Abstract
Zoantharia (or Zoanthidea) is the third largest order of Hexacorallia, characterised by two rows of tentacles, one siphonoglyph and a colonial way of life. Current systematics of Zoantharia is based exclusively on morphology and follows the traditional division of the group into the two suborders Brachycnemina and Macrocnemina, each comprising several poorly defined genera and species. To resolve the phylogenetic relationships among Zoantharia, we have analysed the sequences of mitochondrial 16S and 12S rRNA genes obtained from 24 specimens, representing two suborders and eight genera. In view of our data, Brachycnemina appears as a monophyletic group diverging within the paraphyletic Macrocnemina. The macrocnemic genus Epizoanthus branches as the sister group to all other Zoantharia that are sampled. All examined genera are monophyletic, except Parazoanthus, which comprises several independently branching clades and individual sequences. Among Parazoanthus, some groups of species can be defined by particular insertion/deletion patterns in the DNA sequences. All these clades show specificity to a particular type of substrate such as sponges or hydrozoans. Substrate specificity is also observed in zoantharians living on gorgonians or anthipatharians, as in the genus Savalia (Gerardia). If confirmed by further studies, the substrate specificity could be used as reliable character for taxonomic identification of some Macrocnemina.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The order Zoantharia (= Zoanthidea, Zoanthiniaria) is characterised by colonies of clonal polyps possessing two rows of tentacles, a single ventral siphonoglyph linked together by a coenenchyme. The name Zoantharia is used here to give homogeneity to the order names in subclass Hexacorallia (Actiniaria, Antipatharia, Ceriantharia, etc.). Based on the organisation of septa, the Zoantharia are currently divided in two suborders, Macrocnemina and Brachycnemina (Haddon and Schackelton 1891). The suborders differ by the fifth pair of septa, which is complete in the suborder Macrocnemina (Fig. 1) and incomplete in the Brachycnemina. The separation between both suborders is also supported by the few data available on sexual reproduction. Brachycnemina produce planktonic larvae, called zoanthella or zoanthina depending on the family, whereas no planktonic larvae have been reported for Macrocnemina (Ryland 1997).
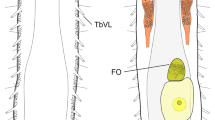
Thin cross-section of Parazoanthus axinellae. The lower part of the pharynx is visible on the bottom right, whereas the rest of the picture is in the upper part of the gastric cavity. The numbers indicate the five first pairs of septa (1,3=incomplete; 2,4,5=complete). Sand grains and sponge spicules are incorporated in the ectoderm. S indicates the siphonoglyph
While the separation between the two suborders of Zoantharia is well accepted, the interpretation of families and genera is not always very clear, especially within Macrocnemina. Identification at the species level is also difficult because most diagnoses are incomplete, and type specimens are often missing. The main characters used for species identification are (1) the number of tentacles or septa, (2) the colour, (3) the shape and position of the sphincter muscle and (4) the size and distribution of different types of nematocysts. The position of the sphincter is useful mainly at the generic level whereas the characters associated with nematocysts seem to be useful at the interspecific level but they necessitate a precise analysis of many samples of nematocysts within and between different polyps of a colony (Ryland et al. 2004). Both analyses of sphincter and nematocysts require high-quality histological preparations.
Because of identification problems, zoantharians are often overlooked in ecological surveys in spite of their abundance and cosmopolitan distribution. Some field guides propose the use of ecological factors, such as an association between Zoantharia and their substratum, as alternative criterion for taxonomic identification. However, their specificity to one or a few clearly defined substrate types has not been investigated for most species.
Current classification of Zoantharia (Table 1) is based exclusively on morphological characters (Ryland and Muirhead 1993; Ryland and Lancaster 2003). It follows the taxonomic scheme presented by Haddon and Shackleton (1891), based mostly on septa characteristics (Fig. 2a). Schmidt (1974) challenged this classification proposing a phylogeny based on cnidome analysis and suggested a derived position of the special microbasic b-mastigophores nematocysts in Parazoanthus and Zoanthus (Fig. 2b). These results were supposed to be supported by sperm structure analyses (Schmidt 1979), however, in this study, none of Zoanthus species was considered. More recently, Haddon and Shackleton’s classification has been supported by a phylogenetic study of allozymes (Burnett et al. 1997) which, however, only focussed on Australian brachycnemic species, using Parazoanthus as an outgroup (Fig. 2c).
In the present study, 24 sequences of mitochondrial 12S and 16S rRNA genes were obtained to infer the molecular phylogeny of the order Zoantharia. These two mitochondrial ribosomal genes were chosen following their successful use in inferring molecular phylogeny of octocorals (Sanchez et al. 2003) and scleractinians (Le Goff-Vitry et al. 2004). In the light of our molecular results, morphological characteristics classically used for the classification of Zoantharia are reassessed and their specificity to different types of substrata is discussed.
Material and methods
Sampling
The Zoantharia examined in this study (Table 2) were collected from Honduras and the Mediterranean Sea, by F. Sinniger, in Sulawesi, by M. Boyer, within the framework of the biodiversity project approved by LIPI (Indonesian Institute of Marine Sciences), and in the Canary, Principe and Cape Verde Islands, by P.Wirtz. Deep-water samples were collected by H. Zibrowius during the cruise CORTI on board of R/V URANIA (chief scientist: Marco Taviani, Bologna). Six samples originating from aquarium shops in Geneva are of uncertain origin. Each sample was divided into two parts, one preserved in 75% ethanol and the other in 4% seawater formalin for histological analysis. Metridium senile was used as outgroup and its corresponding sequences were taken from GenBank (AF000023).
DNA extraction, PCR amplification and sequencing
DNA was extracted using the DNeasy Plant Minikit (QIAGEN) from previously dried ethanol-preserved samples. The following PCR primers were designed: 16Sant1a: 5′-GCCATGAGTATAGACGCACA-3′, 16SbmoH: 5′-CGAACAGCCAACCCTTGG-3′, 12S1a: 5′-TAAGTGCCAGCMGACGCGGT-3′,
12S3r: 5′-ACGGGCNATTTGTRCTAACA-3′. PCR amplifications were performed as follows: 2′ at 94°C, then 40 cycles: 30′′ at 94°C, 1′ at 52°C, 2′ at 72°C, followed by 5′ final elongation at 72°C. Direct sequencing was carried out using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Sequences were runned on an ABI-3100 Avant automatic sequencer. GenBank accession numbers of the obtained sequences are shown in Table 2.
Histology and thin sections
Samples fixed and preserved in 4% seawater formalin were decalcified by chelation and then put into fluorhydric acid to dissolve siliceous sponge spicules and sand grains. Samples were then embedded in paraffin and cut into 8 μmslices. For microscopic observation, the slices were then stained with Masson’s trichrome. To assess the presence or absence of a zoantharian secreted layer on the substrate, the antipatharian on which the Cape Verde species was found was cut into thin cross-sections. The samples were put into Araldite, and then cut at low speed with a circular saw into slices of 0.5 mm. The sections were polished to a thickness of ca. 0.2 mm. In some cases, sections were then stained with toluidine blue to increase contrast, and obtain information on the tissues around the axis. The same protocol was used to cut several polyps of Parazoanthus axinellae (Fig. 1).
Phylogenetic analysis
Sequences were manually aligned using BioEdit 5.0.9 (Hall 1999) and uncertain alignment positions were removed. The methods of maximum likelihood (ML) and Bayesian inference (BI) were used for phylogenetic reconstructions. For ML method, the best evolutionary model was defined using Modeltest (Posada and Crandall 1998). Robustness of the ML trees was tested by bootstrapping (1,000 replicates). The Kishino–Hasegawa (KH) test (Kishino and Hasegawa 1989) was used to determine if Bayesian topology was significantly different from ML topology. ML method and KH tests were performed using Paup*4.0 beta 10 (Swofford 2002). Bayesian analyses were made using Mr. Bayes 3.0 (Huelsenbeck and Ronquist 2001).
Results
Species identification
Mediterranean species were identified by using the morphology of the polyps and of the colonies. The specimens of Parazoanthus from Honduras were initially identified based on their presumed substrate specificity (Crocker and Reiswig 1981) and confirmed later by the morphology of the polyp and the number of septa. Substrate type (anthipatharian) was also used for preliminary identification of Cape Verde and Principe specimens as belonging to Savalia macaronesica (Ocaña and Brito 2003). However, since the main feature distinguishing Savalia from Parazoanthus is the secretion of a scleroprotein layer, this character was examined by thin cross-section analyses. No Zoantharia-secreted axis was observed in specimens from Cape Verde and Principe, their identification as Savalia was not confirmed.
Parazoanthus gracilis was tentatively identified to the species level by histological interpretation and its specificity to the colonised hydrozoan (Plumularia habereri), while Epizoanthus illoricatus was provisionally identified based on the general morphology of the colony and its attachment to a eunicid worm tube. As available literature on Zoantharia contains only few precise descriptions for some of our collection localities, some specimens were not identifiable to the species level.
Phylogenetic analysis
Partial sequences of 12S and 16S mt rDNA were obtained. The sequences of 12S varied in length between 676 bp (Acrozoanthus sp.) and 709 bp (‘‘Yellow polyps”), whereas the sequences of 16S varied between 663 bp (Savalia spp.) and 768 bp (E. paxi, E. vagus). The analyses were performed on both genes separately and then their sequences were concatenated. The concatenated alignment contained 1,210 characters of which 962 were constant and 117 were parsimony informative. The mean base frequencies were A: 0.3076 C: 0.1887 G: 0.2683 T: 0.2454, the homogeneity test of base frequencies showed no significant divergences across taxa (χ2 =37.6941; df=72; P=0.9997).
Since the trees obtained by separate analyses of both genes do not show enough resolution, only the analysis of concatenated sequences is presented here. The phylogeny of Zoantharia inferred using the Bayesian method is shown in Fig. 3. The trees obtained by using Bayesian and ML methods are very similar. They differ only by the relationships between P. puertoricense and Parazoanthus. sp5, which are monophyletic in the Bayesian tree and paraphyletic in the ML tree. The comparison of Bayesian and ML trees using the KH test revealed no significant differences between their topologies (Δln L=0.57557, P= 0.391).
Bayesian tree of the concatenated mitochondrial 12S and 16S rRNA genes. The Actiniaria used as outgroup is Metridium senile. Posterior probabilities are given as the first number; the second number represents bootstrap values, the nodes supported by the variable regions (not included in the analyses) are marked with a black circle. Although not represented as a monophyletic clade on the tree, the species followed by a white square are supported as monophyletic by insertions
The general structure of Zoantharia tree shows paraphyletic Macrocnemina, within which the Brachycnemina form a monophyletic lineage (Fig. 3). Within the Brachycnemina, two relatively well-supported monophyletic groups can be distinguished. The first group comprises Palythoa and Protopalythoa, and the second group includes Isaurus, Zoanthus and Acrozoanthus. Within Macrocnemina, the genus Epizoanthus branches as a sister group to the rest of Zoantharia analysed. The sequences of the genus Parazoanthus form several independent clades or individual branches. The position of some representatives (P. axinellae, P. sp3 from Sulawesi and P. swiftii) are not resolved. The position of P. parasiticus, branching as a sister group of the Cape Verde/Principe species, is also weakly supported. The white form of P. tunicans (P. tunicans W) appears closer to P. gracilis than to the black form of the same species (P. tunicans B). The specimens of S. savaglia from Mediterranean Sea and S. macaronesica from Canaries have identical sequences. The Cape Verde and Principe specimens initially identified as belonging to S. macaronesica branch independently within the Parazoanthus group of sequences.
Indels as phylogenetic markers
To obtain complementary information on phylogenetic relations among Zoantharia, we identified and analysed several insertion/deletions (indels) that were not included in our phylogenetic analyses (Fig. 4). In the 12S alignment, two indels (V1, V2) are variable in length and sequence whereas a third (V3) consists of a 28 bp insertion in the ‘‘Yellow polyps” sequence. The 16S fragment contains five variable regions (V4–V8). V4 is an insertion shared only by P. gracilis and P. tunicans that live on hydrozoans. The second variable region V5 is informative at an intergeneric level (Fig. 4). This indel is located within a region corresponding to the domain V of E. coli 23S (Beagley et al. 1998) and supports the monophyly of the following groups: Epizoanthus (V5a), Savalia (V5b), the two specimens from Cape Verde and Principe (V5c), Parazoanthus sp5 from Sulawesi and P. puertoricense (V5d), Parazoanthus sp3 from Sulawesi, P. axinellae, P. swiftii and P. parasiticus (V5e), the ‘‘Yellow polyps”, P. tunicans and P. gracilis (V5f) and the Brachycnemina (V5g). Despite the identical length of the indel, the two brachycnemic families (Sphenopidae and Zoanthidae) can be distinguished by the sequence variations within the V5g indel. The third variable region V6 is a 51–53 bp insertion in Epizoanthus, which corresponds to a polyC (6–8 bp) in the rest of the Zoantharia (only in the two Acrozoanthus does this polyC start with a single T). The fourth variable region V7 consists of a polyC followed by a polyG and the last variable region V8 is an indel variable in size and sequence within the order. In all these regions (V1-V8), the Brachycnemina have almost identical sequences.
Discussion
Molecular phylogeny versus morphological classification of Zoantharia
Traditional morphology-based classification is only partially confirmed by our molecular data. Although the suborder Brachycnemina was found to be monophyletic, the suborder Macrocnemina appears as a paraphyletic group within which Brachycnemina diverged. Our trees confirm the imperfect status of the fifth septum as a synapomorphic trait of Brachycnemina, but the perfect fifth septum in Macrocnemina appears to be a symplesiomorphic trait. In view of our data, Schmidt’s ‘‘later” and ‘‘earlier” Zoanthidea are not valid groupings, especially concerning the position of the genus Zoanthus, (Fig. 2b). Unfortunately, Schmidt (1979) did not examine the sperm structure of the genus Zoanthus and establish its position exclusively on nematocysts study. However, Schmidt’s aim was to resolve the relationships within the class Anthozoa rather than to establish the phylogeny of Zoantharia.
At a lower taxonomic level, our study confirms the monophyly of the two families of Brachycnemina: the family Zoanthidae characterised by loss of the ability to incorporate sand grains, spicules and other particles in the column wall and the family Sphenopidae as recently characterised by its ability to incorporate sand in the column wall, a zoanthella larva and the absence of b-mastigophore nematocysts in the cnidome (Ryland et al. 2000, 2004). Within Sphenopidae, the low genetic divergence observed between Palythoa and Protopalythoa confirms the suggestion of Burnett et al. (1997) that these two genera could be grouped in a single one.
Within Macrocnemina, two of the three examined genera (Savalia, Epizoanthus) are shown to be monophyletic. Only two species collected from the Mediterranean and Canary Islands are considered as belonging to the genus Savalia. These two species have identical sequences, however, they are characterised by different morphology and ecology (Ocaña and Brito 2003) and therefore, we suspect that the genes examined here are too conserved to resolve their interspecific relationships. As shown by our data, the specimens described as S. macaronesica from Cape Verde and Principe form a separate clade and may represent a new genus.
Concerning the genus Epizoanthus, its extremely divergent position is quite unexpected. Indeed, we do not observe striking morphological differences between Epizoanthus and other zoanthids. Our results confirm the validity of the families Epizoanthidae and Parazoanthidae created by Delage and Hérouard (1901) essentially based on the position of the sphincter, and the presence/absence of a lacuna in the mesoglea of the column. Within the genus Epizoanthus, E. illoricatus branches separately from the three others confirming the doubts concerning the assignment of this species to this genus, expressed already in its original description (Tischbierek 1930).
The phylogeny of species growing on sponges belonging to the genus Parazoanthus is unresolved. This lack of resolution can be explained by the low divergence between analysed sequences of this genus. Interestingly, significant differences observed in Parazoanthus indels indicate close relationships between species according to their substrate. In view of our data, the diagnostic morphological characters of this genus (endodermic sphincter, well-developed canal system in the mesoglea) should be reconsidered. In the future, new genera could be defined based on molecular groupings, substrate specificity in certain cases and, maybe, a detailed revision of their morphology; although no reliable morphological characters resolving relationships within Parazoanthus have been found until now.
Substrate specificity
Epizoism is a well-known trait of the ecology of Zoantharia, but the causes and the range of this association is not well understood. Macrocnemina live generally attached to different animal substrates. Most of the species of the genus Epizoanthus live at great depths (down to 5,000 m) where hard substrates are seldom found and are usually associated with molluscs, pagurids or stems of hyalonematid glass sponges (Ryland et al. 2000). Colonies associated with pagurids create a carcinoecium around the shell inhabited by the crustacean. E. illoricatus grows on the tube secreted by a sedentary polychaete belonging to the Eunicidae. This trait of colonisation of eunicid tubes is shared with the brachycnemic genus Acrozoanthus. In this case, the substrate character can be considered homoplasic. The other brachycnemic species live on rocks or are buried in the sand as the genus Sphenopus. In the genera Parazoanthus and Savalia, some species are currently identified according to their substratum, but their morphological identification is not sufficient to confirm the specificity of such relationships.
By resolving the phylogenetic relations among Zoantharia, our study allows us to test substrate specificity in this group. In the genus Parazoanthus, all monophyletic lineages are associated with well-defined substrates. For example, P. tunicans and P. gracilis, which form a relatively well-supported clade, live specifically on hydrozoans. Substrate specificity is also observed in the clade of Parazoanthus sp5 and P. puertoricense found on the sponge Agelas. The monophyly of this group, characterised by very small polyps, is not well supported by posterior probabilities or bootstrap values but both species share the same V5 and V8 indels (Fig. 4), suggesting a close relationship. Indels also indicate close relationships between the other four Parazoanthus living on sponges and characterised by bigger polyps, despite their unclear position in our tree. The situation is similar in the case of genus Savalia, growing on gorgonians or antipatharians, and the species from Cape Verde and Principe growing exclusively on the antipatharian Tanacetipathes cavernicola. If these data are confirmed by further studies, substrate specificity could be used as a reliable indicator for taxonomic identification of some Macrocnemina.
References
Beagley CT, Okimoto R, Wolstenholme DR (1998) The Mitochondrial genome of the sea anemone Metridium senile (Cnidaria): introns, a paucity of tRNA genes, and a near-standard genetic code. Genetics 148:1091–1108
Burnett WJ, Benzie JAH, Beardmore JA, Ryland JS (1997) Zoanthids (Anthozoa, Hexacorallia) from the Great Barrier Reef and Torres Straits, Australia: Systematics, evolution and a key to species. Coral Reefs 16:55–68
Crocker LA, Reiswig HM (1981) Host specificity in sponge-encrusting Zoanthidea (Anthozoa: Zoantharia) of Barbados, West Indies. Mar Biol 65:231–236
Delage Y, Hérouard E (1901) Traité de Zoologie concrète Volume 2 les Coelentérés. Schleicher Frères, Paris
Gravier C (1918) Note sur une Actinie (* Thoracactis* n. g., * topsenti* n. sp.) et un Annélide Polychète (* Hermadion Fauveli* n. sp.), commensaux d’une Éponge siliceuse (* Sarostegia oculata* Topsent). Bull Inst Oceanogr 344:1–20
Haddon AC, Shackleton AM (1891) Reports on the zoological collections made in Torres Straits by Professor A.C. Haddon, 1888–1889. Actiniae: I. Zoantheae. Sci Trans R Dublin Soc 4:673–701
Häussermann V (2003) Zoanthidea. In: Das Mittelmeer, bestimmungsführer Band II/1 (Hofrichter R.).—Heidelberg: Spektrum, Akad. Verl. pp 501–505
Hall TA (1999) BioEdit: a user-friendly biological sequence alignement editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Herberts C (1972) Etude systématique de quelques zoanthaires tempérés et tropicaux. Téhtys suppl 3:69–156
Huelsenbeck JP, Ronquist F (2001) MrBayes: a program for the Bayesian inference of phylogeny. Available from http://morphbank.ebc.uu.se/mrBayes3/manual.php
Kishino H, Hasegawa M (1989) Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol 29:170–179
Lacaze-Duthiers H (1864) Mémoires sur les antipathaires (Genre Gerardia, L. D.). Ann Sci Nat 2(5):169–239
Le Goff-Vitry MC, Rogers AD, Baglow D (2004) A deep sea slant on the molecular phylogeny of the Scleractinia. Mol Phyl Evol 30:167–177
Ocaña O, Brito A (2003) A review of Gerardiidae (Anthozoa: Zoantharia) from the Macaronesian islands and the Mediterranean sea with the description of a new species. Rev Acad Canar Cienc XV 3–4:159–189
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:419–431
Ryland JS, Muirhead AM (1993) Order Zoanthidea (Class Anthozoa, Zoantharia). In: Mather P, Bennett I (eds) A coral reef handbook, 3rd edn. Surrey Beatty& Son Pty Ltd, Sydney, pp 52–58
Ryland JS (1997) Reproduction in Zoanthidea (Anthozoa: Hexacorallia). Invert Reprod Dev 31:177–188
Ryland JS, de Putron S, Scheltema RS, Chimonides PJ, Zhadan DG (2000) Semper’s (zoanthid) larvae: pelagic life, parentage and other problems. Hydrobiologia 440:191–198
Ryland JS, Lancaster JE (2003) Revision of methods for separating species of Protopalythoa (Hexacorallia: Zoanthidea) in the tropical West Pacific. Invert Syst 17:407–428
Ryland JS, Brasseur MM, Lancaster JE (2004) Use of cnidae in taxonomy: implications from a study of Acrozoanthus australiae (Hexacorallia, Zoanthidea). J Nat Hist 38:1193–1223
Sanchez AJ, Lasker HR, Taylor DJ (2003) Phylogenetic analyses among octocorals (Cnidaria): mitochondrial and nuclear DNA sequences (lsu-rRNA, 16S and ssu-rRNA, 18S) support two convergent clades of branching gorgonians. Mol Phyl Evol 29: 31–42
Schmidt H (1974) On the evolution in the Anthozoa. In: Cameron AM, Campbell BM, Cribb AB, Endean R, Jell JS, Jones OA, Mather P, Talbot FH (eds) The Great Barrier Reef Committee, Brisbane. Proceedings of the 2nd international symposium on coral reefs, vol 1, pp 533–560
Schmidt H (1979) Die Spermien der Anthozoen und ihre phylogenetische Bedeutung. Zoologica 44(129):1–46
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland
Tischbierek H (1930) Zoanthiden auf Wurmröhren. Zool Anz 91:91–95
Veron JEN (1995) Corals in space and time; the biogeography and the evolution of Scleractinia. UNSW Press, Sydney, p 321
Acknowledgments
We are grateful to L. Zaninetti for her enthusiasm and constant support for this study and to H. Zibrowius for his precious naturalist advices and ideas. We thank the divers who collected the samples: M. Boyer and P. Wirtz; J. Wuest and M. Crevecoeur’s group (Geneva), C. Bezac and C. Marschal (Marseille) for their help with the histology, J.D. Reimer for his advice on the manuscript, as well as to P. Schuchert and J. Vacelet for help in identification of hydrozoans and sponges. All the experiments comply with the current laws of the country in which they were conducted.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by O. Kinne, Oldendorf/Luhe
Rights and permissions
About this article
Cite this article
Sinniger, F., Montoya-Burgos, J.I., Chevaldonné, P. et al. Phylogeny of the order Zoantharia (Anthozoa, Hexacorallia) based on the mitochondrial ribosomal genes. Marine Biology 147, 1121–1128 (2005). https://doi.org/10.1007/s00227-005-0016-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00227-005-0016-3