Abstract
Objectives
To investigate the prediction of 1-year survival (1-YS) in patients with metastatic colorectal cancer with use of a systematic comparative analysis of quantitative imaging biomarkers (QIBs) based on the geometric and radiomics analysis of whole liver tumor burden (WLTB) in comparison to predictions based on the tumor burden score (TBS), WLTB volume alone, and a clinical model.
Methods
A total of 103 patients (mean age: 61.0 ± 11.2 years) with colorectal liver metastases were analyzed in this retrospective study. Automatic segmentations of WLTB from baseline contrast-enhanced CT images were used. Established biomarkers as well as a standard radiomics model building were used to derive 3 prognostic models. The benefits of a geometric metastatic spread (GMS) model, the Aerts radiomics prior model of the WLTB, and the performance of TBS and WLTB volume alone were assessed. All models were analyzed in both statistical and predictive machine learning settings in terms of AUC.
Results
TBS showed the best discriminative performance in a statistical setting to discriminate 1-YS (AUC = 0.70, CI: [0.56, 0.90]). For the machine learning–based prediction for unseen patients, both a model of the GMS of WLTB (0.73, CI: [0.60, 0.84]) and the Aerts radiomics prior model (0.76, CI: [0.65, 0.86]) applied on the WLTB showed a numerically higher predictive performance than TBS (0.68, CI: [0.54, 0.79]), radiomics (0.65, CI: [0.55, 0.78]), WLTB volume alone (0.53, CI: [0.40. 0.66]), or the clinical model (0.56, CI: [0.43, 0.67]).
Conclusions
The imaging-based GMS model may be a first step towards a more fine-grained machine learning extension of the TBS concept for risk stratification in mCRC patients without the vulnerability to technical variance of radiomics.
Key Points
• CT-based geometric distribution and radiomics analysis of whole liver tumor burden in metastatic colorectal cancer patients yield prognostic information.
• Differences in survival are possibly attributable to the spatial distribution of metastatic lesions and the geometric metastatic spread analysis of all liver metastases may serve as robust imaging biomarker invariant to technical variation.
• Imaging-based prediction models outperform clinical models for 1-year survival prediction in metastatic colorectal cancer patients with liver metastases.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer is the third most common cancer worldwide [1]. Approximately 50% of patients with colorectal cancer will be diagnosed with metastases either at the time of diagnosis or as part of recurrent disease, whereas the liver is the most common site for metastases [1]. Although surgical resection of hepatic metastases is considered the only curative treatment option, approximately 85% of these patients are ineligible for this treatment due to large tumor burden, multifocal disease, or inadequate liver function [2, 3]. Computed tomography (CT) provides valuable capabilities for non-invasive assessment and quantification of colorectal liver metastases (CRLM) towards the development of predictive quantitative imaging biomarkers (QIBs) [4,5,6]. In recent years, there has been an increased interest to understand survival and response to therapy in tumor patients using the whole tumor burden rather than single lesions [7, 8]. For CRLM patients, the volume of the whole liver tumor burden (WLTB) and the tumor burden score (TBS) were quantified. The TBS is the Pythagorean addition of the lesion number and the diameter of the largest lesion. This measurement was capable to better estimate survival than the number of lesions or the diameter of the largest lesion alone [9, 10]. Although being a natural extension of this concept, the relevance of geometric measures of the WLTB distribution such as distances between various lesions has not yet been evaluated.
Furthermore, texture analysis and machine learning [4,5,6, 11, 12] are playing an increasingly important role in radiology, displacing statistical analysis of QIB. A special branch of this research represents radiomics. This is based on extracting a large number of quantitative features from the images and combining them with machine learning to make the diagnosis, therapy response, and outcome prediction more accurate [13, 14]. In patients with CRLM, radiomics analysis of target lesions was shown to significantly correlate with response to chemotherapy, as well as with survival [4,5,6, 12].
Since the added predictive value of the geometric or radiomics analysis of WLTB is not known, we compare the predictive performance of established clinical and quantitative imaging biomarkers and novel exploratory whole liver tumor burden–based QIBs in CRLM patients by a statistical and also a machine learning approach.
The purpose of our study is therefore to investigate the prediction of 1-year survival in patients with metastatic colorectal cancer with use of a systematic comparative analysis of QIBs based on the geometric and radiomics analysis of WLTB in comparison to predictions based on the TBS, WLTB volume alone, and a clinical model.
Materials and methods
Our retrospective study was approved by and registered with the local institutional review board of the Ludwig-Maximilians-University Munich (approval number: 502-16). Written informed consent was obtained from all subjects.
Study sample
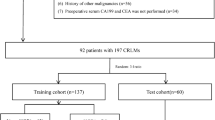
A database of patients with metastatic colorectal cancer from January 2007 to October 2017 was reviewed and 485 patients with metastatic colorectal cancer were identified. Of the 485 enrolled oncological patients, 269 had an available baseline CT scan. Of those, for 220 patients, sufficient clinical data were available. A total of 133 patients of this cohort had colorectal liver metastases (CRLM). Further 15 patients had to be excluded due to limited image quality, native scans, or motion artifacts. Additionally, 15 further patients had to be excluded due to missing information regarding survival status or lack of clinical follow-up information. The final study cohort therefore consisted of 103 patients, of which 82 survived at least 1 year. A flowchart describing the exclusion criteria for patients can be found in Fig. 1.
Imaging studies
Our retrospective study includes baseline CT scans which were acquired using a variety of multidetector-row CT scanners from different manufacturers (see Table 1); default settings were 120 kV tube voltage. Weight-adapted contrast agent was applied intravenously, and images were acquired in portal venous phase and reconstructed using a standard soft tissue kernel. Slice thickness varied between 0.75 and 5 mm.
WLTB segmentations
All CT scans were reviewed independently in a randomized fashion and blinded to the clinical data; only contrast-enhanced CT scans were used for further analysis. All included CT scans were screened for metastases by 2 board-certified radiologists (D.N. and T.H.) with each having 6 years of experience in oncological imaging. WLTB segmentations were performed automatically using custom software based on MeVisLab (MeVis Medical Solutions, Fraunhofer MEVIS) with support of a convolutional neural network [15]. If needed, both radiologists could revise the WLTB segmentations interactively by adding tumors or redefining tumor contours.
Analyzed prognostic models
A general overview of a radiomics workflow can be found in Fig. 2a. After image segmentations, imaging features were extracted by an in-house software that also integrates the PyRadiomics library [16]. Features were grouped into 5 (i to v) prognostic models (Fig. 2b): (i) the imaging prior model, (ii) the clinical prior model, (iii) the Aerts radiomics prior (ARP) model, (iv) the geometric metastatic spread (GMS) model, and (v) the naive model. An overview of the models is shown in Fig. 2b. In detail, 3 of them (i–iii) are based on prior knowledge, one (iv) is our own hypothesis to introduce a novel quantitative imaging biomarker related to the spatial tumor distribution of all liver metastases, and the last model (v) uses all available features in a mechanic standard radiomics model building approach. The prior models are grouped according to their source, e.g., imaging or clinical data.
Basic schemes of our analyses pipelines. a The general radiomics workflow. b The analyzed prior models (i–iii), the GMS model (iv), and the standard radiomics model building (naive, v). c The statistical (I) as well as standard radiomics machine learning (II) model building, and also our machine learning setup based on the GMS hypothesis and prior models (III). d The complexity and effort involved in the respective analyses. Roughly divided, the complexity and/or effort associated with each analysis increases from bottom to top, due to a higher effort to generate WLTB segmentations, the higher model complexity of a non-linear machine learning approach in comparison to a regularized statistical model and the complexity of controlling the impact of scan parameter variation on texture measurements within a radiomics analysis
At first, as a benchmark, we propose an imaging prior (i) model. This model incorporates already discovered discriminative quantitative imaging biomarkers (QIBs) found in oncological imaging such as TBS [9, 17], primary tumor sidedness (PTS) [18,19,20], tumor attenuation [21], and also the whole liver tumor burden volume [22, 23]. PTS was defined as right-sided or left-sided if the tumor arose from the cecum to the hepatic flexure or from the splenic flexure to the rectum, respectively. Those QIBs are also analyzed individually for their prognostic value. In analogy to the imaging prior model, a (ii) clinical prior model is proposed based on current clinical parameters, including data from laboratory and histopathology analysis. This model consists of PTS [19, 20, 22], presence of liver-limited disease (LLD) [20], age, sex, grading, syn-/metachronous metastases [20, 24], histology, and carcinoembryonic antigen (CEA) levels as well as UICC and TNM staging [20]. LLD denotes a specific subgroup of metastatic colorectal cancer patients where the liver is the only metastatic site.
Furthermore—since this model provided good predictive performance when applied on target lesions in multiple oncological imaging studies—we also evaluate the established Aerts radiomics prior (ARP) model (iii) [13, 25,26,27] for the WLTB analysis. This model consists of 4 quantitative image features describing the tumor heterogeneity and compactness.
As described above, we hypothesize that the spatial geometric distribution of the tumors within the liver may also be of diagnostic value and propose a (iv) geometric metastatic spread (GMS) model. This model consists of the maximum distance of liver metastases along x, y, and z CT scanner axes, termed metastatic spread (MSx, MSy, MSz) and the addition of these squared distances (MS). Furthermore, the GMS model integrates a dispersion quantification by 2 features, namely the surface-area-to-volume ratio (SA/V) and the compactness of the spatial metastases distribution. A formal description of the model can be found in Suppl. Mat. B.
Finally, a mechanic construction of a predictive model based on all extractable features (PyRadiomics library + imaging priors + clinical priors + ARP + GMS) solely by machine learning and a minimum redundancy maximum relevance (mRMR) feature selection [28] is tested and termed (v) naive approach. The term naive indicates that no prior knowledge or intuition was used which was formed on the basis of previous study results but a standard radiomics model building process was pursued. Figure 2d describes and ranks the complexity and effort involved in the respective analyses described above.
Data analysis
1-YS and survival time were measured from the date of initial baseline CT at time of the initial diagnosis of metastatic disease to the date of death (if applicable). An overview of the data analysis pipeline can be found in Fig. 2c displaying both our statistical (I) as well as machine learning (II, III) model building. As some biomarkers are rather analyzed statistically while others such as the ARP model are derived from machine learning approaches, we had to include both analyses for a profound comparison.
For statistical model building (Fig. 2c (I)), descriptive statistics, such as means and standard deviation (SD) for continuous variables and frequencies for categorical variables, are used to summarize the data and each feature of prior models and GMS model introduced above. Due to multiple testing corrections, the naive model using all available features (> 1500) is not suitable for statistical analysis. Our statistical pipeline approach is similar to prior studies [29, 30]. Univariable statistics are reported by p value determined via Student’s t test if applicable (Shapiro-Wilk and Levene’s test) or Wilcoxon’s rank-sum test for continuous variables, a Fisher’s exact test for categorical variables with 2 factors, and a chi-square test for > 2 factors. A two-sided p value < 0.05 was considered significant. For multivariable model construction, univariably significant features are selected after false discovery rate Benjamini-Hochberg multiple testing correction. This feature selection is further reduced by a best subset selection according to the Bayesian information criterion. Multivariable models are then fitted by logistic regression for the five best subsets and reported by the statistical AUC with 95% confidence interval (CI) and odds ratios (OR) for features normalized to 1 SD with 95% CI. Additionally, univariable and multivariable Cox proportional hazard (CPH) models are used to determine concordance index (C-index, a generalization of the AUC applicable for survival regression). For the multivariable model, survival differences based on the fitted CPH median survival stratification in high- and low-risk groups are visualized by Kaplan-Meier curve and survival differences are quantified by the log-rank test. For a deeper understanding of the features, a univariable Spearman correlation heatmap with absolute values and dendrogram is generated to quantify associations of clinical with imaging variables.
For the machine learning approach (Fig. 2c (II), (III)), two methods are used to generate derivation and validation data: a temporal 2/3 split, i.e., patients are split according to the date of their baseline scan, and 10 × 10-fold cross-validation (CV), i.e., 10 different random seeds are used for a 10-fold CV. 10 × 10 CV is used for the prediction of 1-YS. The temporal split is introduced for the survival regression to estimate temporal batch effects on the prediction, e.g., a temporal change of doctor in charge, a common effect described by Leek et al [31]. A random forest for 1-YS prediction and a CPH for survival prediction are trained on the derivation data for each of the introduced prognostic models. Predictive performance is evaluated by predictive AUC (random forest), C-index (survival regression), and significance for the models on the validation data. Additionally, a CPH median survival stratification threshold is determined on the derivation data and applied to the validation data. Then, Kaplan-Meier curves are generated and the log-rank test is used to assess the significance of the predicted risk group on the validation data. Data analysis is done with R (version 3.3.2, www.R-project.org) and Lifelines [32].
Results
Demographic data
Demographics of the included patients and used CT scanner types of our study sample are shown in Table 1.
WLTB segmentations
In Fig. 3, four representative patients are shown visualizing patients with varying values for TBS, WLTB volume, and geometric metastatic spread.
Exemplary WLTB segmentations of four patients with CRLM and their according measurements (MSx/MSy [mm]; TBS; volume [cm3]). a A patient with intermediate metastatic spread, low TBS, and low WLTB volume and “no 1-year survival” (1-YS). b A patient with high metastatic spread and TBS, intermediate volume, and also “no 1-YS.” Patients in c and d had “1-YS” with intermediate (c) or low (d) TBS and intermediate (c) or low (d) metastatic spread while their tumor volume was larger than that in a. Patient a appears to be especially interesting, as “no 1-YS” is correctly indicated here by the metastatic spread while the TBS points rather towards “1-YS”
Data analysis and survival prediction
Significant features of the univariable statistical evaluation before multiple testing correction are shown in Fig. 4a. The complete results of univariable analysis are shown in Table 1S of Suppl. Mat. A.
Statistical analyses to assess 1-YS and survival time. a Univariable significant features are shown by boxplots and p values with (without) multiple testing correction. b AUC with CI for the best subset multivariable model consisting of only TBS. c Kaplan-Meier curve of the multivariable model for high- and low-risk groups. Results in c are given as model C-index/score of log-rank test (*p < 0.05, **p < 0.01). TBS tumor burden score, MS metastatic spread, MSx metastatic spread along CT scanner x-axis)
Using univariable statistics, features with the significant discriminative performance were TBS, MS, MSy, MSz, and the compactness of the tumor distribution. The GMS features MS, MSy, MSz, and compactness also showed a significant goodness of fit and C-index between 0.62 and 0.65.
The association heatmap with dendrogram to visualize the univariable associations of imaging and clinical variables is shown in Fig. 5. The highest association between clinical parameters and imaging was found between CEA and WLTB volume and the metastatic spread along the CT y-axis MSy and the M staging.
Regarding multivariable statistics, of the 5 significant features, only the TBS remains in the best multivariable model according to the Bayesian information criterion (Fig. 4b, c; Table 2) and shows a good discriminative performance with a discriminative AUC of 0.70 [0.56, 0.90] for 1-YS. However, the best 3 models achieve similar performance. All 5 multivariable models consist of only one feature.
Results for the machine learning analysis to predict 1-YS for unseen data are shown in Table 3 and Fig. 6a. Here, the QIB TBS achieved also a good predictive performance for 1-YS prediction with a predictive AUC of 0.68 [0.54, 0.79]. The QIBs WLTB volume, attenuation of WLTB, and PTS individually showed inferior performance with AUCs between 0.5 and 0.57. The imaging prior model (i) consisting of all QIBs yields similar results as TBS alone with a predictive AUC of 0.67 [0.54, 0.79], whereas the clinical prior model (ii) achieves 0.56 [0.43, 0.67]. A combination of both prior models achieves again a similar performance with 0.66 [0.54, 0.77] (data not shown). The GMS model (iv) and the ARP model (iii) were numerically superior to both with a predictive AUC of 0.73 [0.602, 0.84] and with 0.76 [0.65, 0.86], respectively. The naive model, i.e., the standard radiomics model building approach using all features, results in an AUC of 0.65 [0.55, 0.78], highlighting the importance of prior knowledge or intuition.
Machine learning analyses to predict 1-YS and survival time for unseen patients. a AUC for 10 × 10 CV (light gray) and C-index (red) for temporal 2/3 split. b Selected Kaplan-Meier curves for unseen patients and predictive performance in temporal 2/3 split. Results in b are given as model: C-index/score of log-rank test (*p < 0.05, **p < 0.01). #1-YS prediction based on a logistic regression model. PTS primary tumor sidedness, TBS tumor burden score, GMS geometric metastatic spread model, ARP Aerts radiomics prior model
Kaplan-Meier curves for the predictive performance on unseen data are shown in Fig. 6b. C-index was highest for the GMS and the ARP model with 0.70 and 0.66. Again, the TBS showed a good performance with a C-index of 0.64.
Discussion
We investigated whether whole liver tumor burden (WLTB), and especially geometric and radiomics analyses of WLTB, extracted from pretreatment CT, could be used as prognostic biomarkers of the 1-YS of patients with colorectal liver metastases (CRLM). We compared established QIB and five different models ((i) imaging prior, (ii) clinical prior, (iii) Aerts radiomics prior (ARP), (iv) geometric metastatic spread (GMS), (v) naive model, i.e., standard radiomics model building), for predicting 1-YS. We therefore analyzed contrast-enhanced CT scans of 103 patients with CRLM scheduled for first-line therapy to assess the prognostic value of each model. Our goal was a systematic comparative analysis of quantitative imaging biomarkers applied on WLTB in metastatic colorectal cancer patients and if potential and robust predictors of patient survival could be identified, which may serve as early imaging biomarkers for risk stratification. The main findings of our study are that geometric but also radiomics WLTB-based measures are significantly associated with the outcome of patients with mCRC. The tumor burden score represents a reliable predictive QIB and shows higher predictive values than all clinical models or the WLTB volume alone. However, the ARP as well as GMS model even shows a numerically higher predictive performance than TBS in a machine learning setting.
Generally, heterogeneity information using radiomics and texture analysis are achieved for one target lesion of a single anatomical site. In prior cancer studies, the prognostic utility of radiomics was used for survival prediction and disease relapse in head and neck as well as lung cancer patients [13, 33,34,35]. A comparable radiomics approach was also used to predict survival and therapy response in patients with nasopharyngeal cancer [36] or glioblastoma [37], the latter based on MRI data. Aerts et al [13] defined a four-feature signature, which represents the ARP model of our study, by focusing on the most robust features for prognostication in a lung dataset, and validated their signature using independent lung and head and neck cancer patient cohorts. Additionally, several previous studies have also analyzed gross whole tumor morphology, including tumor size and number, as important QIBs for survival prediction in mCRC patients [7,8,9,10, 38]. In terms of CRLM, the tumor burden score, incorporating maximum tumor size and number of lesions, was analyzed for survival discrimination in mCRC patients [9] and was outlined as an accurate tool to account for the impact of tumor morphology on long-term survival. As shown previously, TBS-based survival analysis revealed excellent prognostic discrimination for the TBS model and outperformed discrimination based on maximum tumor size and/or total number of lesions as performed in daily clinical routine by use of the established Fong score [9]. Our study therefore assesses and compares the value of a geometric metastatic spread model and radiomics analysis with the TBS as an already established predictive QIB. In our study, TBS reproduced its strong discriminative performance in a regularized logistic regression statistics approach, i.e., model fit and application on the same data, but the GMS and the ARP model in combination with a random forest classifier yielded an enhanced predictive performance for unseen data. The performance of the ARP model appears plausible since this model has often been shown to be of high and reproducible predictive value across various cancer types [13, 16, 25, 26]. Of note, a prior study targeting the vulnerability of radiomics approaches determined that the tumor volume alone in the aforementioned head and neck and lung cancer datasets [13, 33,34,35] has a similar prognostic accuracy as the ARP model [39]. The authors conclude that the ARP model was a surrogate for tumor volume and that intensity and texture values were not pertinent for prognostication [40]. In our study, we analyzed both the ARP model and the volume only approach for prognostication and could clearly show a higher predictive value of the ARP model in a CRLM dataset. To overcome underlying dependencies of intensity and texture-based measures, our study outlines the GMS of liver metastases as newly defined imaging biomarker with a comparable prognostic accuracy, but invariant to technical variation and independent of texture- or intensity-based values. The spread of metastases as quantified by the GMS could be expected to be diagnostically relevant as it might influence the resectability of liver metastases based on their spatial distribution. The GMS model shows strong performance in classification and survival regression and its features were also significant in a univariable statistics analysis.
Thus, our additional flexible machine learning approach using the geometry of the spatial WLTB distribution as well as the Aerts radiomics prior model led to a numerically superior assessment of survival time in comparison to a regularized statistical analysis of TBS.
Furthermore, a clinical model incorporating important clinical baseline parameters, especially primary tumor sidedness [18, 19], yielded an inferior predictive performance than all evaluated imaging-based approaches in our study. As risk stratification of patients with mCRC is nowadays mainly based on traditional prognostic scores including clinical and pathological parameters of the primary tumor and metastases, these results underline the potential of novel imaging-based models and biomarkers for patient risk stratification.
Taking all factors into account such as stability to scan parameter variation, interpretability, and predictive performance averaged over all settings (univariable statistics, 1-year survival classification, and survival regression), the GMS appears to be the most promising and robust model. The reliable and efficient usage of models based on texture features for outcome prediction still remains a very challenging problem. The ARP model is potentially non-robust due to the susceptibility of texture measurements to technical variation [41, 42] (Table 2S of Suppl. Mat. A). This is particularly noteworthy as we used 26 different CT scanner types in 103 patients with baseline scans due to referrals from external physicians. Although there exist approaches to calibrate texture to technical variation [43, 44], a complete absence of the influence of technical variation could fundamentally increase confidence in AI-supported systems. The GMS model showed good predictive and also statistical performance and can trivially be interpreted as a machine learning extension of TBS to integrate more fine-grained and non-linear patterns regarding metastasis distribution. This is in principle similar to the transformation of tumor heterogeneity to the radiomics setting by a machine learning–based assessment of the Aerts features. Although the effort and complexity of the WLTB GMS analysis may be higher than the assessment of the TBS, the good predictive performance, interpretability, and probable robustness to scan parameter variation could justify the effort and should be tested in larger multicenter to provide prospective evaluation as well as external validation. Notably, previous studies have largely focused on texture-based measures. The geometric metastatic spread analysis developed in our study could convert the radiology image into a “spatial map” of liver metastases. This could greatly facilitate and empower comprehensive analysis of spatial distribution, as well as its role in tumor progression and prognostication in future studies. The GMS may prove its usefulness not only in CRLM patients but could also be applied in cross-cancer studies of other gastrointestinal tumors and may be transferred as robust imaging biomarker into the setting of longitudinal studies including CT and MR imaging to assess survival prediction and treatment response. Thus, our study may provide better insights into factors associated with patient survivability by a robust data analytical model. Ultimately, holistic assessment of WLTB and robust predictive parameters such as GMS might directly translate into optimized patient management.
Limitations
Our study has a number of potential limitations. First, the study is only of medium sample size. Second, no external validation cohort was available. Another problem may arise from the variety of different CT scan protocols especially for texture quantifications of the ARP model. However, the GMS is independent from texture- or intensity-based values and is therefore expected to be invariant to technical variability.
Conclusion
Whole liver tumor burden–based measures are significantly associated with the outcome of patients with mCRC. The TBS confirms its importance for risk stratification and shows higher predictive values than clinical models or WLTB volume alone. The ARP as well as GMS model even shows a numerically higher predictive performance than TBS in a machine learning setting. The GMS as a machine learning extension of the TBS concept appears to be the most promising approach, not least due to its invariance to technical variation.
Abbreviations
- 1-YS:
-
1-Year survival
- ARP:
-
Aerts radiomics prior
- CPH:
-
Cox proportional hazards
- CRLM:
-
Colorectal liver metastases
- GMS:
-
Geometric metastatic spread
- LLD:
-
Liver-limited disease
- mCRC:
-
Metastatic colorectal cancer
- MS:
-
Metastatic spread
- MSx(y/z):
-
Metastatic spread along CT scanner x(y/z)-axis
- PTS:
-
Primary tumor sidedness
- QIB:
-
Quantitative imaging biomarker
- TBS:
-
Tumor burden score
- WLTB:
-
Whole liver tumor burden
References
Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66(1):7–30
Rees M, Tekkis PP, Welsh FK, O’Rourke T, John TG (2008) Evaluation of long-term survival after hepatic resection for metastatic colorectal cancer: a multifactorial model of 929 patients. Ann Surg 247(1):125–135
Morris EJ, Forman D, Thomas JD et al (2010) Surgical management and outcomes of colorectal cancer liver metastases. Br J Surg 97(7):1110–1118
Lubner MG, Stabo N, Lubner SJ et al (2015) CT textural analysis of hepatic metastatic colorectal cancer: pre-treatment tumor heterogeneity correlates with pathology and clinical outcomes. Abdom Imaging 40(7):2331–2337
Beckers RCJ, Trebeschi S, Maas M et al (2018) CT texture analysis in colorectal liver metastases and the surrounding liver parenchyma and its potential as an imaging biomarker of disease aggressiveness, response and survival. Eur J Radiol 102:15–21
Beckers RCJ, Lambregts DMJ, Schnerr RS et al (2017) Whole liver CT texture analysis to predict the development of colorectal liver metastases-a multicentre study. Eur J Radiol 92:64–71
Sahu S, Schernthaner R, Ardon R et al (2017) Imaging biomarkers of tumor response in neuroendocrine liver metastases treated with transarterial chemoembolization: can enhancing tumor burden of the whole liver help predict patient survival? Radiology 283(3):883–894
Fleckenstein FN, Schernthaner RE, Duran R et al (2016) 3D quantitative tumour burden analysis in patients with hepatocellular carcinoma before TACE: comparing single-lesion vs. multi-lesion imaging biomarkers as predictors of patient survival. Eur Radiol 26(9):3243–3252
Sasaki K, Morioka D, Conci S et al (2018) The tumor burden score: a new “metro-ticket” prognostic tool for colorectal liver metastases based on tumor size and number of tumors. Ann Surg 267(1):132–141
Sasaki K, Margonis GA, Andreatos N et al (2017) The prognostic utility of the “tumor burden score” based on preoperative radiographic features of colorectal liver metastases. J Surg Oncol 116(4):515–523
De Cecco CN, Ganeshan B, Ciolina M et al (2015) Texture analysis as imaging biomarker of tumoral response to neoadjuvant chemoradiotherapy in rectal cancer patients studied with 3-T magnetic resonance. Invest Radiol 50(4):239–245
Dohan A, Gallix B, Guiu B et al (2019) Early evaluation using a radiomic signature of unresectable hepatic metastases to predict outcome in patients with colorectal cancer treated with FOLFIRI and bevacizumab. Gut. https://doi.org/10.1136/gutjnl-2018-316407
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Bressem KK, Adams LC, Vahldiek JL et al (2020) Subregion radiomics analysis to display necrosis after hepatic microwave ablation-a proof of concept study. Invest Radiol. https://doi.org/10.1097/RLI.0000000000000653
Chlebus G, Schenk A, Moltz JH, van Ginneken B, Hahn HK, Meine H (2018) Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci Rep 8(1):15497
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77(21):e104–e107
Chen Y, Chang W, Ren L et al (2020) Comprehensive evaluation of relapse risk (CERR) score for colorectal liver Metastases: Development and Validation. Oncologist. https://doi.org/10.1634/theoncologist.2019-0797
Kamran SC, Clark JW, Zheng H et al (2018) Primary tumor sidedness is an independent prognostic marker for survival in metastatic colorectal cancer: results from a large retrospective cohort with mutational analysis. Cancer Med. https://doi.org/10.1002/cam4.1558
Modest DP, Stintzing S, von Weikersthal LF et al (2017) Exploring the effect of primary tumor sidedness on therapeutic efficacy across treatment lines in patients with metastatic colorectal cancer: analysis of FIRE-3 (AIOKRK0306). Oncotarget 8(62):105749–105760
Ahmed S, Pahwa P, Le D et al (2018) Primary tumor location and survival in the general population with metastatic colorectal cancer. Clin Colorectal Cancer 17(2):e201–e206
Froelich MF, Heinemann V, Sommer WH et al (2018) CT attenuation of liver metastases before targeted therapy is a prognostic factor of overall survival in colorectal cancer patients. Results from the randomised, open-label FIRE-3/AIO KRK0306 trial. Eur Radiol 28(12):5284–5292
Bester L, Meteling B, Pocock N et al (2012) Radioembolization versus standard care of hepatic metastases: comparative retrospective cohort study of survival outcomes and adverse events in salvage patients. J Vasc Interv Radiol 23(1):96–105
Jakobs TF, Hoffmann RT, Dehm K et al (2008) Hepatic yttrium-90 radioembolization of chemotherapy-refractory colorectal cancer liver metastases. J Vasc Interv Radiol 19(8):1187–1195
Colloca GA, Venturino A, Guarneri D (2020) Different variables predict the outcome of patients with synchronous versus metachronous metastases of colorectal cancer. Clin Transl Oncol. https://doi.org/10.1007/s12094-019-02277-7
Parmar C, Leijenaar RT, Grossmann P et al (2015) Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep 5:11044
Coroller TP, Grossmann P, Hou Y et al (2015) CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol 114(3):345–350
Parmar C, Rios Velazquez E, Leijenaar R et al (2014) Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One 9(7):e102107
Ding C, Peng H (2005) Minimum redundancy feature selection from microarray gene expression data. J Bioinform Comput Biol 3(2):185–205
Muhlberg A, Museyko O, Bousson V, Pottecher P, Laredo JD, Engelke K (2019) Three-dimensional distribution of muscle and adipose tissue of the thigh at CT: association with acute hip fracture. Radiology 290(2):426–434
Bousson VD, Adams J, Engelke K et al (2011) In vivo discrimination of hip fracture with quantitative computed tomography: results from the prospective European Femur Fracture Study (EFFECT). J Bone Miner Res 26(4):881–893
Leek JT, Scharpf RB, Bravo HC et al (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11(10):733–739
Davidson-Pilon C, Kalderstam J, Zivich P (2020) CamDavidsonPilon/lifelines: v0.23.7. Zenodo
Leijenaar RT, Carvalho S, Hoebers FJ et al (2015) External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol 54(9):1423–1429
Leger S, Zwanenburg A, Pilz K et al (2017) A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep 7(1):13206
Vallieres M, Kay-Rivest E, Perrin LJ et al (2017) Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 7(1):10117
Zhao L, Gong J, Xi Y et al (2020) MRI-based radiomics nomogram may predict the response to induction chemotherapy and survival in locally advanced nasopharyngeal carcinoma. Eur Radiol 30(1):537–546
Ingrisch M, Schneider MJ, Norenberg D et al (2017) Radiomic analysis reveals prognostic information in T1-weighted baseline magnetic resonance imaging in patients with glioblastoma. Invest Radiol. https://doi.org/10.1097/RLI.0000000000000349
Vogl TJ, Dommermuth A, Heinle B et al (2014) Colorectal cancer liver metastases: long-term survival and progression-free survival after thermal ablation using magnetic resonance-guided laser-induced interstitial thermotherapy in 594 patients: analysis of prognostic factors. Invest Radiol 49(1):48–56
Welch ML, McIntosh C, Haibe-Kains B et al (2019) Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol 130:2–9
Gevaert O, Mitchell LA, Achrol AS et al (2014) Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273(1):168–174
Kim H, Park CM, Lee M et al (2016) Impact of reconstruction algorithms on CT radiomic features of pulmonary tumors: analysis of intra- and inter-reader variability and inter-reconstruction algorithm variability. PLoS One 11(10):e0164924
Ger RB, Zhou S, Chi PM et al (2018) Comprehensive investigation on controlling for CT imaging variabilities in radiomics studies. Sci Rep 8(1):13047
Muhlberg A, Katzmann A, Heinemann V et al (2020) The Technome - a predictive internal calibration approach for quantitative imaging biomarker research. Sci Rep 10(1):1103
Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127
Funding
This study was funded by the German Federal Ministry of Education and Research (BMBF).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Dominik Nörenberg.
Conflict of interest
All coauthors contributed substantially to the success of our research project, which was part of a consortium and funded by an academic BMBF grant by the German Federal Ministry of Education and Research. A.M., M.S., O.T., and A.K. are employees of Siemens Healthineers and contributed with support of J.M. substantially to the statistical and machine learning analyses. However, no commercial software or product-related application has been tested in the study. The research is not financially supported by industry. Academic radiologists (D.N., T.H., S.M., M.F., E.G.), oncologists (J.H., V.H.), and doctoral students (N.J., L.L.) had full control over the data. There are no potential conflicts of interest.
Statistics and biometry
Several authors have significant statistical expertise (A.M., J.M., A.K., O.T.).
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethical approval
Institutional Review Board approval was obtained. Our retrospective study was approved by and registered with the institutional review board of the Ludwig-Maximilian University, Munich, Germany (approval no. 502-16).
Methodology
• retrospective
• prognostic study
• performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 32 kb)
Rights and permissions
About this article
Cite this article
Mühlberg, A., Holch, J.W., Heinemann, V. et al. The relevance of CT-based geometric and radiomics analysis of whole liver tumor burden to predict survival of patients with metastatic colorectal cancer. Eur Radiol 31, 834–846 (2021). https://doi.org/10.1007/s00330-020-07192-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07192-y