination of both zoanthid species (Reimer et al. 2006a, b) and genera (Reimer et al. 2006c), and aided in the identification of new zoanthid families (Reimer et al. 2007a).
During the preparation of the Galapagos Marine Life Series, the marine fauna of the Galapagos has, in recent years, begun to be investigated and catalogued. The Series now includes echinoderms (Hickman 1998), marine molluscs (Hickman and Finet 1999), and crustaceans (Hickman and Zimmerman 2000). Forthcoming in this series is a field guide to the corals and other benthic cnidarians of Galapagos; it will include, in addition to both hermatypic and ahermatypic corals, anemones, zoanthids, gorgonians, and hydroids. As part of an effort to clarify the diversity of cnidarians of the Galapagos Islands, numerous collected samples of zoanthids were examined both morphologically and molecularly (using mt 16S rDNA, COI and internal transcribed spacer of ribosomal DNA [ITS-rDNA]). Preliminary results of collected zooxanthellate zoanthids in the Galapagos from the genera Zoanthus and Palythoa have recently been presented elsewhere (Reimer and Hickman in press). In this study, molecular data on zoanthids from the Galapagos are reported. Additionally, morphological characteristics and ecological notes of all the observed Galapagos zoanthid genera and species are provided, and a morphological dichotomous key is provided to assist in field identification.
Materials and methods
Specimen collection
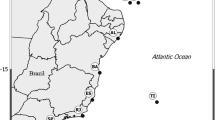
Specimens of zoanthids were collected by hand intertidally or by SCUBA diving from sites at most major islands in the Galapagos (Fig. 1—in bold) between June 2001 and December 2004 (Table 1). Additional sampling was conducted in March 2007 (Fig. 1—in non-bold), but collected specimens were not made available in time for genetic investigation. As samples were collected in situ, photographs were taken to assist in identification and for collection of morphological data (oral disk/polyp diameter, color, polyp form. etc.). After collection specimens were examined and again photographed alive, then stored in 75% alcohol at ambient temperature.
Map of sampling locations in the Galapagos and specimens collected. Locations are indicated with filled target symbols and location names in italics. Other geographic features (islands, etc.) are in regular font. Genera collected from locations are in parentheses after location names. Abbreviations: Zs = Zoanthus cf. sansibaricus, Zv = Z. cf. vietnamensis, Pm = Palythoa cf. mutuki, Pt = P. cf. tuberculosa, A = Parazoanthus sp. G1, P = Parazoanthus sp. G2, H = Parazoanthus sp. G3, u = unknown zoanthid 03-103. Locations and specimen abbreviations in bold indicate specimens for which DNA data are available
Sample nomenclature
Samples were assigned names based on sample year and an assigned sampling number (Table 1). Thus, sample 02-27 is sample 27 from 2002.
DNA extraction, PCR Amplification, cloning, and sequencing
DNA was extracted from samples weighing 5–20 mg using a spin-column Dneasy Animal Extraction protocol (Qiagen, Santa Clarita, CA, USA). PCR amplification using the genomic DNA as a template was performed using HotStarTaq DNA polymerase (QIAGEN, Tokyo, Japan) according to the manufacturer’s instruction. Mitochondrial (mt) 16S rDNA was amplified following procedures outlined in Sinniger et al. (2005). COI was amplified following procedures outlined in Reimer et al. (2004). The ITS-rDNA region was amplified following procedures outlined in Reimer et al. (2007c). The amplified products were visualized by 1.5% agarose gel electrophoresis.
Phylogenetic analyses
New sequences obtained in the present study (Table 1) were deposited in DDBJ and GenBank (accession numbers EU333744-EU333810). By using CLUSTAL X version 1.8 (Thompson et al. 1997), the nucleotide sequences of mt 16S rDNA and COI from samples were aligned with previously published sequences (see Reimer et al. 2007a) from various zoanthid species representing the genera Palythoa, Zoanthus, Savalia, and Parazoanthus. The outgroup sequences for both mt 16S rDNA and COI trees were from the genus Epizoanthus, previously shown to be basal in the order Zoantharia phylogeny for both mt 16S rDNA and COI (Sinniger et al. 2005; Reimer et al. 2007a). These genera, together with Abyssoanthus (Reimer et al. 2007a), represent the full range of described zoanthid genera. New mt 16S rDNA and COI sequences from specimens in this study were clearly divergent when compared with Abyssoanthus sequences, and Abyssoanthus sequences are not included in trees presented in this study. Parazoanthidae ITS-rDNA sequences (particularly ITS-1 and ITS-2 spacers) were highly divergent from other obtained ITS-rDNA sequences, and thus an ITS-rDNA alignment consisting only of Parazoanthidae sequences with sequence AB214161 from Parazoanthus gracilis (Reimer et al. 2007c) as the outgroup was generated. The alignments were inspected by eye and manually edited. All ambiguous sites of the alignments were removed from the dataset for phylogenetic analyses. Consequently, three alignment datasets were generated: (1) 675 sites of 40 sequences (mt 16S rDNA); (2) 280 sites of 46 sequences (COI); and (3) 770 sites of 15 sequences (ITS-rDNA). The alignment data are available on request from the corresponding author.
For the phylogenetic analyses of the three genetic markers, the same methods were independently applied. Alignments were subjected to analyses with the maximum likelihood (ML) with PhyML (Guindon and Gascuel 2003). PhyML was performed using an input tree generated by BIONJ with the general time-reversible model (Rodriguez et al. 1990) of nucleotide substitution incorporating invariable sites and a discrete gamma distribution (eight categories) (GTR + I + Γ). The proportion of invariable sites, a discrete gamma distribution, and base frequencies of the model were estimated from the dataset. PhyML bootstrap trees (500 replicates) were constructed using the same parameters as the individual ML tree. The distances were calculated using a Kimura’s 2-parameter model (Kimura 1980).
Bayesian trees were also reconstructed by using the program MrBayes 3.0 (Ronquist and Huelsenbeck 2003) under GTR + I + Γ. One cold and three heated Markov chain Monte Carlo (MCMC) chains with default-chain temperatures were run for 1,000,000 generations, sampling log-likelihoods (InLs), and trees at 100-generations intervals (10,000 InLs and trees were saved during MCMC). The likelihood plot for mt 16S rDNA, COI, and ITS-rDNA datasets suggested that MCMC reached the stationary phase after the first 40,000 generations all datasets. Thus, the remaining 9,600 trees of mt 16S rDNA, COI, ITS-rDNA were used to obtain clade probabilities and branch-length estimates, respectively.
Results
Phylogeny of mt 16S rDNA
The resulting ML tree for the aligned mt 16S rDNA sequences is shown in Fig. 2. Sample 01-05 (identical to Zoanthus sansibaricus from Japan) was within a moderately supported (ML = 55%; Bayesian posterior probability [B] = 0.81) clade corresponding to the genera Zoanthus and Acrozoanthus. Similarly, samples 02-122 and 01-106 (identical to Palythoa tuberculosa) were within a moderately well-supported clade (ML = 82%; B = 0.92) corresponding to Palythoa. Zoanthid-specific primers for mt 16S rDNA did not successfully amplify any molecule from sample 03-103.
Maximum likelihood tree of mitochondrial 16S ribosomal DNA (mt 16S rDNA) sequences for zoanthid specimens. Values at branches represent ML probabilities (>50%). Bayesian posterior probabilities of >95% are represented by thick branches. New sequences from this study in bold. For sample name abbreviations see Table 1. Sequences/species names from previous studies in regular font
Mitochondrial (mt) 16S rDNA sequences from 01-61, 04-346, 02-09, 03-641, 04-347, 04-345, 02-59, 02-27, and 03-135 were monophyletic with very high support (ML = 98%; B = 0.98; 3/561 positions variable in the alignment of only these sequences = 0.53% sequence variation), and were in a weakly supported clade with Parazoanthus gracilis and Parazoanthus tunicans (ML = 63%; B = 0.93). This clade was basal to other Parazoanthidae clades, although support was not very high (ML = 62% but B = 0.98).
Sequences from 04-348, 03-47, 03-652, 03-290, 03-177, and 04-328 formed a moderately well-supported monophyly (ML = 96% but B = 0.54; sequence variation 1/598 base pairs = 0.17%) derived from Parazoanthus swiftii, and sister to a poorly resolved Parazoanthidae group including the sponge-associated Parazoanthus parasiticus and Parazoanthus puertoricense, as well as a highly supported antipatharian-associated clade (ML = 100%; B = 1.00). Within this antipatharian-associated clade were included Antipathes galapagensis-associated 03-221, 03-549, and 04-341 (sequences identical) as well as the antipatharian-associated “Cape Verde zoanthid” and “Principe zoanthid.”
All acquired sequences were within a large Parazoanthidae/Sphenopidae/Zoanthidae clade, highly supported (ML = 100%; B = 1.00), and separate from Epizoanthidae sequences.
Phylogeny of COI
The resulting ML tree for the aligned COI sequences is shown in Fig. 3. Samples 01-05 (identical to Zoanthus sansibaricus from Japan) and 01-105 (identical to Zoanthus kuroshio and Zoanthus aff. vietnamensis from Japan) were within a well-supported (ML = 94%, B = 1.00) clade corresponding to the genus Zoanthus. Similarly, samples 02-122, 03-709, and 01-106 (identical to Palythoa tuberculosa and P. mutuki) were within a moderately well-supported clade (ML = 92%, B = 0.76) corresponding to Palythoa. Sample 03-103 was seen to be basal to the Palythoa and Zoanthus clade and did not group with any known zoanthid, although support was only moderately high (ML = 62%, B = 1.00) for its phylogenetic position.
Maximum likelihood tree of mitochondrial cytochrome oxidase subunit I (mt COI) sequences for zoanthid specimens. Values at branches represent ML probabilities (>50%). Bayesian posterior probabilities of >95% are represented by thick branches. New sequences from this study in bold. For sample name abbreviations see Table 1. Sequences/species names from previous studies in regular font
COI sequences from 01-61, 03-566, 04-346, 02-09, 03-641, 04-347, 04-641, 04-345, 02-59, 04-343, 02-27, 03-46, and 03-135 were monophyletic with moderately weak support (ML = 84%, B = 0.57; sequence variation 2/280 base pairs = 0.71%), and included in a well-supported clade with Parazoanthus gracilis, Parazoanthus tunicans, and “yellow polyps” (ML = 94%, B = 1.00). This clade was sister to the 03-103/Palythoa/Zoanthus clade (ML = <50% but B = 1.00).
Sequences from 04-348, 03-47, 03-652, 03-290, 03-177, 04-155, and 04-328 were all identical to Parazoanthus swiftii, and part of a poorly resolved Parazoanthidae group basal to the other described sequences from above. Within this poorly resolved group, Antipathes galapagensis-associated 03-549 and 04-341 formed a very highly supported clade (ML = 99.8%, B = 1.00; sequences identical) with the antipatharian-associated “Cape Verde zoanthid.”
All acquired sequences were within a large Parazoanthidae/Sphenopidae/Zoanthidae clade, highly supported (ML = 100%, B = 1.00), and separate from Epizoanthidae sequences.
Phylogeny of ITS-rDNA
As ITS-rDNA sequences for both Zoanthus (Reimer et al. 2007c) and Palythoa (Reimer et al. 2007b) have been shown to be highly divergent from Parazoanthidae sequences, an ML tree for the aligned Parazoanthidae-only ITS-rDNA sequences is shown in Fig. 4.
Maximum likelihood tree of internal transcribed spacer of ribosomal DNA (ITS-rDNA) sequences for zoanthid specimens of the families Parazoanthidae. Values at branches represent ML probabilities (>50%). Bayesian posterior probabilities of >95% are represented by thick branches. New sequences from this study in bold. For sample name abbreviations see Table 1. Sequences/species names from previous studies in regular font
All three observed Parazoanthidae monophylies previously seen with COI and mt 16S rDNA sequences from corresponding Galapagos zoanthid specimens were again seen in the ITS-rDNA tree, and all were very highly supported. Antipathes galapagensis-associated 03-221 and 03-549 formed one clade (ML = 100%; B = 1.00; sequence variation 6/700 base pairs = 0.86%) sister to the sponge-associated clade (ML = 100%; B = 1.00; sequence variation 4/633 base pairs = 0.63%) consisting of 03-47, 04-348, 03-177, and 03-290. The remaining sequences (04-347, 01-61, 02-59, 04-346, 03-641, 03-566, 02-27, and 03-135) formed a moderately well-supported monophyly (ML = 96%; B = 0.75; sequence variation 6/623 base pairs = 0.96%) basal to the other two clades.
Discussion
Utility of molecular identification of zoanthids
This study clearly demonstrates the effectiveness of using molecular DNA markers (mt 16S rDNA, COI, ITS-rDNA) in zoanthid identification, especially with specimens that are difficult to distinguish based on morphological characters. In fact, some samples of Parazoanthidae were initially morphologically identified as Epizoanthidae as these families are notoriously difficult to distinguish (the sphincter muscle being either endodermal [Parazoanthidae] or mesogleal [Epizoanthidae]). Once molecular data were acquired, a re-examination of morphological and ecological data facilitated description of the zoanthid groups seen in this study. Based on the molecular data, all specimens were placed into known zoanthid genera except for sample 03-103.
Nomenclature of Parazoanthidae
As shown previously in Sinniger et al. (2005) and in the three phylogenetic trees in this study (Figs. 2, 3, 4), according to molecular data the family Parazoanthidae is not a valid monophyletic grouping. However, for the purpose of discussing Galapagos zoanthids, all three observed Parazoanthidae monophylies in this study are designated as Parazoanthus species (G1, G2, G3), although supraspecific (generic or higher) assignment for two clades (Parazoanthus sp. G1 and Parazoanthus sp. G3) will likely change in the near future. The remaining observed group (Parazoanthus sp. G2) is likely to remain Parazoanthus as it is closely related to the type species for Parazoanthus, P. axinellae (see Fig. 3 in Sinniger et al. 2005).
Zoanthid diversity in the Galapagos
The preliminary survey of zoanthids in the Galapagos identified three families, three genera, and seven provisional species of zoanthids, and also found one potentially unidentified generic-level group (“03-103” group). The zoanthids found in the Galapagos are: Zoanthus cf. sansibaricus, Zoanthus cf. vietnamensis/kuroshio (both Zoanthidae); Palythoa cf. tuberculosa, Palythoa cf. mutuki (Sphenopidae); Parazoanthus sp. G1, Parazoanthus sp. G2, and Parazoanthus sp. G3 (Parazoanthidae). The Zoanthus and Palythoa spp. from the Galapagos listed above have been discussed in a recent paper (Reimer and Hickman in press), although obtained sequences are also shown Tables and Figures in this study.
While mitochondrial molecular markers (i.e., mt 16S rDNA and COI) have been shown to have generally slow evolution in Anthozoa (Shearer et al. 2002; Huang et al. 2008), mt 16S rDNA and COI together are considered to be generally accurate (∼92–95%) in identification of zoanthid specimens to the species level (i.e., Reimer et al. 2008). Also, previous studies have shown mt 16S rDNA and COI to be accurate markers for separating and identifying zoanthid species of Zoanthus and Palythoa from Japan (Reimer et al. 2004, 2006b, c, 2007b). Additional faster-evolving ITS-rDNA sequences also show highly supported monophylies of the three nominal Parazoanthidae species groups collected in this study (Fig. 4), and thus the grouping of these specimens into three species-level groups can be made with a high level of confidence.
As mentioned earlier in this study, little information exists on zoanthids from the Galapagos. In fact, there is an overall lack of data on zoanthids from the entire Pacific coast of Central and South America, as well as islands of the eastern Pacific. Several specimens of Zoanthus and Palythoa have been recorded from a few locations in Panama, El Salvador, Mexico, Easter Island, and French Polynesia (see zooxanthellate zoanthid literature discussed in Reimer and Hickman [in press]), and Epizoanthus elongatus has been documented from Zorritos, Peru (Verrill 1869). Additionally, there is one record of a larval “Zoanthella decipiens” from a depth of 600 m at a location northwest of Darwin Island (Senna 1907) and another record of larval Zoanthella galapagoensis in Heath (1906), but until Reimer and Hickman (in press) and this research there have been no records of any non-larval Zoantharia from the Galapagos.
Due to this lack of data on zoanthid distribution for the entire eastern Pacific, it is somewhat difficult to assess if the azooxanthellate Parazoanthus spp. specimens collected during the course of this investigation are endemic Galapagos species that are undescribed, or simply range extensions of described species from other regions. However, based on morphological and molecular differences, it is highly likely that all three Parazoanthus spp. described below are undescribed species and formal descriptions of these species are planned.
Based on known distribution patterns of zoanthids from other regions of the world, the Galapagos zoanthid diversity seen in this study is not surprising for the depths sampled in this study (intertidal to ∼30 m). Zooxanthellate zoanthids of the genera Palythoa and Zoanthus are common in infra-littoral and shallow sub-tropical and tropical waters in many regions of the world. Parazoanthidae species are known from Japan (i.e., Reimer et al. 2004), the Mediterranean (i.e., Pax and Muller 1962), New Caledonia (Sinniger 2006), and other locations at similar depths as seen in the Galapagos.
One family of zoanthids not seen in this Galapagos survey despite frequent records from numerous locations worldwide was Epizoanthidae (consisting solely of the genus Epizoanthus). Many records of Epizoanthidae exist from the deep sea (e.g., see Muirhead et al. 1986; Beaulieu 2001), and it may be that Epizoanthidae in the Galapagos remain to be found at depths below those explored in the present study (approximately 35 m).
Species descriptions of azooxanthellate zoanthids of the Galapagos
Note that informal descriptions of all zooxanthellate zoanthids noted from the Galapagos (Zoanthus cf. sansibaricus, Zoanthus cf. vietnamensis/kuroshio (both Zoanthidae); Palythoa cf. tuberculosa, Palythoa cf. mutuki (Sphenopidae)) are found in Reimer and Hickman (in press) and are not presented in this study.
Parazoanthus sp. G1 (Fig. 5)
Parazoanthus sp. G1 belongs to a Parazoanthidae clade that is potentially an undescribed genus, characterized by being epizoic on antipatharians (black coral) (see Sinniger et al. 2005). Parazoanthus sp. G1 has approximately 40 bright yellow and/or red tentacles, with long red, yellow, or cream-colored polyps that extend well clear of the coenenchyme (Fig. 5). Tentacles are almost always longer than the expanded oral disk diameter. Polyps are approximately 6–12 mm in diameter when open, and approximately 6–15 mm in height. Morphologically similar zoanthids (epizoic on antipatharians, similar sizes, yellow in color) have been recorded from Cape Verde, Madagascar, and Japan (specimens in FS and JDR’s respective collections), although these other specimens were found on different antipatharians species than Parazoanthus sp. G1 and were never red or cream in color.
All collected samples from Galapagos were on the black coral Antipathes galapagensis at depths of 12 m to 35 m. Although A. galapagensis is found throughout the archipelago, Parazoanthus sp. G1 colonies were observed only at Santiago, Floreana, Isabela, and Pinzon Islands, and it may be that this genus has a patchy distribution in the Galapagos. They may cover only a portion of a living A. galapagensis black coral colony, or cover the entire colony, thus killing the antipatharian and suggesting this species may be parasitic.
Parazoanthus sp. G2 (Fig. 6)
Parazoanthus sp. G2 polyps have yellow, orange, or cream tentacles, and a red, yellow, or light yellow oral disk, with a light tan, light pink, or cream coenenchyme. Polyps are approximately 3–6 mm in diameter when open, and approximately 2–6 mm in height. Polyps have between 24 and 30 tentacles that are usually longer than expanded oral disk diameter. Although polyps extend clear of the coenenchyme, when contracted the polyps are mere bumps on the surface of the coenenchyme, much more embedded than both Parazoanthus sp. G1 and Parazoanthus sp. G3. While many Parazoanthus species are known to be epizoic on sponges, none of the described species thus far are morphologically similar (color, size, polyp shape; see below) to Parazoanthus sp. G2.
Collected Parazoanthus sp. G2 specimens from Galapagos are often (but not always) associated with a bright yellow-orange or red sponge, possibly Pseudosuberites or Tedania. Parazoanthus sp. G2 colonies often grow in patches over the sponge, or may even cover it entirely, and often extend to surrounding rock substrate. Despite being covered by Parazoanthus sp. G2, the sponge is always alive, suggesting this association may be symbiotic. Similar to Parazoanthus sp. G3, Parazoanthus sp. G2 are found on rock walls, in crevices, or at the base of rocks, and were found from depths of 5 m to ∼30 m, and may extend deeper. Colonies may be very small (a few cm2 in area), or extend to cover large areas over a square meter in area. Parazoanthus sp. G2 were seen at Wolf, Marchena, Isabela, Fernandina, Santa Cruz, San Cristobal, Española, and Floreana Islands, and its range is likely throughout the entire archipelago.
Despite COI sequences of this species being identical to sequences from P. swiftii from the Caribbean (Fig. 3), longer mt 16S rDNA sequences were not identical and P. swiftii was not included in the Parazoanthus sp. G2 monophyly in the mt 16S rDNA tree (Fig. 2). Additionally, due to the morphology of P. swiftii (rarely not found on sponges, relatively shorter tentacles, large [6 mm] diameter polyps that often extend well clear from coenenchyme) and large geographic distances between P. swiftii and Parazoanthus sp. G2 localities, it is very likely that these are two different species groups.
Parazoanthus sp. G3 (Fig. 7)
Parazoanthus sp. G3 belongs to a Parazoanthidae clade that is potentially an undescribed genus or family, characterized by often (but not always) being epizoic on hydrozoans, as well as being phylogenetically distinct from other Parazoanthidae (Figs. 2, 3, 4; see also Sinniger et al. 2005). Parazoanthus sp. G3 has red or red-brown oral disks and the outer surface of polyps are tan to dark brown, with polyps more clear of the coenenchyme than Parazoanthus sp. G2, but less well developed than Parazoanthus sp. G1. To our knowledge, no other described Pacific Parazoanthus species to date are generally non-epizoic and bright red in color. Two specimens (02-09 and 03-560) had white oral disks, but otherwise were similar to red specimens morphologically (polyp dimensions, polyp form, substrate). Additionally, COI and mt 16S rDNA sequences for sample 02-09 were within the well-supported Parazoanthus sp. G3 monophylies (Table 1, Figs. 2 and 3). Thus, based on both morphological and molecular data examined specimens are apparently from a single species. Polyps are approximately 4–12 mm in diameter when open, and rarely more than 20 mm in height. Colonies may reach sizes of over a meter in diameter. Both red and white oral disk forms have 32–40 tentacles that are almost as long as the diameter of the expanded oral disk (Fig. 7).
Collected Parazoanthus sp. G3 specimens in the Galapagos were found on rock substrate in areas of high current (i.e., the base of large rocks, rock walls, etc.). Most known Parazoanthidae species in this clade (i.e., Parazoanthus gracilis in Japan) associate closely or are epizoic on hydrozoans, although some species can be found on rocky substrates, (e.g., not obligate symbionts), strongly suggesting species within this Parazoanthidae clade are not obligate on hydrozoans. Colonies were found at Darwin, Marchena, Genovesa, Isabela, Pinzon, Española, and Floreana Islands, and it is likely Parazoanthus sp. G3 is found throughout the archipelago. This species has been found from the low infra-littoral to depths of over 35 m and is likely to be found at even deeper depths.
Unknown zoanthid sp. “03-103” (Fig. 8)
Currently only one specimen of this unknown zoanthid (specimen 03-103, see Table 1) has been molecularly examined. However, many similar specimens (n = 29) were collected during the 2007 expedition and await molecular examination to confirm if they are the same taxon as 03-103.
This zoanthid is characterized by its encrustation of relatively large sand and detritus (individual particles barely visible to the naked eye—see Fig. 8), particularly on the coenenchyme (stolons) connecting polyps. The oral disk color of specimen 03-103 was clear, and similar 2007 samples had either black or clear oral disks and 24–34 tentacles that are up to approximately twice as long as the expanded oral disk diameter. Both 03-103 and 2007 specimens were very small for zoanthids, with polyp diameters of 1–2 mm, oral disks of 1–3 mm, and polyps no more than 4 mm in height. Polyps are barely connected to each other by a thin coenenchyme, or even occasionally unitary (not connected).
All samples have been found on the underside of flat rocks or dead coral resting on sandy bottoms, at depths of 6 m to 27 m. 03-103 was associated with coral, and while some specimens collected in 2007 were attached to rocks near bryozoans, other specimens were not noted to be associated with any particular fauna. Specimen 03-103 was found at Devil’s Crown, Floreana Island, but in 2007, 29 additional specimens were found at Darwin, Wolf, Marchena, Santiago, North Seymour, San Cristobal, Española, and Floreana Islands, and this zoanthid may be distributed throughout the archipelago. While no zoanthids similar to 03-103 have been described, similar undescribed zoanthids have also been seen in Okinawa, Japan (J. Reimer and T. Fujii, specimens conserved in JDR’s collection). Clearly proper description and further investigation of this potentially novel zoanthid are needed.
Morphological key to zoanthids of the Galapagos
-
1.
Zoanthids do not have sand or other detritus encrusted anywhere in their tissue—go to 3.
-
2.
Zoanthids have sand encrusted in their exoderm and/or endoderm—go to 5.
-
3.
Zooxanthellate, oral disk pale pink or green, polyps often but not always well embedded (“immersae” or “intermediae”) in the coenenchyme, outside of polyps light purple—Zoanthus cf. vietnamensis/kuroshio.
-
4.
Zooxanthellate, oral disk may be green, red, white, blue, often with fluorescence, polyps free and clear of coenenchyme (“liberae”), outside of polyps dark purple—Zoanthus cf. sansibaricus.
-
5.
Zoanthids zooxanthellate, with large (∼1 cm) oral disks, found in shallow water (<10 m to intertidal), tissue generally light tan-dark brown—go to 7.
-
6.
Zoanthids azooxanthellate, with oral disks smaller than ∼1 cm, not found intertidally at any location—go to 9.
-
7.
Polyps free and clear of coenenchyme (“liberae”)—Palythoa cf. mutuki.
-
8.
Polyps embedded or barely protruding from coenenchyme (“immersae” or “intermediae”)—Palythoa cf. tuberculosa.
-
9.
Zoanthids not found growing on antipatharians—go to 11.
-
10.
Zoanthids found growing on Antipathes galapagensis—Parazoanthus sp. G1.
-
11.
Zoanthids not found under rocks on sandy bottoms—go to 13.
-
12.
Zoanthids found under rocks on sandy bottoms—unknown zoanthid sp. “03-103”.
-
13.
Zoanthids never associated with living sponges, polyps free and clear of coenenchyme (“liberae”) even when retracted, tissue and oral disk red or white, 32 to 40 tentacles—Parazoanthus sp. G3.
-
14.
Zoanthids often associated with living sponge (possibly Pseudosuberites or Tedania), polyps barely protrude above coenenchyme when retracted, tissue light pink—tan, 24 to 30 tentacles—Parazoanthus sp. G2.
Effectiveness of molecular identification and future research needs
The work in this study and Reimer and Hickman (in press) reports the presence of several species of Zoantharia in the Galapagos for the first time. Molecular identification techniques combined with morphological and ecological data proved to be highly useful in identifying collected zoanthid specimens, and we recommend the protocols used in this study for any future investigation of zoanthid diversity, especially in areas that have not yet been investigated. As a result of the surveys conducted, three genera and eight nominal species of zoanthids were found in the Galapagos, including potential new species and genera. However, it is likely other zoanthids in the Galapagos await discovery at depths below the range of SCUBA, and this needs to be investigated.
With the recording of the existence of zoanthids in the Galapagos for the first time, it is apparent that Zoantharia species can colonize areas that are relatively isolated. The data in this study suggest that zoanthids may also be present in other isolated and insular understudied regions of the world, and that many new species and genera may await description.
Future research on zoanthids in this region should focus on both more in-depth investigations at different locations and at deeper depths in the Galapagos, and additional surveys should be conducted along the Pacific coast of South and Central America. Only then will the diversity of zoanthids in this region be clarified, helping increase understanding of shallow-water clonal cnidarian communities in the east Pacific. It is hoped the investigations in this study provide a basis for such future research.
References
Beaulieu SE (2001) Life on glass houses: sponge stalk communities in the deep sea. Mar Biol 138:803–817
Breedy O, Guzman HM (2005) A new species of alcyonacean octocoral from the Galápagos archipelago. J Mar Biol Assoc UK 85:801–807
Breedy O, Guzman HM (2007) A revision of the genus Leptogorgia Milne Edwards & Haime, 1857 (Coelenterata: Octocorallia: Gorgoniidae) in the eastern Pacific. Zootaxa 1407:1–90
Breedy O, Hickman CP Jr, Williams G (in press) Octocoral research in the Galapagos Islands. Galapagos Res
Bustamante R, Collins KJ, Bensted-Smith R (2000) Biodiversity conservation in the Galápagos Marine Reserve. Bull Ins Roy Sci Natur Belg Supp 70:31–38
Bustamante RH, Wellington GM, Branch GM, Edgar GJ, Martinez P, Rivera F, Smith F, Witman J (2002) Outstanding marine features. In: Bensted-Smith R (ed) A biodiversity vision for the Galápagos Islands, Charles Darwin Foundation and World Wildlife Fund, Puerto Ayora, pp 38–67
Cairns SD (1991) A revision of the ahermatypic Scleractinia of the Galápagos and Cocos Islands. Smithson Contrib Zool 504:1–44
Cairns SD (2001) A generic revision and phylogenetic analysis of the Dendrophylliidae (Cnidaria: Scleractinia). Smithson Contrib Zool 615:1–75
Calder DR, Mallinson JJ, Collins K, Hickman CP Jr (2003) Additions to the hydroids (Cnidaria) of the Galápagos, with a list of species reported from the islands. J Nat Hist 37:1173–1218
Darwin C (1859) On the origin of species. John Murray, London
Fautin DG (2007) Hexacorallians of the world. http://geoportal.kgs.ku.edu/hexacoral/anemone2/index.cfm
Fautin DG, Hickman CP Jr, Daly M, Molodtsova T (2007) Shallow-water sea anemones (Cnidaria: Anthozoa: Actiniaria) and tube anemones (Cnidaria: Anthozoa: Ceriantharia) of the Galápagos Islands. Pac Sci 61:549–573
Glynn PW (2003) Coral communities and coral reefs of Ecuador. In: Cortes J (ed) Latin American coral reefs. Elsevier Sciences BV, Amsterdam, pp 449–472
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Heath H (1906) A new species of Semper’s larva from the Galapagos Islands. Zool Anz 30:171–175
Hickman CP Jr (1998) A field guide to sea stars and other echinoderms of Galápagos. Sugar Spring Press, Lexington
Hickman CP Jr, Finet Y (1999) A field guide to marine molluscs of Galápagos. Sugar Spring Press, Lexington
Hickman CP Jr, Zimmerman TL (2000) A field guide to crustaceans of Galápagos. Sugar Spring Press, Lexington
Huang D, Meier R, Todd PA, Chou LM (2008) Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol 66:167–174
Kimura M (1980) A simple method for estimating evolutionary rates pf base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Muirhead A, Tyler PA, Thurston MH (1986) Reproductive biology and growth of the genus Epizoanthus (Zoanthidea) from the north-east Atlantic. J Mar Biol Assoc UK 66:131–143
Pax F, Muller I (1962) Die Anthozoenfauna der Adria. Fauna et Flora Adriatica, III. Institut za Oceanografiju i Ribastvo, Split
Reimer JD, Ono S, Takishita K, Fujiwara Y, Tsukahara J (2004) Reconsidering Zoanthus spp. diversity: molecular evidence of conspecificity within four previously presumed species. Zool Sci 21:517–525
Reimer JD, Ono S, Iwama A, Tsukahara J, Maruyama T (2006a) High levels of morphological variation despite close genetic relatedness between Zoanthus aff. vietnamensis and Zoanthus kuroshio (Anthozoa: Hexacorallia). Zool Sci 23:755–761
Reimer JD, Ono S, Iwama A, Tsukahara J, Takishita K, Maruyama T (2006b) Morphological and molecular revision of Zoanthus (Anthozoa: Hexacorallia) from southwestern Japan with description of two new species. Zool Sci 23:261–275
Reimer JD, Ono S, Takishita K, Tsukahara J, Maruyama T (2006c) Molecular evidence suggesting species in the zoanthid genera Palythoa and Protopalythoa (Anthozoa: Hexacorallia) are congeneric. Zool Sci 23:87–94
Reimer JD, Hirano S, Fujiwara Y, Sinniger F, Maruyama T (2007a) Morphological and molecular characterization of Abyssoanthus nankaiensis, a new family, new genus and new species of deep-sea zoanthid (Anthozoa: Hexacorallia: Zoantharia) from a northwest Pacific methane cold seep. Invertebr Systemat 21:255–262
Reimer JD, Takishita K, Ono S, Maruyama T (2007b) Diversity and evolution in the zoanthid genus Palythoa (Cnidaria: Hexacorallia) utilizing nuclear ITS-rDNA. Coral Reefs 26:399–410
Reimer JD, Takishita K, Ono S, Tsukahara J, Maruyama T (2007c) Molecular evidence suggesting intraspecific hybridization in Zoanthus (Anthozoa: Hexacorallia). Zool Sci 24:346–359
Reimer JD, Ono S, Tsukahara J, Iwase F (2008) Molecular characterization of the zoanthid genus Isaurus (Anthozoa: Hexacorallia) and its zooxanthellae (Symbiodinium spp). Mar Biol 153:351–363
Reimer JD, Hickman CP Jr (in press) Preliminary survey of zooxanthellate zoanthids (Cnidaria: Hexacorallia) of the Galápagos and associated symbiotic dinoflagellates (Symbiodinium spp.). Galápagos Res
Rodriguez F, Oliver JL, Marin A, Medina JR (1990) The general stochiatic model of nucleotide substitution. J Theor Biol 142:485–501
Ronquist F, Huelsenbeck JP (2003) Bayesian phylogenetic inference under mixed models. Bioinformatics (Oxf) 19:1572–1574
Senna A (1907) Nuove larve pelagiche di Ceriantidi e di Zoanttidi, Nota preliminare. Monit Zool Ital 18:96–102
Shearer TL, van Oppen MJH, Romano SL, Worheide G (2002) Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol Ecol 11:2475–2487
Sinniger F (2006) Zoantharia of New Caledonia. In: Payri C, Richier De Forges B (eds) Compendium of marine species from New Caledonia. Documents scientifiques et techniques II (7). IRD Editions, Noumea. pp 127–128
Sinniger F, Montoya-Burgos JI, Chevaldonne P, Pawlowski J (2005) Phylogeny of the order Zoantharia (Anthozoa, Hexacorallia) based on mitochondrial ribosomal genes. Mar Biol 147:1121–1128
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Verrill AE (1869) Review of the corals and polyps of the west coast of America. Trans Conn Acad Arts Sci 1:377–558
Williams GC, Breedy O (2004) The Panamic gorgonians genus Pacifigorgia (Octocorallia: Gorgoniidae) in the Galápagos archipelago, with descriptions of three new species. Proc Calif Acad Sci 55:55–88
Acknowledgments
Graham Edgar, Angel Chiriboga, Luis Vinueza, and William C. Ober generously helped with sampling and photography. CPH is indebted to the staff of the Biomarine Laboratory of the Charles Darwin Research Station (Pto. Ayora, Santa Cruz Island) for logistical field help. The captain and crew of the Mabel (December 2004), the Flamingo (January 2003), the Golindrina (November 2003), the San Jose (May 2002), and the Beagle III (June 2001) helped make sampling possible. JDR thanks Dr. Kiyotaka Takishita, as well as Masoru Kawato and Dr. Hirokazu Kuwahara (all JAMSTEC), for research assistance. Specimen collection during the 2006 and 2007 trips were partially funded by the Darwin Initiative Project. JDR was additionally partially financed by a Japan Society for the Promotion of Science post-doctoral fellowship (#P04868) at JAMSTEC (Yokosuka, Japan), the 21st Century Center of Excellence Program (COE) at the University of the Ryukyus, and a grant from the Fujiwara Natural History Foundation (Tokyo, Japan). Three anonymous reviewers greatly helped improve this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Biology Editor Dr. Ruth Gates.
Rights and permissions
About this article
Cite this article
Reimer, J.D., Sinniger, F. & Hickman, C.P. Zoanthid diversity (Anthozoa: Hexacorallia) in the Galapagos Islands: a molecular examination. Coral Reefs 27, 641–654 (2008). https://doi.org/10.1007/s00338-008-0376-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-008-0376-5