Abstract
Purpose
To investigate the value of T2-radiomics combined with anatomical MRI staging criteria from pre-treatment rectal MRI in predicting complete response to neoadjuvant chemoradiation therapy (CRT).
Methods
This retrospective study included patients with locally advanced rectal cancer who underwent rectal MRI before neoadjuvant CRT from October 2011 to January 2015 and then surgery. Surgical histopathologic analysis was used as the reference standard for pathologic complete response. Anatomical MRI staging criteria were extracted from our institutional standardized radiology report. In radiomics analysis, one radiologist manually segmented the primary tumor on T2-weighted images for all 102 patients (i.e., training set); two different radiologists independently segmented 66/102 patients (i.e., validation set). 108 radiomics features were extracted. Then, scanner-independent features were identified and least absolute shrinkage operator analysis was used to extract a radiomics score. Finally, a support vector machine model combining the radiomics score and anatomical MRI staging criteria was compared against both anatomical MRI-only and radiomics-only models using the deLong test.
Results
The study included 102 patients (42 women; median age = 61 years).The radiomics score produced an area under the curve (AUC) of 0.75. Comparable results were found using the validation set (AUCs = 0.75 and 0.71 for each radiologist, respectively). The anatomical MRI-only model had an accuracy of 67% (sensitivity 42%, specificity 72%); when adding the radiomics score, the accuracy increased to 74% (sensitivity 58%, specificity 77%).
Conclusion
Combining T2-radiomics and anatomical MRI staging criteria from pre-treatment rectal MRI may help to stratify patients based on the prediction of treatment response to neoadjuvant therapy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The treatment for locally advanced rectal cancer involves neoadjuvant chemoradiation therapy (CRT) followed by surgery and then adjuvant chemotherapy. After neoadjuvant treatment, approximately 50–60% of patients are downstaged and 15–27% demonstrates a pathologic complete response (pCR) [1]. Hence, a non-operative approach presents an increasingly attractive alternative to surgery for these patients after neoadjuvant treatment [2]. However, the accurate prediction of complete response after neoadjuvant treatment remains a challenge.
To diagnose a pCR after neoadjuvant treatment, digital rectal examination and endoscopy have accuracies of approximately 85% but are limited in extra-luminal assessment and the interpretation varies among surgeons [3, 4]. Meanwhile, rectal MRI is the preferred imaging modality for local staging and restaging of rectal cancer; in clinical practice, it is preferably performed with endoscopy [5]. A limitation in current clinical practice for all these imaging modalities is that they all involve qualitative assessment with variable reproducibility between different readers [6]. For rectal MRI, T2-weighted imaging and diffusion-weighted imaging (DWI) are the two most frequent sequences used to diagnose tumor response [3, 7,8,9]. However, the reproducibility is suboptimal, predominantly fair for T2-weighted imaging and moderate for DWI [10, 11].Thus, there is a critical need for accurate, consistent, and quantitative measures of response. Such measures could help to stratify patients before treatment and allow alternative and more effective individualized therapies to be offered to patients.
Particularly, quantitative measures derived from the computerized analysis of MR images have been shown to predict survival [12, 13]. Thus, they may also allow the identification of risk factors for local recurrence and distant disease, serving as imaging markers to tailor treatment. Such an approach involving computer automated analysis of images is known as radiomics [14, 15].Research involving radiomics analysis in rectal cancer is emerging [16,17,18,19,20], with a focus on predicting and diagnosing treatment response after neoadjuvant CRT [12, 19, 21,22,23,24,25].
The purpose of this study was to investigate the value of T2-radiomics extracted from pre-treatment rectal MRI combined with anatomical MRI staging criteria in predicting complete response after neoadjuvant CRT for rectal cancer. We hypothesized that the non-invasive radiomics classifiers could add value to stratify patients at the time of initial cancer diagnosis based on the predicted treatment response.
Materials and methods
Study population
This retrospective, single-institution study was compliant with the Health and Insurance Portability and Accountability Act and was approved by the institutional review board with a waiver for written informed consent. We examined our retrospective database for consecutive patients who underwent pre-treatment rectal MRI before starting neoadjuvant CRT from October 2011 to January 2015 followed by surgery (a and b).The most common treatment at our institution during the study period was radiation (50.4 GY) with preferably Capecitabine over 5 weeks [26, 27]. We did not separate our analyses based on drugs or doses received, since it was not a scope of our study. All 102 patients in this study have been previously analyzed in a previous study [19]. In the previous study, the same patients were used to analyze the added value of radiomic features on restaging rectal MRI performed after neoadjuvant treatment, whereas here we focused on the value of radiomic features on the baseline rectal MRI performed before neoadjuvant treatment.
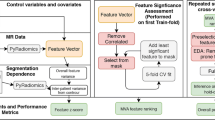
The exclusion criteria were: (a) unavailable baseline MRI scan, (b) mucinous tumor, (c) poor image quality, (d) incomplete tumor coverage on MRI, and (e) rectal perforation. (See Fig. 1 for the flowchart for patient inclusion.)
MRI protocol
Of 102 rectal MRI examinations, 93/102 (91%) were performed at our institution while the remaining 9/102 (9%) examinations were performed at outside institutions. MRI examinations were performed using different GE Healthcare System platforms (Discovery 2MR750, Optima MR450w, Signa EXCITE, and Signa HDxt; Waukesha, WI) with a phased-array coil. MRI parameters at our institution are summarized in Table 1. The minimum requirements for the inclusion of MRI sequences were: (a) high-spatial-resolution axial oblique T2-weighted imaging through the tumor, (b) section thickness of 2–4 mm, and (c) field of view of 180–220 mm. DWI and dynamic contrast-enhanced imaging were included in the qualitative assessment when available.
Qualitative MRI assessment
Anatomical MRI staging criteria were collected from institutional standardized reports for rectal MRI used in clinical practice routinely. The standardized reports were made by our group of specialized radiologists with 5–15 years of experience; in cases of discrepancy or lack of information, they were resolved by a radiologist with training in oncologic imaging (EO).
The following anatomical MRI staging criteria were assessed to complete the standardized reports: (a) tumor location, (b) tumor length, (c) tumor T stage, (d) extramural depth of invasion, and (e) extramural vascular invasion (See Fig. 2).Tumor location was specified according to the distance from the anal verge as low (0–5 cm), middle (5.1–10 cm), or high (10.1–15 cm) [28]. Tumor stage was determined according to the American Joint Committee on Cancer Staging Manual of TNM staging [29] as follows: T1 (tumors are confined to mucosal/submucosal layer), T2 (tumors invade muscularis propria), T3 (tumors invade the mesorectum), and T4 (tumors invade the peritoneum or adjacent organs). Extramural vascular invasion was defined as present when there was expansion or irregularity of vessel wall, loss of normal vascular flow void, and intraluminal tumor signal within a vessel contiguous with the tumor [30, 31].
Quantitative MRI texture analysis
Image segmentation
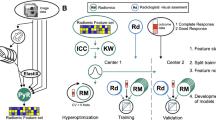
For the training set, a radiologist with training in oncologic imaging (EO) manually segmented the volume of interest (VOI) on all slices on all 102 high resolution axial oblique T2-weighted images using open-source software (ITK-SNAP, version 3.4.0; http://itksnap.org). Tumor deposits, desmoplastic reaction, and extramural vascular invasion were excluded from the segmentation. For the validation set, two different radiologists (JGP and VP) with 5 years of experience in rectal MRI, respectively, independently reviewed all images and segmented tumors on all slices on 66/102 T2-weighted images following the same guidelines as above.
Quantitative texture analysis
Prior to radiomics feature extraction, all images and segmentations were resampled to obtain isotropic voxels of 1 mm3 to compute radiomics features in 3D. A total of 108 radiomics features were extracted using the CERR software which is compliant with the Imaging Biomarker Standardization Initiative (IBSI) standards [32]. These features were divided into the following categories: 20 features from the intensity histogram, 26 features from the gray-level co-occurrence matrix (GLCM), 15 from the gray-level size-zone matrix (GLSZM), 7 shape features, and 40 edges descriptors. GLCM and GLSZM were constructed with a bin size of 128, where the normalized MRI signal intensities were grouped by their values into those bins. Then, a merging strategy was used to compute the GLCM measures, whereby the co-occurrence matrices computed using directional offsets in all directions (N = 24) were combined by averaging [33]. Edge descriptors metrics were obtained with first order statistics (mean, standard deviation, skewness, and kurtosis) of edge maps computed with Sobel, Laplacian of Gaussian, and Gabor (2 bandwidths (σ)—2, \(2\sqrt 2\) and 4 angles (θ)—0°, 30°, 45°, 90°) edge filters.
Reference standard
The reference standard was the standardized histopathologic report of the surgical resection specimens from total mesorectal excision. No additional pathologic analysis was done for our project. pCR was defined as stage ypT0N0.
Statistical analysis
All statistical analyses were performed using R studio with R version 3.5.1 and pROC and caret packages. For selecting radiomics features robust to the magnetic field strength, the Wilcoxon test was performed with a significance threshold of p < 0.05 after Bonferroni adjustment for multiple comparisons. Radiomics features that were not dependent on the considered scanner parameters were deemed stable and used in the subsequent analysis.
From the stable radiomics features, Least Absolute Shrinkage and Selection Operator (LASSO) with fivefold stratified cross-validation was computed to select the most relevant radiomics features for classification. Stratified cross-validation considers that the outcomes (or class labels) are imbalanced, such that all cross-validation folds in the model computation have a similar prevalence of classes. For this study, this strategy maintained the ratio of the number of patients who had a pCR to those who did not. The selected most relevant radiomics features and their coefficients were used to compute a radiomics score [34] which was the weighted sum of features according to their relevance as determined by the LASSO classifier. The capacity of this radiomics score to discriminate pCR patients was investigated with the Wilcoxon test and receiver operating characteristic (ROC) curves. The radiomics score was also computed from all three different radiologist’s segmentations to evaluate its inter-rater robustness.
Finally, a support vector machine (SVM) model with a radial basis function combining the radiomics and anatomical MRI staging criteria was computed. The two models, namely, the radiomics only and anatomical MRI-radiomics based models were then compared using the deLong test of their ROC curves. The SVM classifier was again trained using cross-validation and the accuracy was computed by evaluating the classifier model in each fold using data that were never used in the model training.
The radiomics features were extracted using the open-source CERR software (https://github.com/cerr/CERR/wiki/Radiomics). All code including the R code used in the machine learning analysis are available through the github repository.
Results
Patient characteristics
Our final study population included 102 patients, 60 (59%) men and 42 (41%) women, with a median age of 61 years (range, 31–91 years). Eighty-three patients (81%) achieved pathologic partial response (pPR) at pathologic examination of surgical specimens and 18 (19%) achieved pCR. The average time between end of the CRT and surgery for this study population (n = 84, 18 no date available of end of radiation) was 91.86 days (interquartile range (IQR), 30.5 days). Among those with a complete response (n = 16), the average time between end of CRT and surgery was 85.69 days (IQR, 25 days).Among those with a partial response (n = 68), the average time between end of CRT and surgery was 93.32 days (IQR, 32.5 days). We conducted a t test to compare the means of the complete and partial responders: the p-value was 0.72 (no significant difference between the two groups).
Qualitative MRI evaluation
Anatomical MRI staging criteria were available for all patients. The median tumor length was 4.65 cm (range 2.6–9.4 cm). Most patients had tumor in the mid rectum (47/102,45.7%), T3 stage, and no extramural vascular invasion (80/102, 78.4%). Table 2 summarizes the anatomical MRI staging criteria.
Radiomic classification of pCR
Of 108 radiomics features, 35 were not robust to the magnetic field strength and were removed for the rest of the analysis (See Supplemental Information). Non-robust features mostly comprised those describing global statistics (16/20 features); edge features from Sobel (4/4); and some GLCM, GLSZM, and Gabor edge features (8/25, 3/15, and 4/40, respectively). Of the remaining 73 features, 6 were selected by LASSO: 2 shape features [surface area (coeff = 0.28) and compactness (coeff = − 0.02)], 1 GLCM feature [difference variance (coeff = − 0.0005)], 1 GLSZM feature [size-zone low-gray level emphasis (coeff = 0.10)] and 2 edges descriptors [standard deviation of Gaborσ=2, θ=30°(coeff = 0.11) and kurtosis of Gaborσ=2√2, θ=30° (coeff = 0.05)].
The radiomics score computed as a weighted sum of these 6 radiomics features produced an area under the curve (AUC) of 0.75 (95% CI: 0.63–0.87) for distinguishing patients by pCR. The best cut-off value of 1.39 was associated with a specificity of 0.74 and a sensitivity of 0.70. The Wilcoxon test using the radiomics score showed a significant association with pCR (p = 0.001), where those with pCR had a lower radiomics score (median = 1.30, IQR = 0.35) when compared with patients with partial response (median = 1.51, IQR = 0.44) (See Fig. 3a).
Receiver operating curves showing the specificity and sensitivity of the radiomics score to identify pathologic complete response (pCR) patients (on the left) and boxplots showing the distribution of the radiomics score for pathologic partial response and pCR patients using the best cut-off value (on the right). a Results based on manual segmentations of one radiologist for the entire cohort (n = 102). b, c Results based on manual segmentations of two radiologists for a subset of patients (n = 66) performed independently of each other
Robustness of radiomics features from different segmentations for classifying pCR
Two independently computed radiomics classifiers using the features extracted from the segmentations from the two radiologists classified pCR with an AUC of 0.75 (95% CI 0.59–0.91) and 0.71 (95% CI 0.52–0.90), respectively (See Fig. 3b, c). The radiomics score cut-off value identified on the full training set was also applied to split the patients into two groups, i.e., pCR vs. not complete response (nCR), using the radiomics scores computed from features using segmentations from the two radiologists. These splits were also associated with pCR as determined using the Wilcoxon test (R1 p = 0.01, R2 p = 0.03) and resulted in a specificity of 0.73 and 0.82 and sensitivity of 0.69 and 0.64, respectively.
Support vector machine classifier model for pCR prediction
Using only the anatomical MRI staging criteria, the most important clinical features selected by the SVM model, in order, were the tumor length, the depth of extramural infiltration, a mid-rectum location, and presence/absence of extramural vascular invasion (Fig. 4a). This anatomical MRI-only model achieved an accuracy of 67% (sensitivity: 42%, specificity: 72%).The same model built in combination with the radiomics score showed an increase in accuracy to 74% (sensitivity 58%, specificity 77%), and the most important features were the radiomic score followed by the same anatomical MRI staging criteria from the other SVM model (Fig. 4b).
Discussion
To allow non-operative management following neoadjuvant chemotherapy, the accurate prediction of complete response is essential but remains a challenge. In our study, we were able to compute a radiomics score using T2-weighted images from baseline rectal MRI scans that distinguished patients with pCR from patients with pPR after neoadjuvant chemotherapy (AUC = 0.75). The predictive value of this score was robust, i.e., not affected when computed using the segmentations of three different radiologists. When the radiomics score was used in combination with anatomical MRI staging criteria, the prediction of pCR was improved (74% for the combined model vs. 67% for the clinical-only model).To our knowledge, this is the first study to date using a combined approach including radiomics and anatomical MRI staging criteria to predict complete response after neoadjuvant therapy in rectal cancer patients.
A prior study using CT scans showed that texture analysis can predict five-year overall survival in colorectal cancer patients [35]. However, rectal MRI is the gold standard imaging technique for preoperative staging and has been proven to allow preoperative stratification of patients and permit better targeted therapy [36]. In a recent study, De Cecco et al. demonstrated the potential of T2-based texture parameters to act as predictive tumoral markers of response to neoadjuvant CRT in rectal cancer; however, this study was limited by a small number of patients [21]. In another recent study, Nie et al. assessed the relationship between pre-treatment MRI values and pCR to build predictive models, showing encouraging results [24]. In our study, the computed radiomics score (AUC 0.75) is in line with those of other recently published baseline radiomic studies in rectal cancer; for example, Vandendorpe et al. reported an AUC of 0.70 in predicting downstaging of locally advanced rectal cancer from neoadjuvant CRT using baseline CT images [37].
Our approach combined both T2-radiomics and anatomical MRI staging criteria in one model. Using this combination, compared with using an anatomical MRI-only or radiomics-only model, we were able to improve the accuracy to predict tumor response and stratify patients at baseline to complete responders or partial responders. Our results are in line with the current movement towards individualized treatment in oncology in general and locally advanced rectal cancer specifically. In addition, our results are in line with the need to reliably assess response to treatment earlier. The development of tumor markers based on images such as anatomical and functional MRI may offer additional guidance and the applied field of radiomics such as that used in our study warrants further investigation. Important considerations when performing radiomics analysis is the reproducibility of the extracted features and accounting for the sources of variability starting from the volumetric segmentation of the tumor [38]. To evaluate reproducibility in our study, we used segmentations from three different radiologists with different skill levels and found no difference in the predictive value of the radiomics score. We validated if the model we built is robust to the segmentation by using the segmentation from two other radiologists. While we were not able to split the data into a training and testing set because of the imbalanced dataset (only 19 complete responders), we were able to validate the robustness of the radiomics signature to segmentation.
The present study suggests that combined visual and quantitative characterization of rectal tumors may potentially enhance the discrimination between complete responders and partial responders to neoadjuvant treatment at the time of diagnosis. Further studies and correlation with pathologic material will be important to better understand the potential interplay between combined radiomics features calculated in the radiomics score, and tumor stromal elements. We postulate that tumor biology and stromal elements are different between complete responders and partial responders, and radiomics assessment may provide valuable information in a form of virtual biopsy to aid current diagnostic methods. Knowledge of the predicted response after CRT using radiomic features may influence the treatment decisions of the multidisciplinary team, thereby allowing a more personalized treatment and improving the cost-effectiveness of rectal cancer treatment.
Our results should be interpreted in light of the limitations associated with our study design. First, the number of patients was limited due to the retrospective nature of this study. Second, we did not include patients on nonsurgical management which may have introduced selection bias into the study. However, this decision was made so as to have histopathologic reference standard for all patients as well as to achieve a homogeneous dataset to develop a radiomics algorithm. Third, we were limited to an imbalanced dataset between patients with partial response (n = 83) and patients with complete response (n = 19). However, we note that the percentage of patients with complete response in our cohort is in the range of what is described in the literature [1]. Therefore, the limitation is not that our data is imbalanced compared with what we see in clinical practice and what is described in the literature but rather that the limitation is statistical in nature. Fourth, we did not separate our analyses based on drugs or doses received. Our study was designed as a pilot study, mainly to see if complete responders have radiomics features that may be identified at baseline. For partial responders, further studies are needed to investigate different drugs and doses on a larger dataset. Fifth, the anatomical MRI staging criteria was based on available clinical reports which were made inconsensus; we did not evaluate the inter-reader agreement of the anatomical MRI staging criteria assessment since it was not an aim in our study. Sixth, radiomics was performed on only T2-weighted images to minimize variability in acquisition parameters. Further studies exploring performance of other imaging sequences may improve radiomics analysis. Seventh, manual segmentation of volume of interest is a time-consuming process; hence, there is a great need to develop easy-to-use automated or semi-automated tools to encourage real-world application of radiomics in daily clinical practice. Finally, our study did not have external validation. We performed internal validation using segmentations from three different radiologists. Hence, further studies are needed to resolve these limitations and validate our data to provide generalization of the results.
In conclusion, our preliminary results suggest that radiomics features derived from T2-weighted MR images of rectal cancer have the potential to serve as imaging markers of tumoral response to neoadjuvant chemotherapy at time of initial diagnosis of rectal cancer, and when combined with standardanatomical MRI staging criteria, improve accuracy. Further studies with larger sample sizes are warranted for additional evaluation and validation of our preliminary data.
Abbreviations
- AUC:
-
Area under the curve
- CRT:
-
Chemoradiation therapy
- DWI:
-
Diffusion-weighted imaging
- GLCM:
-
Gray-level co-occurrence matrix
- GLSZM:
-
Gray-level size-zone matrix
- IQR:
-
Interquartile range
- LASSO:
-
Least Absolute Shrinkage and Selection Operator
- nCR:
-
Not complete response
- pCR:
-
Pathologic complete response
- pPR:
-
Pathologic partial response
- ROC:
-
Receiver operating curve
- SVM:
-
Support vector machine
References
Maas M, Nelemans PJ, Valentini V, Das P, Rodel C, Kuo LJ, Calvo FA, Garcia-Aguilar J, Glynne-Jones R, Haustermans K, Mohiuddin M, Pucciarelli S, Small W, Jr., Suarez J, Theodoropoulos G, Biondo S, Beets-Tan RG, Beets GL (2010) Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. The Lancet Oncology 11 (9):835-844. https://doi.org/10.1016/s1470-2045(10)70172-8
Renehan AG, Malcomson L, Emsley R, Gollins S, Maw A, Myint AS, Rooney PS, Susnerwala S, Blower A, Saunders MP, Wilson MS, Scott N, O’Dwyer ST (2016) Watch-and-wait approach versus surgical resection after chemoradiotherapy for patients with rectal cancer (the OnCoRe project): a propensity-score matched cohort analysis. The Lancet Oncology 17 (2):174-183. https://doi.org/10.1016/s1470-2045(15)00467-2
Maas M, Lambregts DM, Nelemans PJ, Heijnen LA, Martens MH, Leijtens JW, Sosef M, Hulsewe KW, Hoff C, Breukink SO, Stassen L, Beets-Tan RG, Beets GL (2015) Assessment of Clinical Complete Response After Chemoradiation for Rectal Cancer with Digital Rectal Examination, Endoscopy, and MRI: Selection for Organ-Saving Treatment. Annals of surgical oncology 22 (12):3873-3880. https://doi.org/10.1245/s10434-015-4687-9
van der Sande ME, Beets GL, Hupkens BJ, Breukink SO, Melenhorst J, Bakers FC, Lambregts DM, Grabsch HI, Beets-Tan RG, Maas M (2019) Response assessment after (chemo)radiotherapy for rectal cancer: Why are we missing complete responses with MRI and endoscopy? European journal of surgical oncology : the journal of the European Society of Surgical Oncology and the British Association of Surgical Oncology 45 (6):1011-1017. https://doi.org/10.1016/j.ejso.2018.11.019
Horvat N, Veeraraghavan H, Pelossof RA, Fernandes MC, Arora A, Khan M, Marco M, Cheng CT, Gonen M, Golia Pernicka JS, Gollub MJ, Garcia-Aguillar J, Petkovska I (2019) Radiogenomics of rectal adenocarcinoma in the era of precision medicine: A pilot study of associations between qualitative and quantitative MRI imaging features and genetic mutations. European journal of radiology 113:174-181. https://doi.org/10.1016/j.ejrad.2019.02.022
Sassen S, de Booij M, Sosef M, Berendsen R, Lammering G, Clarijs R, Bakker M, Beets-Tan R, Warmerdam F, Vliegen R (2013) Locally advanced rectal cancer: is diffusion weighted MRI helpful for the identification of complete responders (ypT0N0) after neoadjuvant chemoradiation therapy? European radiology 23 (12):3440-3449. https://doi.org/10.1007/s00330-013-2956-1
Patel UB, Brown G, Rutten H, West N, Sebag-Montefiore D, Glynne-Jones R, Rullier E, Peeters M, Van Cutsem E, Ricci S, Van de Velde C, Kjell P, Quirke P (2012) Comparison of magnetic resonance imaging and histopathological response to chemoradiotherapy in locally advanced rectal cancer. Ann Surg Oncol 19 (9):2842-2852. https://doi.org/10.1245/s10434-012-2309-3
Sclafani F, Brown G, Cunningham D, Wotherspoon A, Mendes LST, Balyasnikova S, Evans J, Peckitt C, Begum R, Tait D, Tabernero J, Glimelius B, Roselló S, Thomas J, Oates J, Chau I (2017) Comparison between MRI and pathology in the assessment of tumour regression grade in rectal cancer. Br J Cancer 117 (10):1478-1485. https://doi.org/10.1038/bjc.2017.320
Siddiqui MR, Gormly KL, Bhoday J, Balyansikova S, Battersby NJ, Chand M, Rao S, Tekkis P, Abulafi AM, Brown G (2016) Interobserver agreement of radiologists assessing the response of rectal cancers to preoperative chemoradiation using the MRI tumour regression grading (mrTRG). Clin Radiol 71 (9):854-862. https://doi.org/10.1016/j.crad.2016.05.005
Aker M, Boone D, Chandramohan A, Sizer B, Motson R, Arulampalam T (2018) Diagnostic accuracy of MRI in assessing tumor regression and identifying complete response in patients with locally advanced rectal cancer after neoadjuvant treatment. Abdom Radiol (NY). https://doi.org/10.1007/s00261-018-1627-8
Sathyakumar K, Chandramohan A, Masih D, Jesudasan MR, Pulimood A, Eapen A (2016) Best MRI predictors of complete response to neoadjuvant chemoradiation in locally advanced rectal cancer. Br J Radiol 89 (1060):20150328. https://doi.org/10.1259/bjr.20150328
Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C (2016) Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 281 (3):947-957. https://doi.org/10.1148/radiol.2016152234
Kim JH, Ko ES, Lim Y, Lee KS, Han BK, Ko EY, Hahn SY, Nam SJ (2017) Breast Cancer Heterogeneity: MR Imaging Texture Analysis and Survival Outcomes. Radiology 282 (3):665-675. https://doi.org/10.1148/radiol.2016160261
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ (2012) Radiomics: extracting more information from medical images using advanced feature analysis. European journal of cancer (Oxford, England : 1990) 48 (4):441-446. https://doi.org/10.1016/j.ejca.2011.11.036
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278 (2):563-577. https://doi.org/10.1148/radiol.2015151169
Shi L, Zhang Y, Nie K, Sun X, Niu T, Yue N, Kwong T, Chang P, Chow D, Chen JH, Su MY (2019) Machine learning for prediction of chemoradiation therapy response in rectal cancer using pre-treatment and mid-radiation multi-parametric MRI. Magnetic resonance imaging 61:33-40. https://doi.org/10.1016/j.mri.2019.05.003
Shu Z, Fang S, Ding Z, Mao D, Cai R, Chen Y, Pang P, Gong X (2019) MRI-based Radiomics nomogram to detect primary rectal cancer with synchronous liver metastases. Scientific reports 9 (1):3374. https://doi.org/10.1038/s41598-019-39651-y
Zhou X, Yi Y, Liu Z, Cao W, Lai B, Sun K, Li L, Zhou Z, Feng Y, Tian J (2019) Radiomics-Based Pretherapeutic Prediction of Non-response to Neoadjuvant Therapy in Locally Advanced Rectal Cancer. Annals of surgical oncology 26 (6):1676-1684. https://doi.org/10.1245/s10434-019-07300-3
Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I (2018) MR Imaging of Rectal Cancer: Radiomics Analysis to Assess Treatment Response after Neoadjuvant Therapy. Radiology 287 (3):833-843. https://doi.org/10.1148/radiol.2018172300
Giannini V, Mazzetti S, Bertotto I, Chiarenza C, Cauda S, Delmastro E, Bracco C, Di Dia A, Leone F, Medico E, Pisacane A, Ribero D, Stasi M, Regge D (2019) Predicting locally advanced rectal cancer response to neoadjuvant therapy with (18)F-FDG PET and MRI radiomics features. European journal of nuclear medicine and molecular imaging 46 (4):878-888. https://doi.org/10.1007/s00259-018-4250-6
De Cecco CN, Ganeshan B, Ciolina M, Rengo M, Meinel FG, Musio D, De Felice F, Raffetto N, Tombolini V, Laghi A (2015) Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Investigative radiology 50 (4):239-245. https://doi.org/10.1097/rli.0000000000000116
Cusumano D, Dinapoli N, Boldrini L, Chiloiro G, Gatta R, Masciocchi C, Lenkowicz J, Casa C, Damiani A, Azario L, Van Soest J, Dekker A, Lambin P, De Spirito M, Valentini V (2018) Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. La Radiologia medica 123 (4):286-295. https://doi.org/10.1007/s11547-017-0838-3
Meng Y, Zhang C, Zou S, Zhao X, Xu K, Zhang H, Zhou C (2018) MRI texture analysis in predicting treatment response to neoadjuvant chemoradiotherapy in rectal cancer. Oncotarget 9 (15):11999-12008. https://doi.org/10.18632/oncotarget.23813
Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, Niu T, Sun X (2016) Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. Clinical cancer research : an official journal of the American Association for Cancer Research 22 (21):5256-5264. https://doi.org/10.1158/1078-0432.Ccr-15-2997
Sun Y, Hu P, Wang J, Shen L, Xia F, Qing G, Hu W, Zhang Z, Xin C, Peng W, Tong T, Gu Y (2018) Radiomic features of pretreatment MRI could identify T stage in patients with rectal cancer: Preliminary findings. Journal of magnetic resonance imaging : JMRI. https://doi.org/10.1002/jmri.25969
Cercek A, Roxburgh CSD, Strombom P, Smith JJ, Temple LKF, Nash GM, Guillem JG, Paty PB, Yaeger R, Stadler ZK, Seier K, Gonen M, Segal NH, Reidy DL, Varghese A, Shia J, Vakiani E, Wu AJ, Crane CH, Gollub MJ, Garcia-Aguilar J, Saltz LB, Weiser MR (2018) Adoption of Total Neoadjuvant Therapy for Locally Advanced Rectal Cancer. JAMA oncology 4 (6):e180071. https://doi.org/10.1001/jamaoncol.2018.0071
Smith JJ, Strombom P, Chow OS, Roxburgh CS, Lynn P, Eaton A, Widmar M, Ganesh K, Yaeger R, Cercek A, Weiser MR, Nash GM, Guillem JG, Temple LKF, Chalasani SB, Fuqua JL, Petkovska I, Wu AJ, Reyngold M, Vakiani E, Shia J, Segal NH, Smith JD, Crane C, Gollub MJ, Gonen M, Saltz LB, Garcia-Aguilar J, Paty PB (2019) Assessment of a Watch-and-Wait Strategy for Rectal Cancer in Patients With a Complete Response After Neoadjuvant Therapy. JAMA oncology 5 (4):e185896. https://doi.org/10.1001/jamaoncol.2018.5896
Nougaret S, Reinhold C, Mikhael HW, Rouanet P, Bibeau F, Brown G (2013) The use of MR imaging in treatment planning for patients with rectal carcinoma: have you checked the “DISTANCE”? Radiology 268 (2):330-344. https://doi.org/10.1148/radiol.13121361
American Joint Committee on Cancer (2017) AJCC Cancer Staging Manual. 8 edn. Springer International Publishing,
Smith NJ, Shihab O, Arnaout A, Swift RI, Brown G (2008) MRI for detection of extramural vascular invasion in rectal cancer. AJR American journal of roentgenology 191 (5):1517-1522. https://doi.org/10.2214/ajr.08.1298
Beets-Tan RGH, Lambregts DMJ, Maas M, Bipat S, Barbaro B, Curvo-Semedo L, Fenlon HM, Gollub MJ, Gourtsoyianni S, Halligan S, Hoeffel C, Kim SH, Laghi A, Maier A, Rafaelsen SR, Stoker J, Taylor SA, Torkzad MR, Blomqvist L (2018) Magnetic resonance imaging for clinical management of rectal cancer: Updated recommendations from the 2016 European Society of Gastrointestinal and Abdominal Radiology (ESGAR) consensus meeting. European radiology 28 (4):1465-1475. https://doi.org/10.1007/s00330-017-5026-2
Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E, Thor M, Um H, Veeraraghavan H, Oh JH, Shukla-Dave A, Deasy JO (2018) Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Medical physics. https://doi.org/10.1002/mp.13046
Zwanenburg A, Leger S, Vallières M, Löck SJapa (2016) Image biomarker standardisation initiative.
Zheng BH, Liu LZ, Zhang ZZ, Shi JY, Dong LQ, Tian LY, Ding ZB, Ji Y, Rao SX, Zhou J, Fan J, Wang XY, Gao Q (2018) Radiomics score: a potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC cancer 18 (1):1148. https://doi.org/10.1186/s12885-018-5024-z
Ng F, Ganeshan B, Kozarski R, Miles KA, Goh V (2013) Assessment of primary colorectal cancer heterogeneity by using whole-tumor texture analysis: contrast-enhanced CT texture as a biomarker of 5-year survival. Radiology 266 (1):177-184. https://doi.org/10.1148/radiol.12120254
Taylor FG, Quirke P, Heald RJ, Moran B, Blomqvist L, Swift I, Sebag-Montefiore DJ, Tekkis P, Brown G (2011) Preoperative high-resolution magnetic resonance imaging can identify good prognosis stage I, II, and III rectal cancer best managed by surgery alone: a prospective, multicenter, European study. Annals of surgery 253 (4):711-719. https://doi.org/10.1097/SLA.0b013e31820b8d52
Vandendorpe B, Durot C, Lebellec L, Le Deley MC, Sylla D, Bimbai AM, Amroun K, Ramiandrisoa F, Cordoba A, Mirabel X, Hoeffel C, Pasquier D, Servagi-Vernat S (2019) Prognostic value of the texture analysis parameters of the initial computed tomographic scan for response to neoadjuvant chemoradiation therapy in patients with locally advanced rectal cancer. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology 135:153-160. https://doi.org/10.1016/j.radonc.2019.03.011
Tixier F, Um H, Young RJ, Veeraraghavan H (2019) Reliability of tumor segmentation in glioblastoma: Impact on the robustness of MRI-radiomic features. Medical physics 46 (8):3582-3591. https://doi.org/10.1002/mp.13624
Acknowledgements
We thank Joanne Chin, Ye Choi, and Natalie Gangai for their editorial support on this manuscript.
Funding
This study was funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748 and the Colorectal Cancer Research Center CC50367 at Memorial Sloan Kettering Cancer Center.
Author information
Authors and Affiliations
Contributions
Conceptualization: IP, HV, FT, EJO; Methodology: IP, HV, FT, EJO; Software:FT, HV; Data Analysis and Interpretation and/or Image Analyses: IP,HV, FT, EJO, JSGP, VP, DDB, NH, JF, MJG, JG-A; Investigation:IP, HV, EJO, FT; Data Curation: EJO, IP, FT, HV, NH, JSGP, VP, JS; Writing—Original Draft Preparation:IP, HV, FT, EJO; Writing—Review & Editing:IP, HV, FT, EJO, JSGP, VP, DDB, NH, JF, MJG, JG-A, JS; Supervision: IP, HV; Final Approval of the Manuscript: IP, HV, FT, EJO, JSGP, VP, DDB, NH, JF, MJG, JG-A, JS.
Corresponding author
Ethics declarations
Conflict of interest
Dr. Garcia-Aguilar has received honoraria from Medtronic, Johnson & Johnson, and Intuitive Surgical. The remaining authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee (institutional review board at Memorial Sloan Kettering cancer Center, IRB #16-1630) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
For this type of study formal consent is not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Petkovska, I., Tixier, F., Ortiz, E.J. et al. Clinical utility of radiomics at baseline rectal MRI to predict complete response of rectal cancer after chemoradiation therapy. Abdom Radiol 45, 3608–3617 (2020). https://doi.org/10.1007/s00261-020-02502-w
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-020-02502-w