Abstract
Low temperature is one of the major environmental challenges that Antarctic bacteria must face. Detailed studies of cold shock responses of cold-adapted microorganisms are still insufficient. Here, we cloned three cold shock protein (CSP) genes (Csp1137, Csp2039, and Csp2531) in the Antarctic bacterium Psychrobacter sp. G and their regulatory sequences were identified. The three CSPs were highly conserved with other known CspAs. qRT-PCR was performed to evaluate their expression characteristics under stress conditions, and the potential influence of regulatory sequences also was analyzed. The expression of Csp1137 was enhanced both by low (0, 10 °C) and high temperature (30 °C). The expression of Csp2039 was enhanced by low temperature (0 °C), but was lower than that of Csp1137. This can be explained by the absence in Csp2039 of the AT-rich UP element. Different from Csp1137, the expression of Csp2531 was inhibited by low temperature (0 °C), even with the presence of AT-rich UP element, and it was not sensitive to high temperature (30 °C). The expression of Csp1137 was enhanced by high salinity (90, 120), whereas that of Csp2531was enhanced by low salinity (0, 15). At 0 °C and a salinity of 15, the expression of Csp1137 was repressed initially, but then it increased greatly during the next 10 h. The expressions of Csp2039 and Csp2531 were repressed significantly under four different combinations of stress conditions. Our results showed that the role of the upstream regulation sequences were much more complex than previously thought. Also, gene expressions were also affected by the environmental salinity. These are helpful in further clarification of the adaptation mechanism of Psychrobacter sp. G.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Antarctic ecosystems are cold, low nutrient environments that experience large temperature fluctuations. A sudden temperature downshift is one of the pivotal environmental stress factors encountered by bacteria in the polar regions of the Earth. Accordingly, they have developed adaptation mechanisms that successfully enable them to thrive in this harsh environment (Margesin and Schinner 1994; Pearce 2008). Low temperature results in a significant decrease in cell membrane fluidity and disturbs virtually all intracellular processes, including transcription and translation (Russell et al. 1995; Jones and Inouye 1996). Cellular proteins, particularly regulatory proteins, require adjustments to cope with the temperature downshift and to allow balanced growth at the new temperature (Berger et al. 1996).
The cold shock response includes repression of many proteins synthesized under normal conditions and the transient induction of a subset of proteins called cold-induced proteins (CIPs) (Graumann and Marahiel 1996). Among the CIPs, members of a family of small proteins (mostly acidic proteins, 7.4 kDa) called cold shock proteins (CSPs) are the most strongly induced in many bacteria when exposed to cold stress (Jones et al. 1987; Graumann et al. 1996; Mayr et al. 1996; Berger et al. 1997). However, not all CSPs are induced by cold shock; some are involved in other cellular processes (Graumann and Marahiel 1998). For example, Blattner et al. (1997) reported that only three (CspA, CspB, and CspG) of the nine CSPs in Escherichia coli were induced by cold stimulation. CspD was induced by starvation and it appeared to play a role in the nutrient-stress response (Yamanaka and Inouye 1997). CspC and CspE reportedly play a role in cell division (Yamanaka et al. 1994; Hu et al. 1996). Thus, CSPs in E. coli are involved in different physiological processes. Although regulated differently, they likely share the same general mechanism of action (Yamanaka et al. 1994).
Members of the CspA family consist of 70 amino acid residue peptides with five anti-parallel sheets that form a β-barrel with no helical structure (Newkirk et al. 1994). CspAs include two RNA binding motifs, ribonucleoprotein 1 (RNP1) and ribonucleoprotein 2 (RNP2), which suggests that CspA may unfold tightly compacted molecules inaccessible for translation or prevent their translation while protecting them from degradation; CspA also may act as an RNA chaperone protein (Jones and Inouye 1994).
Although heat shock responses have been well documented at the molecular level, detailed studies of cold shock responses are still at the early stages. Previous studies of cold responses were focused mainly on the mesophilic bacterium E. coli (Phadtare and Inouye 2004; Qing et al. 2004; White et al. 2008). Relatively, fewer studies have been conducted on cold shock responses of cold-adapted microorganisms (i.e., psychrotrophs and psychrophiles) (Bakermans et al. 2007; Kawamoto et al. 2007). Thus, information about the structure and function of CSPs in these bacteria is lacking.
To better understand the adaptation mechanisms of the Antarctic bacterium Psychrobacter sp. G, three CSP genes (Csp1137, Csp2039, and Csp2531) were cloned and their sequences were analyzed. The expression characteristics of these CSPs under different stress conditions (i.e., temperature, salinity, and combinations of temperature and salinity) also were investigated using qRT-PCR. Regulatory sequences, such as the AT-rich UP element, cold box, and downstream box (DB), were identified and their potential influence on the expression of the three CSP genes was analyzed.
Materials and Methods
Bacteria strain, medium, and growth conditions
The Antarctic bacterium Psychrobacter sp. G was isolated and preserved in our laboratory (Lin et al. 2010). E. coli DH5α (D9057, TaKaRa) and Vector pMD18-T (D101A, TaKaRa) were used in gene cloning. The component of culture medium used to culture Psychrobacter sp. G is listed in Table 1. The seawater was taken from the coast of Qingdao (Shandong, China) and its salinity was approximately 31. The strain was cultured at 20 °C with a rotating speed of 150 r/min.
Growth characteristics of Psychrobacter sp. G
Psychrobacter sp. G was cultured at different temperatures (0, 5, 10, 15, 20, 25, 30, and 35 °C) and salinities (0, 15, 30, 45, 60, 90, 120, and 160) to determine the optimal condition for its growth. Cell growth was monitored spectrophotometrically by measuring the optical density at 600 nm (OD600). The maximum specific growth rate (SGR), which is a reflection of metabolic activity, was evaluated under different culture conditions based on the biomass of the culture (OD600) according to the method described by Rosso et al. (1995).
Cloning of Csp1137, Csp2039, and Csp2531 genes and their regulatory sequences
The genomic DNA of Psychrobacter sp. G was isolated using a Genomic DNA Prep Kit (DP302, Tiangen Biotech) following the manufacturer’s instructions. To obtain the full sequences of the three CSP genes and all of their regulatory sequences, nucleotide sequences from −600 to −550 bp upstream and 50 bp downstream of the open reading frame (ORF), according to the genomic draft of Psychrobacter sp. G (data not published), were used to design the primers. Primers were designed using the Primer premier 5.0 program and are listed in Table 2. The three gene fragments were amplified by 35 PCR cycles (95 °C for 1 min; 42 °C for 30 s; and 72 °C for 1 min). The objective fragments were excised and then purified using an agarose gel DNA fragment recovery kit (DP209-2, Tiangen Biotech). The purified fragments were subcloned into pMD-18T vector (D101A, TaKaRa) and sequenced subsequently by Sunny Biotechnology (Shanghai, China).
The amplified CSP genes were compared and aligned with the nucleotide sequence database in GenBank (http://www.ncbi.nlm.nih.gov) using the Basic Local Alignment Search Tool (BLAST). A nucleotide similarity search was performed using the BLASTN program (http://www.ncbi.nlm.nih.gov). The regulatory sequences (i.e., −10 region, −35 region, ribosomal binding site (RBS), DB, and ORF) were analyzed using the Softberry (http://linux1.softberry.com/berry.phtml) (Panicker et al. 2010) and Neural Network Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html) (Datta and Bhadra 2003) programs. Multiple sequence alignments were performed using MegAlign available in the Lasergene sequence analysis package (v.1.02; DNASTAR Inc.). Theoretical isoelectric points (pI) and molecular weights (MW) of the three CSPs were determined using Lasergene-EditSeq (v.1.02; DNASTAR Inc.). The phylogenetic tree was constructed using the program Mega 4.0 (Kumar et al. 2008), and the reliability of the tree was analyzed using bootstrap probabilities.
Stress treatments
The Psychrobacter sp. G was initially cultured at its optimal growth temperature (20 °C) and salinity (45). As OD600 of the culture reached 0.5 (approximate logarithmic phase), the stress treatments were conducted as follows: (1) In the temperature stress treatments, cultures were then kept at different temperatures for different periods of time (0, 10, and 30 °C for 2, 6, and 12 h, respectively). (2) In the osmotic stress treatments, cultures were then centrifuged at 8,000×g for 5 min at 20 °C. The pellets were collected, and the same volume of medium with a final salinity of 0, 15, 90, and 120 was added. Cells were resuspended thoroughly by gentle inverting. The cultures were then kept at 20 °C for 2, 6, and 12 h. (3) In the combined stress treatments, cultures were then kept under the following conditions: (i) a final salinity of 15 at 0 °C; (ii) a final salinity of 15 at 30 °C; (iii) a final salinity of 90 at 0 °C; and (iv) a final salinity of 90 at 30 °C. After 2, 6, and 12 h of treatment, cultures were centrifuged at 8,000×g for 5 min at their own culture temperature, and the pellets were collected and stored at −80 °C until used for RNA extraction. Psychrobacter sp. G cultured under the optimal conditions (20 °C with a salinity of 45) was used as the control for qRT-PCR analysis.
Expression analysis of the three CSP genes by qRT-PCR
Total RNA was extracted using the RNAiso Plus Kit (D9108D, TaKaRa) following the manufacturer’s instructions. The purity of the prepared RNA was evaluated by measuring absorption at 260 nm and the ratio of absorption at 260:280 nm (A260/A280).
The PrimeScript RT Reagent Kit (DRR037A, TaKaRa) was used to perform the reverse transcription of RNA into cDNA. The cDNA for qRT-PCR was prepared using 500 ng of total RNA and Random 6 primers. The SYBR PrimeScriptTM RT-PCR Kit (DRR041A, TaKaRa) was used for qRT-PCR analysis.
Table 3 lists the primers targeted against sequences of the three CSP genes and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene of Psychrobacter sp. G. qRT-PCR was performed on the Stratagene Mx3000P qPCR System for 40 cycles (95 °C for 5 s; 44 °C for 15 s; and 72 °C for 20 s). All tests were conducted in three independent biological replicates. Quantification of mRNA was based on threshold cycle (Ct) values. The Ct values of the three CSP genes were normalized using the Ct value of the GAPDH gene. The efficiency of each qRT-PCR was also calculated. Data analysis was performed using the comparative Ct (2−ΔΔCt) method (Livak and Schmittgen 2001). Data obtained from qRT-PCR analysis were subjected to analysis of variance to evaluate differences in the mean values among the treatments. Differences were considered significance at P < 0.05 and highly significance at P < 0.01. Statistical analysis was performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA).
Results
The optimal growth temperature and salinity of Psychrobacter sp. G
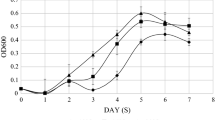
The optimal growth temperature and salinity of Psychrobacter sp. G were 20 °C and 45, respectively. Under such conditions, the SGR of the strain was 0.18 and 0.2 h−1, respectively (Fig. 1).
Sequence analysis of the three CSP genes and their regulatory sequences
Figure 2a shows the nucleotide sequence information for Csp1137. The putative promoter sequences −35 region (5′-TTGCGA-3′) and −10 region (5′-CCTTATAAT-3′) were identified. The putative transcription initiation site (TIS) was located 6 bp downstream from the −10 region. A typical RBS (5′-AGGA-3′) was found 9 bp upstream from the initiation codon, and an ORF of 210 bp between ATG and TAA also was identified. Within the 5′-untranslated region (UTR), an 11 bp cold box element (5′-TGCACCAAATC-3′) was identified 13 bp downstream from the TIS. An AT-rich UP element (5′-AAATCTAAGATAATA-3′), which plays an important role in the transcription of CspA, CspB, CspG, and CspI in E. coli at low temperature (Mitta et al. 1997; Wang et al. 1999), was found 1 bp upstream of the –35 region. A 14 bp DB (5′-CAAAAAGGTACAGT-3′) was located 10 bp downstream of the initiation codon. Mitta et al. (1997) demonstrated that the 14 bp DB was essential for mRNA translation during the lag phase immediately after cold shock.
Sequence analysis of the three CSP genes and their regulatory sequences. a Csp1137; b Csp2039; and c Csp2531. The putative promoter elements (–35 region and –10 region), the cold box element, RBS. and the initiation codon ATG are in boldface and underlined. The putative TIS are in boldface and marked with an arrow. The putative AT-rich UP element and DB are boxed
Figure 2b shows the nucleotide sequence data for Csp2039. The putative promoter sequences −35 region (5′-TTTTTT-3′) and −10 region (5′-ATATATAAT-3′) were identified. The putative TIS was 12 bp downstream from the −10 region. The RBS was 7 bp upstream from the initiation codon, and an ORF of 213 bp between ATG and TAA was also identified. A 14 bp DB (5′-GTTGAAGGCACTGT-3′) was located 10 bp downstream of the initiation codon. Interestingly, no AT-rich UP element or cold box element was found in Csp2039.
The nucleotide sequence information for Csp2531 is shown in Fig. 2c. The putative promoter sequences −35 region (5′-TTGTGT-3′) and −10 region (5′-TGTTATGAT-3′) were identified. The putative TIS was located 11 bp downstream from the −10 region. The RBS was 10 bp upstream from the initiation codon, and an ORF of 216 bp between ATG and TAA was found. An AT-rich UP element located 5 bp upstream of the −35 promoter sequence also was found. A 14 bp DB (5′-GAGCAAGGTATCGT-3′) was found 10 bp downstream from the translation initiation codon. No cold box element was found.
The nucleotide and deduced amino acid sequences of the Csp1137, Csp2039, and Csp2531 genes were submitted to GenBank with accession numbers JN656166, JN656167, and JN656168, respectively. Table 4 lists the occurrences of the regulatory sequences in these CSP genes.
Amino acid sequence analysis of Csp1137, Csp2039, and Csp2531
The ORFs of Csp1137, Csp2039, and Csp2531 were 210, 213, and 216 bp in length and encoded a protein consisting of 69, 70, and 71 amino acid residues with a molecular mass of 7.41, 7.45, and 8.08 kDa and a calculated pI of 5.09, 5.09, and 4.82, respectively (Fig. 3). The deduced amino acid sequences of Csp1137, Csp2039, and Csp2531 in Psychrobacter sp. G then were compared with other known CSPs of representative bacteria using MegAlign software. Multiple sequence alignment revealed that the deduced amino acid sequences of Csp1137, Csp2039, and Csp2531 were highly conserved and homologous with the amino acid sequences of other known CspAs (Fig. 3). Csp1137, Csp2039, and Csp2531 also had RNP1 and RNP2 motifs that are known to be involved in binding to single-stranded nucleic acids (Landsman 1992; Jones and Inouye 1996). Five anti-parallel β-strands and five amino acid residues that form the hydrophobic core in the β-barrel structure also were identified (Fig. 3).
Sequence alignment of the three CSPs with other representative bacterial CSP. The CSP amino acid sequences analyzed in this study were Enhydrobacter aerosaccus CspA (ZP_05620656), Moraxella catarrhalis CspA (YP_003627692), Acinetobacter radioresistens CspA (ZP_05360199), Escherichia coli CspA (CAP78016), E. coli CspB (AAB61739), E. coli CspG (ACI85380), E. coli CspI (AE016765_31), Bacillus subtilis CspB (CAA42235), B. subtilis CspC (AAC45646), and B. subtilis CspD (ZP_06871986). RNP1 and RNP2 are boxed. The five bars indicate the five β-strands. The five amino acid residues are indicated with a spot above the sequences. The total amino acid residue length, the calculated pIs, and the MWs also are shown. Multiple sequence alignments were performed by MegAlign available in the Lasergene sequence analysis package (v.1.02; DNASTAR Inc.)
Representative bacterial CSPs were selected to construct a phylogenetic tree using the program Mega 4.0 (Fig. 4). Csp1137, Csp2039, and Csp2531 were clustered closely together with CSPs of Psychrobacter arcticus 273-4. Phylogenetic analysis showed that Csp1137 and Csp2039 had a relatively closer relationship when compared with Csp2531. Csp1137, Csp2039, and Csp2531 were relatively close to Pseudoalteromonas haloplanktis TAC125 CspA (YP_341869), Moraxella catarrhalis RH4 CspA (YP_003627692), and Enhydrobacter aerosaccus SK60 CspA (ZP_05620656) but were relatively distant from E. coli CspA (AAA23617).
Expression characteristics of Csp1137, Csp2039, and Csp2531 in response to temperature stress
The expressions of the three CSPs under optimal conditions were analyzed over time (2, 6, and 12 h). qRT-PCR analysis showed that there are no significant differences (P < 0.05) between these samples (data not shown). So their expressions in control (20 °C with a salinity of 45) were considered as changelessness over time in the following studies. The strains used in the control were collected at 0 h, when the OD600 of the culture reached 0.5.
Figure 5 shows the expression of Csp1137, Csp2039, and Csp2531 in response to temperature stress. qRT-PCR analysis showed that the expression of Csp1137 was significantly enhanced in the 0 °C treatment group; expression was 9.2-fold higher than that of control after 2 h of treatment, although it dropped significantly during the next 10 h. Similarly, the expression of Csp1137 at 10 °C was 2.5-fold higher than that of control after 2 h and then dropped slowly during the next 10 h. The high temperature (30 °C) treatment enhanced the expression of Csp1137 in the first 6 h and its expression dropped during the next 6 h.
The expression of Csp2039 was 2.2-fold higher than that of the control after 2 h of exposure to 0 °C, and the highest expression level occurred after 6 h of treatment. However, the expression of Csp2039 was not enhanced when the strain was cultured at 10 and 30 °C.
The expression of Csp2531 was inhibited by low temperature (0 °C); after 12 h of treatment, expression reached its minimum value, which was 0.28-fold that of the control. The expression of Csp2531 was not affected significantly by temperature of 10 and 30 °C.
Expression characteristics of Csp1137, Csp2039, and Csp2531 in response to osmotic stress
The responses of Psychrobacter sp. G to different salinities over time are shown in Fig. 6. qRT-PCR analysis showed that the expression of Csp1137 was repressed by low salinity (0, 15) and enhanced by high salinity (90, 120). Expression was highest (4.5-fold greater than the control after 12 h) when the salinity of the culture medium was 120. Both low (15) and high (90, 120) salinity repressed the expression of Csp2039. In contrast to Csp1137, expression of Csp2531 was significantly increased under low salinity (0, 15). When the salinity of the culture medium was 0, the expression of Csp2531 was significantly increased; the maximum value was 2.4-fold higher than that of the control after 12 h of treatment. When the salinity of the culture medium was 15, the expression of Csp2531 reached its maximum of 2.6-fold higher than that of the control after 12 h of treatment.
Expression characteristics of Csp1137, Csp2039, and Csp2531 in response to combined temperature and osmotic stress
When Psychrobacter sp. G was cultured at 0 °C and a salinity of 15 shifted from its optimal growth condition, expression of Csp1137 was repressed initially, but then it increased significantly during the next 10 h. The maximum value of 3.4-fold higher than that of the control occurred after 12 h of treatment. The expression of Csp1137 was repressed under the following two conditions: 30 °C with a salinity of 15 and 30 °C with a salinity of 90. The expression of Csp2039 and Csp2531 was inhibited significantly under all four combinations of stress conditions (Fig. 7).
Discussion
The Antarctic bacterium Psychrobacter sp. G was isolated from Antarctic seawater sample. Its optimal growth temperature was 20 °C and maximum growth temperature was 30 °C (Fig. 1), suggesting that it is a psychrotroph. 16S rDNA analysis indicated that it belongs to the genus Psychrobacter (Lin et al. 2010). Based on the genomic draft of Psychrobacter sp. G (data not published), three CSPs (Csp1137, Csp2039, and Csp2531) were found. They were identified as members of the CspA family based on alignment analysis of the nucleotide acid sequences and the putative amino acid sequences. Phylogenetic analysis (Fig. 4) showed that Csp1137, Csp2039, and Csp2531 clustered together with other known CspAs and this finding was consistent with the results of multiple sequence alignment (Fig. 3).
The amino acid sequence alignment among Csp1137, Csp2039, Csp2531, and other representative bacterial CSPs exhibited relatively high sequence similarity in the conserved RNA binding motifs RNP1 and RNP2, which are known to be involved in binding to single-stranded nucleic acids (Landsman 1992; Jones and Inouye 1996). This finding suggests that Csp1137, Csp2039, and Csp2531 might play a similar role in cold adaptation. As indicated by the three-dimensional structure of E. coli CspA (Newkirk et al. 1994; Schindelin et al. 1994), CspA forms five β-barrels, on the surface of which are two lysine residues and eight aromatic residues that are exposed to solvent. All of these features, together with the presence of RNP1 and RNP2, suggest that CspA may act as an RNA chaperone (Yamanaka et al. 1998).
Although nine CSP family proteins have been found in E. coli, only three of them have been shown to be induced by cold shock (Goldstein and Drlica 1984; Jiang et al. 1996; Jones and Inouye 1996; Wang et al. 1999). They were regulated at transcription (Mitta et al. 1997; Fang et al. 1998), post-transcription (Etchegaray et al. 1996; Mitta et al. 1997; Graumann and Marahiel 1998), mRNA stability (Nakashima et al. 1996), and translation (Yamanaka et al. 1998) levels. Each of the three cold-inducible CSPs had an AT-rich sequence immediately upstream of the −35 region, which has been shown to enhance transcription as an UP element (Etchegaray et al. 1996). In the current study, no AT-rich sequence was found in Csp2039; this might explain why expression of Csp2039 was not as greatly enhanced as that of Csp1137 when cultured at 0 °C. Even with the presence of an AT-rich sequence, expression of Csp2531 was inhibited at low temperature (0 °C) and an explanation for this result requires further study.
Csp1137, Csp2039, and Csp2531 all had a DB located downstream of the initiation codon, which has been shown to play an important role in cold shock induction at the level of translation (Mitta et al. 1997). A cold box element was also present in the mRNA 5′-UTR of Csp1137, but it was not found in Csp2039 and Csp2531. In E. coli, overproduction of the 5′-UTR of CspA mRNAs caused derepression of the CspA genes; the derepression function was abolished when the cold box region was deleted from the CspA 5′-UTR (Fang et al. 1998). Although cold box sequences were not found in Csp2039 and Csp2531, they had much longer 5′-UTRs than Csp1137 (Fig. 2); thus, they might contain repressor binding sites (Fujii et al. 1999). Panicker et al. (2010) suggested that differences in cold box elements and 5′-UTRs, which result in different mRNA secondary structures, might play important roles in regulation.
qRT-PCR was performed to describe the expression characteristics of the three CspA genes in Psychrobacter sp. G exposed to different stress conditions. Expression of Csp1137 increased immediately after exposure to 0 °C and reached a maximum value of 9.2-fold higher than that of the control, then dropped significantly during the next 10 h (Fig. 5). Etchegaray et al. (1996) also found that expression of CspA in E. coli tended to exhibit transient expression immediately following a downshift in temperature. However, expression of Csp1137 was enhanced not only by low temperature but also by high temperature (30 °C). Etchegaray et al. (1996) also reported that CspA can be induced at a broad range of temperatures. Enhanced expression of Csp2039 occurred relatively slowly compared to Csp1137 and its expression level was not as great as that of Csp1137 at 0 °C. This could be due to the absence of an AT-rich UP element and a cold box in the 5′-UTR of the Csp2039 gene, as previous studies had shown that the cold box plays an important role in derepression of the negative effort of low temperature (Etchegaray et al. 1996).
Csp2531 also lacks a cold box and its expression was inhibited significantly by low temperature (0 °C) despite the presence of an AT-rich UP element (Fig. 5). This finding demonstrates the importance of the cold box in the cold response.
In Antarctica where Psychrobacter sp. G occurs, sea ice freezes and thaws during different seasons, which in turn affects the salinity of the offshore seawater (Liu et al. 2010). For this reason, the effects of osmotic stress on the expression of Csp1137, Csp2039, and Csp2531 also were investigated. qRT-PCR analysis showed that high salinity (90, 120) enhanced the expression of Csp1137, whereas the expression of Csp2531 was enhanced by low salinity (0, 15). Stübs et al. (2005) also found that some CSPs were responsive to osmotic change and Schmid et al. (2009) reported that direct growth of Listeria monocytogenes EGD-e under osmotic stress led to a significant induction of CspA and CspD gene expression. In contrast to Csp1137, expression of Csp2531 increased significantly under low salinity (0, 15) conditions. The differences in the expression of the three genes in response to osmotic stress might be due to the different 5′-UTRs present in Csp1137, Csp2039, and Csp2531. CSPs seem functionally required for efficient cold and osmotic stress adaptation responses (Schmid et al. 2009). The CSP functional contributions to osmotic stress adaptation were not yet clear. One possibility was that CSP chaperones might also promote the increased production of sodium ion extrusion transporter proteins through their effects in facilitating transcription and translation processes (Schmid et al. 2009).
In nature, Psychrobacter sp. G often faces more than one environmental stress factor at the same time (e.g., freezing tolerance is often accompanied by tolerance to osmotic stress caused by a lack of free water) (Thomas and Dieckmann 2002). Accordingly, the combined effects of temperature and salinity on the expression of the three CSP genes were investigated. At 0 °C and low salinity (15), the expression of Csp1137was slower and the quantity expressed also was reduced compared to the expression of Csp1137 at 0 °C and a salinity of 45; this finding suggests that the expression of Csp1137 was affected by more than one factor. Expression of Csp2039 and Csp2531 was inhibited significantly under all of the four combinations of temperature and salinity. The mechanism that underlies this phenomenon is unknown and requires further study. The expression characteristics of the three CSPs under the combined effects of the two factors were much different than the characteristics observed when Psychrobacter sp. G was under the stress of a single factor.
To date, most of the available knowledge about physiological and molecular responses of bacteria to cold shock stems from the studies of mesophilic microorganisms, such as E. coli and Bacillus subtilis (Phadtare and Inouye 2004; White et al. 2008; Schmid et al. 2009). The understanding of the cold shock response in psychrophiles/psychrotrophs is still in its infancy (Bakermans et al. 2007; Kawamoto et al. 2007; Bergholz et al. 2009). To our knowledge, few studies have been conducted to characterize the response of three CSP genes to temperature, salinity, and the combined effects of the two factors. Further studies at the translation and hetero-expression levels are needed to clarify the mechanisms employed by Psychrobacter sp. G to acclimate to the Antarctic environment.
References
Bakermans C, Tollaksen SL, Giometti CS, Wilkerson C, Tiedje JM, Thomashow MF (2007) Proteomic analysis of Psychrobacter cryohalolentis K5 during growth at subzero temperatures. Extremophiles 11:343–354
Berger F, Morellet N, Menu F, Potier P (1996) Cold shock and cold adaptation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J Bacteriol 178:2999–3007
Berger F, Normand P, Potier P (1997) CapA, a cspA-like gene that encodes a cold acclimation protein in the psychrotrophic bacterium Arthrobacter globiformis SI55. J Bacteriol 179:5670–5676
Bergholz PW, Bakermans C, Tiedje JM (2009) Psychrobacter arcticus 273-4 uses resource efficiency and molecular motion adaptations for subzero temperature growth. J Bacteriol 191:2340–2352
Blattner FR, Plunkett G, Bloch CA et al (1997) The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462
Datta PP, Bhadra RK (2003) Cold shock response and major cold shock proteins of Vibrio cholerae. Appl Environ Microbiol 69:6361–6369
Etchegaray JP, Jones PG, Inouye M (1996) Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells 1:171–178
Fang L, Hou Y, Inouye M (1998) Role of the cold-box region in the 5′ untranslated region of the cspA mRNA in its transient expression at low temperature in Escherichia coli. J Bacteriol 180:90–95
Fujii S, Nakasone K, Horikoshi K (1999) Cloning of two cold shock genes, cspA and cspG, from the deep-sea psychrophilic bacterium Shewanella violacea strain DSS12. FEMS Microbiol Lett 178:123–128
Goldstein E, Drlica K (1984) Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci USA 81:4046–4050
Graumann P, Marahiel MA (1996) Some like it cold: response of microorganisms to cold shock. Arch Microbiol 166:293–300
Graumann P, Marahiel MA (1998) A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci 23:286–290
Graumann P, Schröder K, Schmid R, Marahiel MA (1996) Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol 178:4611–4619
Hu KH, Liu E, Dean K, Gingras M, DeGraff W, Trun NJ (1996) Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics 143:1521–1532
Jiang W, Fang L, Inouye M (1996) Complete growth inhibition of Escherichia coli by ribosome trapping with truncated CspA mRNA at low temperature. Genes Cells 1:965–976
Jones PG, Inouye M (1994) The cold shock response-a hot topic. Mol Microbiol 11:811–818
Jones PG, Inouye M (1996) RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol 21:1207–1218
Jones PG, VanBogelen RA, Neidhardt FC (1987) Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol 169:2092–2095
Kawamoto J, Kurihara T, Kitagawa M, Kato I, Esaki N (2007) Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins. Extremophiles 11:819–826
Kumar S, Nei M, Dudley J, Tamura K (2008) A biologistcentric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Landsman D (1992) RNP-1, an RNA-binding motif is conserved in the DNA-binding cold shock domain. Nucleic Acids Res 20:2861–2864
Lin XZ, Cui SS, Xu GY, Wang S, Du N, Shen JH (2010) Cloning and heterologous expression of two cold-active lipases from the Antarctic bacterium Psychrobacter sp. G Polar Res 29:421–429
Liu S, Zhang P, Cong B, Liu C, Lin X, Shen J, Huang X (2010) Molecular cloning and expression analysis of a cytosolic Hsp70 gene from Antarctic ice algae Chlamydomonas sp. ICE-L. Extremophiles 14:329–337
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(–Delta Delta C (T)) method. Methods 25:402–408
Margesin R, Schinner F (1994) Properties of cold-adapted microorganisms and their potential role in biotechnology. J Biotechnol 33:1–14
Mayr B, Kaplan T, Lechner S, Scherer S (1996) Identification and purification of a family of dimeric major cold shock protein homologs from the psychrotrophic Bacillus cereus WSBC 10201. J Bacteriol 178:2916–2925
Mitta M, Fang L, Inouye M (1997) Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol 26:321–335
Nakashima K, Kanamaru K, Mizuno T, Horikoshi K (1996) A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol 178:2994–2997
Newkirk K, Feng W, Jiang W, Tejero R, Emerson SD, Inouye M, Montelione GT (1994) Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc Natl Acad Sci USA 91:5114–5118
Panicker G, Mojib N, Nakatsuji T, Aislabie J, Bej AK (2010) Occurrence and distribution of capB in Antarctic microorganisms and study of its structure and regulation in the Antarctic biodegradative Pseudomonas sp. 30/3. Extremophiles 14:171–183
Pearce DA (2008) Climate change and the microbiology of the Antarctic peninsula region. Sci Prog 91:203–217
Phadtare S, Inouye M (2004) Genome-wide transcriptional analysis of the cold shock response in wild-type and cold-sensitive, quadruple-csp-deletion strains of Escherichia coli. J Bacteriol 186:7007–7014
Qing G, Ma LC, Khorchid A et al (2004) Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol 22:877–882
Rosso L, Lobry JR, Bajard S, Flandrois JP (1995) Convenient model to describe the combined effects of temperature and pH on microbial growth. Appl Environ Microbiol 61:610–616
Russell NJ, Evans RI, Steeg PF, Hellemons J, Verheul A, Abee T (1995) Membranes as a target for stress adaptation. Int J Food Microbiol 28:255–261
Schindelin H, Jiang W, Inouye M, Heinemann U (1994) Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA 91:5119–5123
Schmid B, Klumpp J, Raimann E, Loessner MJ, Stephan R, Tasara T (2009) Role of Cold Shock Proteins in Growth of Listeria monocytogenes under Cold and Osmotic Stress Conditions. Appl Environ Microbiol 75:1621–1627
Stübs D, Fuchs TM, Schneider B, Bosserhoff A, Gross R (2005) Identification and regulation of cold-inducible factors of Bordetella bronchiseptica. Microbiology 151:1895–1909
Thomas DN, Dieckmann GS (2002) Antarctic Sea ice—a habitat for extremophiles. Science 295:641–644
Wang N, Yamanaka K, Inouye M (1999) CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol 181:1603–1616
White CA, Um S, Pérez NM, Berns AL, Malhowski AJ, Young S (2008) Low temperature (23 degrees C) increases expression of biofilm-, cold-shock- and RpoS-dependent genes in Escherichia coli K-12. Microbiology 154:148–166
Yamanaka K, Inouye M (1997) Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J Bacteriol 179:5126–5130
Yamanaka K, Mitani T, Ogura T, Niki H, Hiraga S (1994) Cloning, sequencing, and characterization of multicopy suppressors of a mukB mutation in Escherichia coli. Mol Microbiol 13:301–312
Yamanaka K, Fang L, Inouye M (1998) The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol 27:247–255
Acknowledgments
This work was supported financially by the National Natural Science Foundation of China (41176174). We thank the International Science Editing for the help they offered in language modification.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Song, W., Lin, X. & Huang, X. Characterization and expression analysis of three cold shock protein (CSP) genes under different stress conditions in the Antarctic bacterium Psychrobacter sp. G. Polar Biol 35, 1515–1524 (2012). https://doi.org/10.1007/s00300-012-1191-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-012-1191-6