Abstract
Objectives
To establish and validate a radiomics model based on multiparametric magnetic resonance imaging (MRI), and to predict microsatellite instability (MSI) status in rectal cancer patients.
Methods
A total of 199 patients with pathologically confirmed rectal cancer were included. The MSI status was confirmed by immunohistochemistry (IHC) staining. Clinical factors and laboratory data associated with MSI status were analyzed. The imaging data of 100 patients from one of the hospitals were used as the training set. The remaining 99 patients from the other two hospitals were used as the external validation set. The regions of interest (ROIs) were delineated from T1-weighted imaging (T1WI), T2-weighted imaging (T2WI), diffusion-weighted imaging (DWI), and contrast-enhanced T1WI (CE-T1WI) sequence to extract the radiomics features. The Tree-based approach was used for feature selection. The models were constructed based on the four single sequences and a combination of the four sequences using the random forest (RF) algorithm. The external validation set was used to verify the generalization ability of each model. The receiver operating characteristic (ROC) curves and the area under the curve (AUC) were plotted to evaluate and compare the predictive performance of each model.
Results
In the four single-series models, the CE-T1WI model performed the best. The AUCs of the T1WI, T2WI, DWI, and CE-T1WI prediction models in the training set were 0.74, 0.71, 0.71, and 0.78, respectively, while in the external validation set, the corresponding AUCs were 0.67, 0.66, 0.70, and 0.77. The prediction and generalization performance of the combined model of multi-sequences was comparable to that of the CE-T1WI model and it was better than that of the remaining three single-series models, with AUC values of 0.78 and 0.78 in the training and validation sets, respectively.
Conclusion
The established radiomics models based on CE-T1WI or multiparametric MRI have similar predictive performance. They have the potential to predict MSI status in rectal cancer patients.
Key Points
• A radiomics model for the prediction of MSI status in patients with rectal cancer was established and validated using external validation.
• The models based on CE-T1WI or multiparametric MRI have better predictive performance than those based on single unenhanced sequence images.
• The radiomics model has the potential to suggest MSI status in rectal cancer patients; however, it is not yet a substitute for histological confirmation.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
According to 2021 statistics, colorectal cancer (CRC) is the third most common cancer and is the third leading cause of death among all types of cancer, accounting for about 9% of total cancer mortality in men and 8% in women [1]. Microsatellite instability (MSI) status offers a predictive value in response to adjuvant chemotherapy and overall prognosis in CRC cancer [2, 3]. MSI high (MSI-H) colorectal cancer has a better prognosis, in part because it is more likely to benefit from immunotherapy. The National Comprehensive Cancer Network (NCCN) guidelines recommend MSI testing in all CRC patients [4].
MSI status is usually determined by pathological examination of tumor samples. However, the heterogeneity of MSI status within the tumor tissue or between different sites of the tumor, as well as the heterogeneity during different stages of the tumor treatment process, leads to limitations in histological examination methods. Furthermore, histological evaluation of MSI status is rather complex and may not be achieved in some centers without delay, including the transfer of samples to a specialized center. Around 2% of rectal tumors are MSI-H [5]. It might be helpful to develop a relatively non-invasive and convenient surrogate biomarker to predict MSI status.
Radiomics studies in rectal cancer, using magnetic resonance imaging (MRI) mainly reported about the prediction of treatment efficacy [6, 7], tumor stage [8], tumor metastasis [9, 10], and patient survival [11]. There are few MRI-based radiomics reports predicting MSI status in rectal cancer [12, 13]. These studies were conducted in single centers and lacked external validation. The superiority of the multi-sequence MRI in the prediction of MSI status in CRC patients remains to be confirmed. Therefore, the aim of our study was to develop models based on a single sequence and multiple MRI sequences and to evaluate the generalizability using external validation.
Materials and methods
Patients
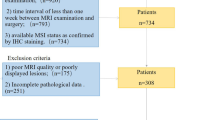
From May 2017 to May 2020, a total of 100 rectal cancer patients from hospital I were enrolled as the training dataset and 99 patients from hospital II and hospital III enrolled as the external verification dataset in our study. The inclusion criteria were as follows: (1) histopathological diagnosis of rectal adenocarcinoma; (2) absence of any type of anti-cancer treatments before MRI examination; (3) time interval of less than one week between MRI examination and surgery; (4) complete MRI examination sequences, including T1WI, T2WI, DWI, and CE-T1WI, in particular, DWI sequences with same or similar b values; (5) available MSI status as confirmed by IHC staining. The exclusion criteria were as follows: (1) incomplete clinical data; (2) insufficient MRI image quality or incomplete sequences; (3) lesions whose identification was difficult on MRI images.
Clinical data, including age, gender, location of the tumor (upper, middle, and low), carcinoembryonic antigen (CEA) (normal or abnormal), and carbohydrate antigen-199 (CA199) status (normal or abnormal), were recorded.
MSI status assessment
The MSI status was confirmed by the expression levels of four MMR proteins, namely MLH1, MSH2, MSH6, and PMS2, which were identified by IHC staining. Specimens were fixed by formaldehyde, and they were paraffin-embedded and routinely stained with hematoxylin-eosin (HE). The normal intestinal mucosa and inflammatory cells present in each section were used as internal controls to interpret the IHC staining results. The results were read and confirmed by two experienced pathologists specializing in CRC diagnosis for 10 years. Based on the expression of MMR proteins, the patients were divided into the following two groups: the MS-low/stable (MS-L/S) group (positive staining of four MMR proteins) and the MSI-H group (any one of the MMR proteins was negative).
MRI protocol, preprocessing, and segmentation
MRI protocol
All MRI examinations on the enrolled patients were performed using a 1.5-T scanner (Optima or Signa, GE Medical Systems). The imaging sequences included fast spin echo T1-weighted sequence, T2-weighted sequence, DWI, and contrast enhancement. Imaging acquisitions of the sequences used in the research are described in Appendix E1.
Tumor segmentation
The Dr. Wise Multimodal Research Platform (https://keyan.deepwise.com,V1.6.2) (Beijing Deepwise & League of PHD Technology Co., Ltd.) was used for radiomics analysis, which included image annotation, feature extraction and selection, and model construction. Among them, the feature extraction module was implemented with the open-source PyRadiomics library.
(https://github.com/Radiomics/pyradiomics). The workflow of the radiomics model construction and validation is shown in Fig. 1.
All images were processed in the original DICOM format and transferred to a post-processing workstation, where two experienced abdominal radiologists manually annotated the lesion in the original image to obtain a 3D volumetric map of the ROI for feature extraction and quantification. Tumors were delineated along the border of the lesion in the images of each sequence (Fig. 2), covering the surrounding burrs while excluding the bowel contents and adjacent mesenteric fat tissue, as well as the non-invaded rectal wall. Both radiologists were blinded to the MSI status. A discrepancy was then resolved by discussion and consensus. Additionally, a total of 30 patients were randomly selected to evaluate the intra-reader and inter-reader agreement. Reader 1 performed the segmentation of the 30 tumors twice with a 1-month interval. Reader 2 performed the segmentation of the 30 tumor features once. Features with an inter-class correlation coefficient > 0.75 and an intra-reader correlation coefficient > 0.75 were considered to be satisfactory agreement and then were selected for further analysis.
Feature extraction
For images with different resolutions, resampling was performed for image pre-processing using B-spline interpolation and all images were resampled to the same resolution [1]. The image gray-scale values were normalized. The normalization procedure was based on the following mathematical formula:
where f(x) is the normalized image density, x is the original image density, μ(x) is the mean image density value, σ(x) is the SD of image density, and s is the scaling factor, which was 100 in this study. The absolute gray-level discretization with fixed bin size (FBS) was set to 5 as previous studies suggested [14].
To emphasize the imaging characteristics, image filters such as log (Laplacian of Gaussian), gradient, and lbp-2d/3d, and four common point-level transforms were applied prior to feature extraction. In addition, wavelet decomposition was applied at each channel for images to obtain all possible combinations in high-pass or low-pass filters (LLH, LHL, LHH, HLL, HLH, HHL, HHH, LLL). The radiomics features of each sequence image were extracted, and they included the following features: first-order features based on the pixel values of the images, shape features describing the morphology of the tumor, and a set of features describing the internal and surface textures of the lesion, Gray Level Co-occurrence Matrix (GLCM), Gray Level Run Length Matrix (GLRLM), Gray Level Zone Size Matrix (GLSZM), and Gray Level Dependence Matrix (GLDM). A total of 6420 radiomics features were extracted for each region of interest (ROI) in all the sequences and the z-score was normalized. The clinical variables (age, gender, tumor location, CEA status, and CA19-9 status) were subjected to statistical tests.
Dimensionality reduction and establishment of the model
Feature reduction, model construction, and external validation of the model were conducted based on a single sequence, including T1WI, T2WI, DWI, and CE-T1WI, and combined multiple sequences. Feature correlation analysis was first performed based on Pearson correlation analysis with a feature correlation threshold of 0.9. When the linear correlation coefficient between any two independent variables in the training set was greater than this threshold, one of the features was removed to alleviate redundancy between features. Priority was given to retaining the features with a higher linear correlation coefficient with the dependent variable. The tree-based approach was used for feature selection, where a tree model was built on the training data and impurity-based feature importance was used to select the important features and obtain the optimal feature subset. A random forest (RF) classifier was used to build the model. The internal validation of the model was completed using a 10-fold cross-validation method, which was a stronger method of model validation as the model was trained on 100% of the data. The generalization ability of the model was evaluated using an external validation set consisting of two hospitals. The ROC curves were used to assess model performance. The ROC curve is an indicator of the rate of false positives and true positives for continuous variables. The evaluation indicators mainly included AUC, sensitivity, specificity, and accuracy. The AUC was the main evaluation indicator of model performance.
Statistical analysis
SPSS (version 16.0) software was used for the statistical analysis of clinical parameters. For quantitative variables, the independent-samples t-test was used to detect differences between groups. For categorical variables, the X2 test was used to detect differences between groups. The Scikit learning package (version 0.20.3) was used to build the classification models. The ROC curves were plotted using Matplotlib (version 3.1.0).
Results
Clinical and laboratory characteristics
Based on the inclusion and exclusion criteria, 199 patients from three institutions were included in the study. There were 100 patients from hospital I who were used as the training set, including 36 MSI-H and 64 MS-L/S CRC patients. Furthermore, 99 patients from hospital II and hospital III were used as the validation set, including 32 MSI-H and 67 MS-L/S CRC patients. A total of 113 males and 86 females were included (average age: 57 years; range: 25–90 years).
The clinical and histopathological characteristics of rectal cancer patients included in our research are provided in Table 1. In both the training and external validation datasets, patient age, tumor location, gender, CA199, and CEA status were comparable between groups (p > 0.05). The clinical characteristics and laboratory features did not differ significantly between the training dataset and the validation dataset (p > 0.05).
Predictive performance and external validation of the radiomics signature
After feature dimensionality reduction, the 20 most significant features were selected to construct the combined model based on multiple sequences of images. These included 6 features in DWI, 4 features in T2WI, 5 features in T1WI, and 5 features in CE-T1WI (Fig. 3).
Plot of feature importance F1: shape_Maximum2DDiameterSlice-CE-T1WI, F2: shape_MinorAxisLength-DWI, F3: shape_MinorAxisLength- CE-T1WI, F4: glrlm_GrayLevelNonUniformity-T2WI, F5: shape_Elongation-DWI, F6: gldm_DependenceEntropy-T1WI, F7: glcm_Idmn-T1WI, F8:original_glcm_Imc2-DWI, F9: glrlm_GrayLevelNonUniformity-T1WI, F10: gldm_LargeDependenceHighGrayLevelEmphasis-T2WI, F11: gldm_SmallDependenceLowGrayLevelEmphasis-DWI, F12: shape_Maximum2DDiameterSlice-DWI, F13: shape_Maximum2DDiameterColumn-DWI, F14: shape_SurfaceVolumeRatio-T1WI, F15:original_shape_Elongation- CE-T1WI, F16: gldm_SmallDependenceLowGrayLevelEmphasis-T2WI, F17: shape_Maximum2DDiameterRow- CE-T1WI, F18: shape_SurfaceVolumeRatio-T2WI, F19: glcm_ClusterShade- CE-T1WI, F20: glcm_ClusterProminence-T1WI
In total, we obtained four models based on four independent sequences and a combined model based on multiple sequences images. The accuracy, sensitivity, specificity, and AUC value of the training set were listed in Tables 2 and 3. The CE-T1WI model had a comparable AUC value of 0.78 to the multi-sequence model, whose AUC value was higher than that of the remaining single sequence models (Table 3). After external validation, the AUC values of the T1WI model and T2WI model decreased significantly, from 0.74 to 0.67 and from 0.71 to 0.66, respectively, while the AUC values of the remaining models did not show a significant decline (Table 3). The ROC curves of the radiomics signature are shown in Fig. 4.
Discussion
In this study, the clinical and laboratory characteristics were not included for model construction, as they were not statistically significant. Five radiomics models were established based on T1WI, T2WI, DWI, CE-T1WI, and multiple sequences (Fig. 4). The prediction models based on CE-T1WI and multiple sequences performed the best, with the highest AUC values and no significant decrease in the external validation set, suggesting that the developed predictive models were reliable and reproducible. They may be able to provide complementary information to determine MSI status in patients with rectal cancer to guide targeted therapy.
In clinical practice, MSI testing has the following important implications for CRC tumors: (1) MSI testing is used as a screening method for Lynch syndrome [15]; (2) MSI is a prognostic factor for stage II CRC [16]; (3) MSI is a predictor of the efficacy of adjuvant chemotherapy for stage II CRC [17]; (4) For the treatment of patients with stage IV CRC, MSI status helps to screen the population for whom the immune checkpoint inhibitor therapy is indicated. But it is important to note that there is a difference in MSI status between the colon and the rectum.
The frequency of MSI-H in a statistically linear fashion from the rectum to the ascending colon [18]. Given the low incidence of MSI rectal cancers, it is a challenge to study the effect of MSI on rectal cancer. Studies with a limited sample size predominate the literature. In Hasan et al’s study, only 8% of rectal cancers had their MSI status determined which may result in the selection bias issue, perhaps testing was reserved for cases that did not respond well to conventional chemotherapy, or perhaps it was only routinely being conducted at academic centers [3]. MSI has been consistently associated with both improved survival in colon cancer and decreased survival in rectal cancer [19, 20]. Locally advanced MSI rectal cancer was significantly associated with lower rates of pCR in response to neoadjuvant chemoradiotherapy [3]. Previous studies have shown the value of a CT image–based model for identifying the MSI status of CRC [21, 22]. It is well-known that MRI has a higher soft tissue resolution compared to CT, and it is the preferred imaging modality for rectal cancer [23]. It already has a key role in triaging patients for pre-operative chemoradiotherapy [24, 25]. it may be used as an imaging biomarker of MSI status.
As each sequence has a different role, it is difficult to fully characterize a tumor by relying on a single sequence alone. T1WI and T2WI provide basic morphological features of the tumor, while the inhomogeneity of DWI images reflects tumor heterogeneity [23, 26, 27]. Previous studies were based on only one or two non-enhanced sequences of MRI, whereas our research included all sequences of rectal MRI in the study. Compared to non-enhanced sequences, the model based on CE-T1WI was able to obtain an optimal predictive value compared to that of the model based on multiparametric images, with better generalization performance. This may be related to the histopathological characteristics of MSI-H tumors. MSI-H tumors are usually larger in size; tend to have mixed morphological features of glandular, mucinous, and solid components; and have a higher microvascular density [28, 29]. The enhancement sequence reflects not only the differences in tumor perfusion but also the differences in pixels in the mucinous and tumor cell-rich areas of the tumor.
The model developed by Zhang et al based on T2WI images accurately predicted the MSI status of rectal cancer with an AUC of 0.895 [12]. Li et al combined T2WI and ADC images to build the model, which had an AUC of 0.908 [30]. Although we expanded the sample size, the AUC values in the training set were still lower than those in their study. In our study, the AUC values of the model constructed based on T1WI and T2WI sequence images showed a significant decrease in the external validation set, demonstrating poor reproducibility of the model. Therefore, models derived from T1WI and T2WI sequences that have not been validated for repeatability may not be reliable. This is the problem of single-center research. It is difficult to compare and reproduce these results in studies at different institutions due to large differences in scan equipment and parameter settings, image pre-processing, the method of ROI segmentation, and the process of feature extraction [31]. The application of multi-center external validation provides a diverse range of imaging information to better assess the reproducibility of the model.
However, it should be emphasized that the model in this study has moderate predictive efficacy for the prediction of MSI status, and it may complement, but not replace histological MSI detection.
There are some limitations to our study. First, manual segmentation of the ROI used in this study is highly subjective and time-consuming, and further applications of semi-automatic or automatic segmentation are needed. Second, there were difficulties in outlining the lesions on T1WI, we referred to the enhanced images to determine the extent of lesions on T1WI. This may have implications for modelling, but the T1WI sequence was still included in this study to make it more comprehensive. Third, in order to retain as much image information as possible, all sequences were resampled to 1, 1, 1 mm isotropic voxels which may result in heavy interpolation. Fourth, although patients in our research were from three institutions, the number of patients with MSI-H rectal cancer was small; thus, the sample size of this study was relatively small. Further investigation with a larger sample size is still required.
In summary, the prediction and generalization abilities of the models based on CE-T1WI and multiparametric images are better than those of the model based on single unenhanced sequence images, which were moderately associated with the MSI mutation status of rectal cancer. However, this requires further validation with more samples before it can be used in clinical applications.
Abbreviations
- AUC:
-
Area under the curve
- CA199:
-
Carbohydrate antigen-199
- CEA:
-
Carcinoembryonic antigen
- CE-T1WI :
-
Contrast-enhanced T1WI
- CRC :
-
Colorectal cancer
- DWI:
-
Diffusion-weighted imaging
- IHC:
-
Immunohistochemistry
- MRI:
-
Magnetic resonance imaging
- MSI:
-
Microsatellite instability
- MSI-H:
-
MSI high
- MS-L/S:
-
MS-low/stable
- NCCN:
-
National Comprehensive Cancer Network
- RF:
-
Random forest
- ROC:
-
Receiver operating characteristic
- ROI:
-
Regions of interest
- T1WI:
-
T1-weighted imaging
- T2WI:
-
T2-weighted imaging
References
Siegel RL, Miller KD, Fuchs HE et al (2021) Cancer statistics, 2021. CA Cancer J Clin 71(1):7–33
O'Connell E, Reynolds IS, McNamara DA et al (2020) Microsatellite instability and response to neoadjuvant chemoradiotherapy in rectal cancer: a systematic review and meta-analysis. Surg Oncol 34:57–62
Hasan S, Renz P, Wegner RE et al (2020) Microsatellite Instability (MSI) as an Independent Predictor of Pathologic Complete Response (PCR) in locally advanced rectal cancer: a National Cancer Database (NCDB) analysis. Ann Surg 271(4):716–723
Benson AB, Venook AP, Al-Hawary MM et al (2018) Rectal cancer, version 2.2018, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 16(7):874–901
Song Y, Wang L, Ran W et al (2020) Effect of tumor location on clinicopathological and molecular markers in colorectal cancer in Eastern China patients: an analysis of 2,356 cases. Front Genet 11:96
Cheng Y, Luo Y, Hu Y et al (2021) Multiparametric MRI-based Radiomics approaches on predicting response to neoadjuvant chemoradiotherapy (nCRT) in patients with rectal cancer. Abdom Radiol (NY) 46(11):5072–5085
Wan L, Peng W, Zou S et al (2020) MRI-based delta-radiomics are predictive of pathological complete response after neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Acad Radiol Suppl 1:S95–S104
Shu Z, Mao D, Song Q et al (2021) Multiparameter MRI-based radiomics for preoperative prediction of extramural venous invasion in rectal cancer. Eur Radiol 32(2):1002–1013
Shu Z, Fang S, Ding Z et al (2019) MRI-based Radiomics nomogram to detect primary rectal cancer with synchronous liver metastases. Sci Rep 9(1):3374
Liu H, Zhang C, Wang L et al (2019) MRI radiomics analysis for predicting preoperative synchronous distant metastasis in patients with rectal cancer. Eur Radiol 29(8):4418–4426
Chen F, Ma X, Li S et al (2020) MRI-based radiomics of rectal cancer: assessment of the local recurrence at the site of anastomosis. Acad Radiol Suppl 1:S87–S94
Zhang W, Huang Z, Zhao J et al (2021) Development and validation of magnetic resonance imaging-based radiomics models for preoperative prediction of microsatellite instability in rectal cancer. Ann Transl Med 9(2):134
Zhang W, Yin H, Huang Z et al (2021) Development and validation of MRI-based deep learning models for prediction of microsatellite instability in rectal cancer. Cancer Med 10(12):4164–4173
Zwanenburg A, Vallieres M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2):328–338
Pathak SJ, Mueller JL, Okamoto K et al (2019) EPCAM mutation update: Variants associated with congenital tufting enteropathy and Lynch syndrome. Hum Mutat 40(2):142–161
Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23(3):609–618
Ribic CM, Sargent DJ, Moore MJ et al (2003) Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med 349(3):247–257
Yang SY, Cho MS, Kim NK (2018) Difference between right-sided and left-sided colorectal cancers: from embryology to molecular subtype. Expert Rev Anticancer Ther 18(4):351–358
Samowitz WS, Curtin K, Wolff RK et al (2009) Microsatellite instability and survival in rectal cancer. Cancer Causes Control 20(9):1763–1768
Samowitz WS, Curtin K, Ma KN et al (2001) Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol Biomarkers Prev 10(9):917–923
Li Z, Zhong Q, Zhang L et al (2021) Computed tomography-based radiomics model to preoperatively predict microsatellite instability status in colorectal cancer: a multicenter study. Front Oncol 11:666786
Cao Y, Zhang G, Zhang J et al (2021) Predicting microsatellite instability status in colorectal cancer based on triphasic enhanced computed tomography radiomics signatures: a multicenter study. Front Oncol 11:687771
Beets-Tan RGH, Lambregts DMJ, Maas M et al (2018) Magnetic resonance imaging for clinical management of rectal cancer: updated recommendations from the 2016 European Society of Gastrointestinal and Abdominal Radiology (ESGAR) consensus meeting. Eur Radiol 28(4):1465–1475
Bulens P, Couwenberg A, Haustermans K et al (2018) Development and validation of an MRI-based model to predict response to chemoradiotherapy for rectal cancer. Radiother Oncol 126(3):437–442
Petkovska I, Tixier F, Ortiz EJ et al (2020) Clinical utility of radiomics at baseline rectal MRI to predict complete response of rectal cancer after chemoradiation therapy. Abdom Radiol (NY) 45(11):3608–3617
Kudou M, Nakanishi M, Kuriu Y et al (2020) Value of intra-tumor heterogeneity evaluated by diffusion-weighted MRI for predicting pathological stages and therapeutic responses to chemoradiotherapy in lower rectal cancer. J Cancer 11(1):168–176
Cai C, Hu T, Gong J et al (2021) Multiparametric MRI-based radiomics signature for preoperative estimation of tumor-stroma ratio in rectal cancer. Eur Radiol 31(5):3326–3335
Maby P, Tougeron D, Hamieh M et al (2015) Correlation between density of CD8+ T-cell infiltrate in microsatellite unstable colorectal cancers and frameshift mutations: a rationale for personalized immunotherapy. Cancer Res 75(17):3446–3455
De Smedt L, Lemahieu J, Palmans S et al (2015) Microsatellite instable vs stable colon carcinomas: analysis of tumour heterogeneity, inflammation and angiogenesis. Br J Cancer 113(3):500–509
Li Z, Dai H, Liu Y et al (2021) Radiomics analysis of multi-sequence MR images for predicting microsatellite instability status preoperatively in rectal cancer. Front Oncol 11:697497
Zhao B, Tan Y, Tsai WY et al (2016) Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 6:23428
Funding
The authors state that this work has not received any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Feng Chen.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 110 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Z., Zhang, J., Zhong, Q. et al. Development and external validation of a multiparametric MRI-based radiomics model for preoperative prediction of microsatellite instability status in rectal cancer: a retrospective multicenter study. Eur Radiol 33, 1835–1843 (2023). https://doi.org/10.1007/s00330-022-09160-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-09160-0