Abstract
Key message
Overexpression of FvC5SD improves drought tolerance in soybean.
Abstract
Drought stress is one of the most important abiotic stress factors that influence soybean crop quality and yield. Therefore, the creation of drought-tolerant soybean germplasm resources through genetic engineering technology is effective in alleviating drought stress. FvC5SD is a type of C-5 sterol desaturase gene that is obtained from the edible fungus Flammulina velutipes. This gene has good tolerance to the effects of stresses, including drought and low temperature, in yeast cells and tomato. In this study, we introduced the FvC5SD gene into the soybean variety Shennong9 through the Agrobacterium-mediated transformation of soybean to identify drought-tolerant transgenic soybean varieties. PCR, RT-PCR, and Southern blot analysis results showed that T-DNA was inserted into the soybean genome and stably inherited by the progeny. The ectopic expression of FvC5SD under the control of a CaMV 35S promoter in transgenic soybean plants enhanced the plant’s tolerance to dehydration and drought. Under drought conditions, the transgenic plants accumulated lower levels of reactive oxygen species and exhibited higher activities and expression levels of enzymes and cell than wild-type soybean. iTRAQ analysis of the comparative proteomics showed that some exogenous genes coding either functional or regulatory proteins were induced in the transgenic lines under drought stress. FvC5SD overexpression can serve as a direct and efficient target in improving drought tolerance in soybean and may be an important biotechnological strategy for trait improvement in soybean and other crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean [Glycine max (L.) Merrill] is a leguminous plant whose seeds are rich in protein and is one of the most widely grown economic crops. Soybeans contain essential amino acids, dietary minerals, vitamins, etc., and eating soybean seeds can enhance immune function, prevent hardening of the arteries, improve bowel movement, and exert hypoglycemic and lipid-lowering effects. The growth, metabolism, and yield of plants are subjected to various conditions, including biotic and abiotic stresses (Le Gall et al. 2015). Drought stress is the most common and threatening problem that seriously affects the growth, development, and reproduction of plants. It is also responsible for the reduction of crop yield. During drought, soybean yield losses can reach up to 40%, and these losses are greater when moisture stress occurs during the vegetative and reproductive stages (Valliyodan et al. 2017). Therefore, the use of biotechnological methods to cultivate new drought-tolerant varieties that increase soybean yield is of great significance for the production and cultivation of soybeans.
Genetic engineering or transgenic technology is a versatile and alternative technology that is used to overcome the limitations of classical and modern breeding technologies (Kamthan et al. 2016). Genetically modified (GM) soybean is one of the earliest introduced GM crops for commercial cultivation and the largest GM crop in terms of acreage planted worldwide. RoundUp Ready soybean cultivars are a successful example of transgenic soybean and have been planted in many soybean fields worldwide since 2004 (ISAAA, http://www.isaaa.org/) (Yamada et al. 2012). Considerable progress has been reported in improving plant drought stress responses, and a large number of drought-tolerant genes have been identified in soybean. These genes include the transcription factor (TF) DREB gene family members that regulate the expression of several genes related to abiotic stress defense responses (de Paiva Rolla et al. 2014). Soybean plants overexpress AtAREB1 gene to induce drought tolerance (Leite et al. 2014). LOS5/ABA3 overexpression can improve drought tolerance in transgenic soybean via the enhanced abscisic acid (ABA) accumulation (Li et al. 2013). AtABF3 gene overexpression enhances drought and salt tolerance in major soybean crops, especially under low-water growth conditions (Kim 2018a, b). The GmFDL19 gene enhances tolerance to drought and salt stress in soybean (Li et al. 2017). The overexpression of ER-resident molecular chaperone binding protein (BiP) in tobacco and soybean leads to delayed leaf senescence during drought (Valente et al. 2009). The virus-induced downregulation of GmERA1A and GmERA1B genes enhances the stomatal response to ABA and drought resistance in soybean (Ogata et al. 2017). AtDREB1D TF overexpression improves drought tolerance in soybean (Guttikonda et al. 2014). The expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean (Weber et al. 2014). OsDREB2A, a rice TF, considerably affects salt tolerance in transgenic soybean (Zhang et al. 2013).
The FvC5SD gene encodes C-5 sterol desaturase and is isolated from an edible fungus Flammulina velutipes (Kamthan et al. 2012). The C-5 sterol desaturase present in most eukaryotic cells belongs to the fatty acid hydroxylase superfamily of integral membrane proteins that bind an Fe cofactor via a 3-histidine motif (HXXXXH, HXXHH, and HXXHH) (Miyazaki et al. 1999). Many studies have proven that the mechanism of temperature adaptation in an organism includes several cellular functions and components, and the fatty acid composition of the membrane, which determines its fluidity, also plays a key role (Suutari et al. 1990). FvC5SD plays an important role under extreme temperatures and low-pH stress (Kamthan et al. 2017). In plants, FvC5SD overexpression can be attributed to improved tomato drought tolerance, pathogen resistance, and nutritional quality (Kamthan et al. 2012).
To cultivate soybeans with drought resistance and high yield, the engineering of regulatory genes can be used to breed new varieties. In the present study, the Δ7-sterol-C5 (6) desaturase (FvC5SD) from edible fungus F. velutipes was expressed in soybean to develop transgenic plants. To verify the drought resistance of the gene (FvC5SD) in soybean, we transferred this gene into soybean and obtained several different transgenic families. We investigated the drought resistance of soybean from the genotype, drought-tolerant phenotype, and physiological data of GM soybeans. At the same time, we also analyzed the changes in the types and quantities of proteins caused by the expression of foreign genes before and after drought treatment. These results can help elucidate the role of FvC5SD in soybean drought resistance and provide new target genes for molecularly breeding drought-tolerant varieties in soybeans.
Materials and methods
FvC5SD overexpression vector construction and soybean transformation
The complete CDS of FvC5SD (GenBank no. JN696291.1) from F. velutipes was cloned into the pEASY-T1 Cloning Vector (Transgene, China) and sequenced. Then, the corrected FvC5SD fragment was subcloned into the SpeI and SacI sites of the pTF101.1 vector (Fig. 1a). The pTF101 binary vector contained a kanamycin-resistant gene for the bacterial selection and an enhanced CaMV 35S promoter driving bar expression for bialaphos resistance. The Agrobacterium tumefaciens line EHA101 was used in this study.
Construction and Agrobacterium-mediated cotyledonary node transformation of FvC5SD in soybean. a The T-DNA region of the FvC5SD overexpression vector. LB left border, RB right border, bar phosphinothricin acetyl transferase gene, P35S CaMV double 35S promoter, T35S CaMV 35S terminator; b the experimental process of Agrobacterium-mediated transformation by using half-seed soybean explants
The soybean cultivar Shennong9 was utilized for Agrobacterium-mediated transformation as described by Paz et al. (2006) with modifications (Paz et al. 2006). The regenerated plantlets were transplanted into a greenhouse at 28 °C/24 °C with a light/dark photoperiod of 16 h/8 h until maturity. The progeny of each independent soybean transformation was referred to as a line, and the seed of this transgene line was named T0 seed. Each plant that germinated from one T0 seed was called a line, and line designation continued throughout generations.
Molecular analysis of putative transgenic plants
First, soybean plants from independent transformation events (T0) were selected on the basis of herbicide resistance [segregated wild types (WTs) were killed]. Half (along the midrib) of the leaf (upper surface) was painted with 135 mg/L of glufosinate by using a swab and drawn on the other half of the leaf to mark it as the control. Approximately 7 days later, if the leaf half with glufosinate treatment was the same as the control, then the plant was tolerant to the herbicide. Thus, the plant is considered positive. The expression of the resistance gene (bar) was also confirmed using LibertyLink strip (EnviroLogix Inc., Portland, ME, USA) according to the manufacturer’s instructions.
For the PCR analysis of bar and FvC5SD, the total genomic DNA was extracted from the young leaves of transgenic plants by using a simple and quick DNA extraction method developed by Edwards et al. (1991). In our study, the detection primers were designed in accordance with the bar and FvC5SD sequences. For bar, the forward primer (bar-F) was 5′-ATGAGCCCAGAACGACGCCCGGCCG-3′, and the reverse primer (bar-R) was 5′-ATCTCGGTGACGGGCAGGACCGGAC-3′; the length of the product was 548 bp. For FvC5SD, the forward primer (FvC5SD-F) was 5′- ATGCATAGCTCACTGGAGACCAC-3′, and the reverse primer (FvC5SD-R) was 5′-TCAGTTCTTTGAACTAGTGTTTCGC-3′. The length of the product was 891 bp. PCR amplification was conducted with 2X Taq PCR MasterMix (Tiangen Co. Ltd., Beijing, China) according to the manufacturer’s protocols. Transgenic plants from the T0 to T2 generations were all screened using the three methods above.
Southern blot hybridization analysis was carried out to confirm transgene integration into the trifoliate orange genome and estimate the transgene copy number. Southern blot hybridization was performed using a digoxigenin (DIG)-High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany). Genomic DNA was extracted from enlarged soybean leaves by using a modified high-salt CTAB method (Attitalla 2011). The 891-bp PCR-generated fragment of FvC5SD (primers described above for FvC5SD-F/R) was labeled with DIG-high prime and used as the probe. Chemical staining was carried out at room temperature with BCIP/nitroblue tetrazolium (NBT) as substrate until the signal was clearly detected (Labeling and Detection Starter Kit I, 11745832910; Roche Applied Science, USA).
Plant materials, growth conditions, and drought stress treatments
The cultivated variety Shennong9 was the WT in this study. The T0 transgenic lines were grown in a greenhouse at the Jilin Academy of Agricultural Sciences, Gongzhuling, during a 16 h photoperiod at 30 °C and 25 °C daytime and nighttime temperatures, respectively. The T0 seeds were germinated per independent line observing 100% germination. All T0 plants were PCR+, and T1 seeds from these plants were used to grow a large number of plants to obtain sufficient seed material for full characterization (T2 generation). For the soybean drought treatments, the T2 seeds of WT soybean and transgenic lines were planted in plastic pots (35 cm × 25 cm) filled with soil compounds, soil nutrients, and tillaged soil at a 1:1:1 ratio. To screen transgenic lines for drought tolerance, we used 20 seedlings from each transgenic line and WT to serve as control and the four-leaf-stage soybean seedlings for all stress treatments.
To measure root parameters, we harvested the plants carefully without disturbing the root system from soil pots after the stress period. After oven drying at 60 °C for 48 h, the root dry weights were recorded for transgenic and WT plants under controlled and drought treatments.
Physiological parameter measurement under drought stress
The 4-week-old seedlings of T2 homozygous transgenic lines (i.e., L05, L19, L22, and L52) and WT plants were exposed to drought for 7 days, and 0.2 g of leaf tissue was used to measure superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) activity and free proline content. SOD activity was determined by monitoring its ability to inhibit the photochemical reduction of NBT at 560 nm. POD activity was determined using the guaiacol oxidation method. CAT activity was assayed following the method of Yang et al. (2008). Proline content was assessed following the method of Irigoyen et al. (1992). Soluble sugar content was assayed using the phenol-sulfuric acid method. All experiments were repeated three times. The chlorophyll content of leaves from the entire WT and transgenic plants were determined at days 0 (before drought treatment) and 7 (after drought treatment) as described by Arnon (1949) with minor modifications (Arnon 1949). Approximately 0.1 g (fresh weight) of leaves was incubated in 5 mL of ethanol and acetone mixture (1:2, v/v) for 48 h in the dark. Then, the absorbance of the supernatant was analyzed using an ultraviolet spectrophotometer (UV-5100 Spectrophotometer, METASH, Shanghai China) scanning at 665 and 649 nm. The experiment was repeated three times. Each replicate contained five seedlings.
Expression analysis in soybean by using RT-PCR and qRT-PCR
Total RNAs were extracted using TRIzol reagent (Invitrogen, America) according to the manufacturer’s instructions. Reverse transcription reactions were performed using PrimeScript RT reagent kit with gDNA Eraser (Takara, China) following the manufacturer’s instructions. qRT-PCR was performed using a SYBR Green Master Mix Kit (Roche, Germany) with specific primers (Supplementary File 5: Table S4) on an ABI 7300 system. Three separate biological replicates were carried out for the qRT-PCR experiments. Transcript levels were calculated using the formula 2−ΔΔCt for the expression levels relative to the GmActin gene (NM 001289231). All primers used for RT-PCR and qRT-PCR are provided in Supplementary File 5: Table S4.
iTRAQ analysis
To investigate the global effects of FvC5SD overexpression on the soybean endogenous metabolism further, we chose the transgenic line L19–16 (Fig. 3), which showed significant changes under drought stress, to study the changes in protein expression. The analysis of the total proteins and protein expression changes in line 19–16 (normal condition and drought stress) was performed using iTRAQ-based proteomics with three biological replicates. Four fully expanded leaf FvC5SD transgenic seedlings under normal and drought conditions were rapidly cut, frozen, and stored. The gene ontology (GO) annotation for functional analysis was performed using the DAVID Bioinformatics Resources 6.7 (https://david.ncifcrf.gov/), and the proteins were classified on the basis of their molecular function, biological process, and cellular components. The STRING database was used for protein–protein interaction (PPI) analysis.
Statistical analysis
A two-sided variance (ANOVA) test analysis (*P < 0.05, **P < 0.01, ***P < 0.001) was used to determine the significance of data from each trait. Standard errors are provided in all tables and figures as appropriate. The SPSS v22.0 software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Construction of expression vectors and soybean stable transformation
The BLAST analysis results indicated that FvC5SD had 34% similarity to Δ(7)-sterol-C5 (6)-desaturase of soybean (Supplementary File 1: Figure S1). Meanwhile, FvC5SD was the most closely related to sterol desaturase of the fungus Aspergillus fumigatus (54%) and Cryptococcus neoformans (53% identity) (Kamthan et al. 2012). Soybean was transformed using Agrobacterium-mediated cotyledonary node method (Paz et al. 2006), which has been used to successfully produce stable transgenic lines in this species (Fig. 1b). A total of 415 soybean explants from the cultivar Shennong9 were infected with the pTF101-FvC5SD vector containing the FvC5SD and phosphinothricin acetyl transferase gene (bar gene, which shows resistance to the herbicide phosphinothricin) under 35S constitutive promoters (Fig. 1a). A total of 29 independent transformation events were confirmed on the basis of resistance to the herbicide (LibertyLink strip analysis), thereby showing an overall transformation efficiency of 7.2% (29/415) (Supplementary File 2: Table S1).
Verification of FvC5SD-overexpressing transgenic soybean plants
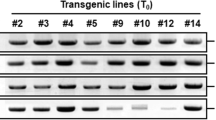
To detect the transgenic soybean plants, we used three methods (i.e., leaf painting, LibertyLink strip analysis, and PCR) to confirm the bar-positive transgenic plants (Fig. 2a–c). After in vitro selection, the plantlets were transplanted in the soil to develop T0 line seeds under the greenhouse condition. During T0 plant development, we performed PCR analysis to determine FvC5SD (Fig. 2d). Finally, we harvested nine different T0 line seeds in Shennong9. The WT and T0 PCR+ plants had the same visual phenotype under normal conditions. We selected four T0 lines with more than 20 seeds for further drought resistance analysis. Southern blot analysis was carried out in four independent T1 transgenic lines by using FvC5SD as the probe. The result showed that the transgene lines 22, 52, 19, and L05 contained three, two, two, and one transgene copies, respectively (Fig. 2e).
Verification of FvC5SD-overexpressing transgenic soybean plants. a Herbicide (glufosinate) painting on soybean leaves. Half of the leaf in the transgene line was marked with a black line in the middle. The left side served as the control, and the right side was painted with 135 mg/L glufosinate. b LibertyLink® strip detection. WT was a NT plant. 1–14 were transgenic soybean plants. c The PCR analysis of putative transgenic soybean plants by using bar gene primers. GmActin gene was used to check the genome DNA of plants, and the length was 403 bp. The length of bar was 548 bp (M, Trans2K® Plus II DNA marker; +, plasmid DNA; and −, nontransformed soybean; 1–14). The events of the PCR positive lines were randomly selected and tested using LibertyLink® strips. d The PCR analysis of putative transgenic soybean plants was performed using FvC5SD gene primers. The length of the FvC5SD was 891 bp. e Southern blot analysis of FvC5SD-overexpressing transgenic lines. L05, L19, L22, and L25 represent four independent FvC5SD overexpressing lines. The genomic DNA of 4-week-old T1 transgenic seedlings and the nontransformed recipient soybean genotype Williams 82 were extracted and digested with EcoRI and HindIII, respectively. The 891 bp FvC5SD was DIG-labeled and used as the probe for analysis
We also observed the growth between GM soybeans and recipients. The measured agronomic traits included plant height, branch number, node number, pod number, seed number, seed yield, and 100-seed weights. Statistical results showed no difference in agronomic traits between GM soybeans and recipients (Table 1).
Physiological changes in soybean plants under drought treatment
The role of FvC5SD in regulating drought response was studied using 4-week-old transgenic (> 1-month-old) soybean seedlings (T2 progenies of the sublines L05–14, L19–16, L22–1718, and L52–20) transformed with FvC5SD and WT plants. A total of 20 positive plants for each class were used for drought tolerance experiments under natural dry treatment. We examined the lines with minimal phenotypic differences compared with those of WT plants to perform drought treatment. All plants were divided in two groups; one group had normal growth conditions in soil with normal water for 7 days, and the other group had suspended water supply for 3 and 7 days. As shown in Fig. 3a, c, e, the difference in phenotype for the transgenic plants compared with the WT plants under normal conditions was insignificant. Meanwhile, during drought tolerance experiments, all WT plants showed clearly shrunken leaves and wilting after 3 days of cutting the water supply. The whole plant also exhibited signs of dwarfism (Fig. 3b), whereas all transgenic plants kept their whole plant turgor and showed delayed wilting. After 7 days without water, when the transgenic plants became dehydrated, all the WT plants showed severe leaf wilting and finally leaf shedding. The whole plant nearly died (Fig. 3d). Then, we started watering the plants again. After 3 days, the transgenic line plants recovered with less inhibition from wilting by rehydration than the WT plants (Fig. 3f). These results further confirmed that FvC5SD overexpression can improve drought tolerance in transgenic soybean seedlings.
FvC5SD overexpression in soybean indicates drought tolerance. The phenotypes of transgenic soybean plants under drought stress at the 4-week-old stage. Four independent homozygous transgenic T2 lines (L05-14, L19-16, L22-1718, and L52-20) and non-transgenic control plants were grown in the soil for 4 weeks and exposed to drought for 3 and 7 days, followed by rewatering. The values are expressed as mean ± SE (n = 10). a The phenotypic effect of watering with water for 3-days. b The phenotypic effect of suspending water supply for 3-days. c The phenotypic effect of watering with water for 7-days. d The phenotypic effect of suspending water supply for 7-days. e The phenotypic effect of watering with water for 10-days. f The phenotypic effect of suspending water supply for 7-days, followed by 3-day recovery period
To investigate the effect of root biomass between WT plants and transgenic lines, we calculated the root dry weight under controlled and drought treatments. As shown in Fig. 4, transgenic soybean showed significantly more roots than the WT plants (Fig. 4a). Under drought treatment, the transgenic lines L05–14, L19–16, L22–1718, and L52–20 showed increased dry root mass (29, 41, 56, and 25%, respectively; Fig. 4b).
SOD activity, POD activity, and chlorophyll content measurement
Reactive oxygen species (ROS) can induce oxidative stress under drought stress. Therefore, we measured the proline content and analyzed the antioxidant activities of enzymes (i.e., SOD, POD, and CAT) between transgenic and WT plants under drought or normal conditions. The whole antioxidant activities of enzymes, proline content, and soluble sugar content in WT and transgenic plants under drought treatment were higher than those under normal condition (Fig. 5a–d). The differences in proline content, soluble sugar content, and antioxidant enzyme activities were insignificant between WT and transgenic plants under normal conditions (Fig. 5). For example, a significantly high proline content level was detected in all plants under drought condition, which was nearly six- to sevenfold higher than that under normal condition. Drought-stressed transgenic plants also possessed higher proline and soluble sugar content than WT plants (Fig. 5d, e). Similar results were also found in the antioxidant enzyme activity during drought treatment (Fig. 5a–c). For example, the SOD activity in the transgenic lines increased by approximately 1.3- to 1.5-fold under drought treatment, and the highest increase was observed in the transgenic line L22–1718, with a 1.5-fold increase in response to drought treatment. A significantly high POD level was detected in transgenic plants under drought condition (nearly 2.4-, 2.27-, 2.32-, and 1.88-fold increase in POD content for L22–1718, L05–14, L19–16, and L52–20, respectively). The whole level of CAT activity in the transgenic plants increased by as much as 1.19- to 1.5-fold under drought condition than under normal condition. These results suggested that FvC5SD may enhance the antioxidant enzyme activity and proline accumulation, thereby decreasing ROS and increasing drought tolerance in soybean.
Physiological parameter analysis in FvC5SD-transgenic plants under drought stress. a SOD activity. b POD activity. c CAT activity. d Proline content of soybean leaves. e Soluble sugar content of soybean leaves. f Chlorophyll content of soybean leaves. Data are shown as the mean ± SE (n = 20). Statistical analysis was performed using ANOVA (P < 0.05), and significant differences between NT and transgenic lines are indicated by *P < 0.05, **P < 0.01, and ***P < 0.001. All values were measured after 7 days of exposure to drought stress
To investigate whether the cuticle properties of the transgenic plants were altered further, we detected the chlorophyll content between WT and FvC5SD-transgenic plants (Fig. 5f). The chlorophyll content leaching from transgenic leaves significantly decreased, which indicated a change in cuticular permeability.
RT and qRT-PCR analyses of transgenic lines
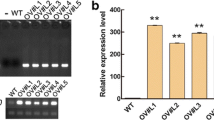
Under normal condition, FvC5SD validation was confirmed in four different T0-positive transgenic soybean lines (i.e., L05, L19, L22, and L52) by RT-PCR and qRT-PCR analyses. FvC5SD was unexpressed in WT plants (Fig. 6a). The FvC5SD expression level in the four lines was increased compared with that in WT Shennong9. In the four different lines, L22 exhibited the highest expression with 408.8-fold increase, followed by L19 with 40.6-fold increase, L05 with 14.9-fold increase, and L52 with 1.43-fold increase in expression (Fig. 6b).
Root phenotype under drought stress. a The semiquantitative RT-PCR analysis of the FvC5SD gene expression in wild-type (Shennong9) and T0 transgenic lines. b qRT-PCR revealed the elevated FvC5SD expression in the T0 transgenic lines (L22, L52, L05, and L19). c The relative expression levels of FvC5SD in four independent transgenic lines (i.e., L22, L52, L05, and L19) under drought conditions. d RT and qRT-PCR analyses showing the transcript levels of FvC5SD. GmActin was used as the reference gene. Data are shown as the mean ± SE of ten biological and four technical replicates
To examine the FvC5SD expression level under drought conditions, we watered the transgenic soybean plants well (control), in which water was withheld for 14 days (drought treatment). FvC5SD expression increased by up to 3.58-fold in L05–14, 2.34-fold in L19–16, and 1.97-fold in L22–1718 drought-stressed transgenic soybean plants. However, the change in FvC5SD expression was insignificant in drought-stressed L52–20 lines. This finding further demonstrated that drought stress can also influence FvC5SD expression (Fig. 6c).
To analyze the tissue-specific expression pattern of FvC5SD, we grew soybean plants in the field to obtain different tissues. At the jointing stage, the total RNA from roots, stems, and leaves was isolated separately at the 4-week-old seedling stage. The flowers and seeds were sampled at the fruit stage. As shown in Fig. 6d, FvC5SD expression was higher in pistils and leaves but lower in seeds, stamens, and roots (Fig. 6d).
Induced differentially expressed proteins (DEPs) by overexpressing FvC5SD in transgenic plants
Some genes that code either functional or regulatory proteins were induced in the transgenic lines under drought stress to analyze the FvC5SD target. We performed iTRAQ analyses on the leaves of transgenic line plants grown under normal conditions and drought stress. A total of 69,722 spectra can be matched to the database, thereby resulting in 11,326 peptides, which were assembled into 2223 nonredundant protein groups. We identified 81 DEPs on the basis of the following two criteria: (i) P < 0.05 and (ii) a fold change in the expression of > 1.5 or < 0.66 (Supplementary File 3: Table S2). The results showed that 43 DEPs were upregulated, and 38 DEPs were downregulated. A total of 56 protein species were identified as unknown proteins (Supplementary File 3: Table S2).
To reveal the functions of DEPs between normal and drought conditions in the transgene plant further, we performed GO analysis to confirm the cellular component, biological process, and molecular function (Supplementary File 4: Table S3). Eleven out of the 81 identified proteins were classified into three large groups containing 27 subgroups on the basis of their functional annotation (Fig. 7). From the enrichment results of cell components, the protein is rich in ten classes, of which nine DEPs are enriched in cells (GO: 0005623), cell components (GO: 0044464), and organelles (GO: 0043226). The macromolecular complex (GO:0032991) and the organelle component (GO:0044422) were enriched in five proteins, and the remaining types were enriched in two proteins. Overall, DEPs are enriched in cells and organelles under drought stress, which is consistent with the location of important enzymes involved in photosynthesis and carbon fixation.
GO classification of the identified DEPs. GO analysis was performed using WEGO software to reveal the functions of the identified 34 DEPs of the transgene plant between normal and drought conditions. The 8 proteins among the 34 identified DEPs were available and then classified into 3 main categories, including cellular component, biological process, and molecular function with 23 subgroups. The number of genes denotes that of proteins with GO annotations
The DEPs enriched in biological processes are divided into 13 categories, covering many aspects, including cell processes, signal transduction, metabolic processes, stress, etc. The largest type of biological process is cellular process (GO: 0009987), enriched in 10 proteins, followed by single tissue process (GO: 0044699) and a metabolic process (GO: 0008152), each enriched in nine proteins, and biological regulation (GO: 0065007), enriched in eight differential proteins. The stimulating (GO: 0050896) and cellular component tissues (GO: 0071840) are enriched in six DEPs; multicellular processes (GO: 0032501) and developmental processes (GO: 0032502) are enriched in four DEPs; localization (GO: 0051179) and signal (GO: 0023052) are enriched in three and two DEPs, respectively; and growth (GO: 0040007), immune system progression (GO: 0040007), and obsolete death (GO: 0016265) are enriched in only one DEP. In general, differential proteins are mainly enriched in processes related to metabolism and stress, while proteins are enriched in developmental immunity.
The DEPs enriched in molecular function are mainly divided into four categories, namely, surface binding of discarded eukaryotic cells (GO: 0043499), binding (GO: 0005488), transporter activity (GO: 0005215), and catalytic activity (GO: 0003824). Among them, the number of DEPs combined with enrichment is 11, followed by catalytic activity enrichment of nine DEPs. The surface binding and transporter activities of discarded eukaryotic cells are enriched by one DEP. From the perspective of molecular function, DEPs are mainly related to catalysis and binding. From the results of GO analysis, the DEPs between transgenic soybean and WT plants after drought stress may be related to the catalytic combination of a certain metabolic process in the cell.
A total of 81 differential proteins were introduced into the STRING database to identify the interaction of these proteins. The protein interaction network was constructed and visualized using the STRING 11.0 database. Of the 81 differential proteins, 32 are involved in PPI. These 32 proteins form three functional modules that are tightly linked (Fig. 8). Nodes of different colors belong to three large groups. The thickness of the line represents the strength of the association, and the stronger the line, the thicker the line. In module 1 (red node), twelve proteins are related to each other; they are glutamine synthetase (GS: GLYMA13G28180.4, GLYMA13G28180.5), asparagine synthetase (GLYMA11G27480.1), lipoxygenase (GLYMA11G13880.2, GLYMA08G20250.1), thiamine thiazole synthase (GLYMA10G39740.1) and ATP synthase and functionally unknown proteins. This finding indicates that amino acid metabolism, fatty acid biosynthesis, and energy supply are closely linked. In addition, the gene psbA encoding photosystem II and the gene psaB encoding photosystem I in photosynthesis are linked to module 2 (green node), containing ferredoxin (GLYMA12G29100.1) and glyceraldehyde-3-phosphate dehydrogenase (LOC732571), which are involved in respiration. This finding suggests that these DEPs play a role in photosynthesis, respiration, and energy metabolism. Finally, DEPs involved in protein folding (GLYMA10G27990.1) and some proteins with unknown functions are assigned to Module 3 (blue node).
PPI network of DEPs in transgene soybean under normal and drought conditions on the basis of STRING analysis. A total of 34 differentially abundant proteins represented by 8 unique proteins from soybean are shown in the PPI network. Nodes in different colors belong to two main groups. Strong associations are represented by thick lines. Among them, the nine upregulated proteins are marked with asterisk, and one downregulated protein is marked with hash
Discussion
Drought stresses substantially influence the growth, development, and productivity of soybean and other crop plants. Therefore, drought-tolerant genes should be explored for the genetic improvement of crops. To combat drought stress in soybean, an increasing number of molecular biology tools have been applied to identify and transfer genes responsible for tolerance to drought and other stresses (Deshmukh et al. 2014; Manavalan et al. 2009; Thao and Tran 2012). Different types of GM soybean, such as the herbicide-tolerant transgenic soybean RoundUp Ready (Funke et al. 2006), have been developed to overcome different biotic and abiotic stresses (Yamada et al. 2012).
In this study, FvC5SD was transferred into soybean. This gene was integrated into the soybean genome and confirmed to be stably expressed via PCR, Southern blot, and RT-PCR analyses. Our results showed that FvC5SD overexpression improved drought tolerance compared with WT plants, which can be used to improve soybean biotechnology. FvC5SD plants exhibited decreased and delayed drought-induced damage compared with control plants and recovered rapidly after rehydration (Fig. 3).
The C-5 sterol desaturase is involved in phytosterol and brassinosteroid biosynthesis in plants (Darnet and Rahier 2004; Taton and Rahier 1996). FvC5SD was significantly upregulated at an extreme temperature and low pH. In yeast, the overexpression of the FvC5SD heterologous gene in Schizosaccharomyces pombe increases tolerance to extreme temperatures, high ethanol concentration, and low pH (Kamthan et al. 2017). The expression of a fungal sterol desaturase improves tomato drought tolerance, pathogen resistance, and nutritional quality (Kamthan et al. 2012). These results showed that FvC5SD plays an important role in plants under extreme temperature and low-pH stress conditions.
In plants, some changes and interaction between physiological indicators are important for studying the response of plants to abiotic stress. Plants are affected by drought stress and cause oxidative stress mediated by ROS. SOD, POD, and CAT work together to prevent excessive levels of ROS in the cells, collectively referred to as the protective enzyme system; proline and soluble sugar are important osmotic adjustment substances in plant cells (Mansour and Ali 2017). In our study, changes in plant physiological indicators were used to determine whether plants are drought-resistant. The experimental results showed that SOD, POD, and CAT contents increased after drought stress, and this trend was more obvious in transgenic soybeans (Fig. 5a–c). These results indicated that the antioxidant capacity of transgenic soybeans was higher than that of WT plants. Similarly, the soluble sugar and proline content of plants increased after drought stress, and the increase was even greater in transgenic plants (Fig. 5d, e). These results indicate that transgenic plants enhance their drought resistance by accumulating soluble sugars and proteins. Changes in the results of these physiological indicators indicated that FvC5SD can increase ROS levels and enhance the drought resistance of plants.
FvC5SD overexpression in tomato can increase the amount of wax deposition in transgenic tomato, thereby inducing drought tolerance and pathogen resistance (Kamthan et al. 2012). Several studies have shown that the modification of the cuticular wax layer in plants is one of the strategies to improve drought tolerance by reducing transpirational water loss (Aharoni et al. 2004; Islam et al. 2009; Zhang et al. 2005). Although we did not perform GC–MS analysis for the fatty acid content in transgenic soybean in this study, we identified at least four DEPs (i.e., I1KUQ6, K7LPI4, Q38IX0, and C6T9R8) that are associated with the fatty acid biosynthetic pathway via TRAQ-based comparative proteomic analysis (Supplementary File 3: Table S2). The expression of these DEPs was significantly changed in WT and FvC5SD-expressing transgenic soybean plants under drought stress condition, which was consistent with the finding that FvC5SD was directly affected and regulated by mutations in other enzymes of the ergosterol biosynthesis pathway. The mechanism of plant adaptation to stress conditions in an organism also includes several cellular functions and components (Bourdenx et al. 2011). Similar results were also found in the Fe pathway. FvC5SD is an Fe-BiP, and its expression increases the total Fe content in transgenic tomato (Kamthan et al. 2012). Two ferredoxin DEPs (C6T1J0 and I1LTF4) were upregulated by 1.54- and 1.75-fold in transgenic soybean under drought stress via iTRAQ analysis. These findings indicated that FvC5SD overexpression in these crops can be a potentially useful strategy to increase the total Fe content.
iTRAQ data analysis also revealed that most represented DEPs in transgenic soybean are associated with photosynthetic and chlorophyll metabolism pathways. Chloroplasts are particularly vulnerable to ROS-induced damage (Berkowitz 1987). Many studies have shown that salt stress affects mRNA editing in chloroplasts (Zhao et al. 2018; Rodrigues et al. 2017). Here, some DEPs (e.g., Q2PMU2, P69195, I1JWU0, and P02957) that are associated with the chlorophyll metabolism pathway were identified. This finding was consistent with the observation that the leaves of drought-stressed WT soybean plants were more affected by chlorosis and contained less chlorophyll than those of FvC5SD transgenic plants (Fig. 5f). Hence, FvC5SD overexpression may prevent chlorophyll from incurring ROS damage to some extent in plants.
PPI analysis results revealed that the DEPs between WT plants and transgenic lines under drought condition were mainly involved in the photosynthetic pathway, N metabolism, and fatty acid biosynthesis (Fig. 8). GS is the key enzyme in plant nitrogen metabolism (Miflin and Habash 2002). The overexpression of the OsGS gene modulates oxidative stress response in rice after exposure to cadmium stress (Lee et al. 2013). In our study, DEP (I1M170), which encodes the GS gene (GLYMA13G28180.4), was up-regulated in soybean leaves under drought stress and also interacted with glyceraldehyde-3-phosphate dehydrogenase (LOC732571), which is involved in fatty acid biosynthesis. The Calvin cycle, which is the primary pathway for photosynthetic C fixation in C3 plants and takes place in the chloroplast stroma, includes three major steps: carboxylation of ribulose-1, 5-bisphosphate (RuBP), reduction of 3-phosphoglycerate, and regeneration of the CO2 acceptor RuBP (Gerhardt et al. 1987; van Leeuwen et al. 2015). These data showed that the DEPs related to photosynthesis, N metabolism, and fatty acid biosynthesis were up-regulated, thereby resulting in the high photosynthesis ability in transgenic soybean line, which requires further evidence to be confirmed. PPI analysis indicated that protein metabolism and photosynthesis collectively functioned to re-establish cellular homeostasis under drought stress.
iTRAQ results indicated that various pathways, such as photosynthesis organisms and photosynthesis, glyoxylate and dicarboxylate metabolism pathway, and energy metabolism, may be involved in FvC5SD-mediated enhanced tolerance to drought in transgenic soybean plants after exposure to drought stress. Gene overexpression may also cause other related synergistic actions.
Conclusion
FvC5SD overexpression can serve as a direct and efficient target for the improvement of drought tolerance in soybean. FvC5SD overexpression in soybean may also provide a nutritional quality (fatty acid and Fe) advantage under water deficit, which will be further studied and confirmed in our transgenic soybean lines. This biotechnological strategy should be of considerable value for trait improvement in soybean and other crops.
Author contribution statement
YSD and LZ developed and framed the research questions. TL analyzed proteomics data. YW were involved in sample collection. LZ drafted and revised the manuscript. All authors read and approved the final manuscript.
References
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480. https://doi.org/10.1105/tpc.104.022897
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in beta vulgaris. Plant Physiol 24:1–15
Attitalla IH (2011) Modified CTAB method for high quality genomic DNA extraction from medicinal plants. Pak J Biol Sci 14:998–999
Berkowitz GA (1987) Chloroplast acclimation to low osmotic potential. Plant Cell Rep 6:208–211. https://doi.org/10.1007/BF00268481
Bourdenx B et al (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156:29–45. https://doi.org/10.1104/pp.111.172320
Darnet S, Rahier A (2004) Plant sterol biosynthesis: identification of two distinct families of sterol 4alpha-methyl oxidases. Biochem J 378:889–898. https://doi.org/10.1042/bj20031572
de Paiva Rolla AA et al (2014) Phenotyping soybean plants transformed with rd29A:AtDREB1A for drought tolerance in the greenhouse and field. Transgenic Res 23:75–87. https://doi.org/10.1007/s11248-013-9723-6
Deshmukh R et al (2014) Integrating omic approaches for abiotic stress tolerance in soybean. Front Plant Sci 5:244. https://doi.org/10.3389/fpls.2014.00244
Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19:1349
Funke T, Han H, Healy-Fried ML, Fischer M, Schonbrunn E (2006) Molecular basis for the herbicide resistance of Roundup Ready crops. Proc Natl Acad Sci USA 103:13010–13015. https://doi.org/10.1073/pnas.0603638103
Gerhardt R, Stitt M, Heldt HW (1987) Subcellular metabolite levels in spinach leaves: regulation of sucrose synthesis during diurnal alterations in photosynthetic partitioning. Plant Physiol 83:399–407
Guttikonda SK et al (2014) Overexpression of AtDREB1D transcription factor improves drought tolerance in soybean. Mol Biol Rep 41:7995–8008. https://doi.org/10.1007/s11033-014-3695-3
Irigoyen J et al (1992) Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativa) plants. Physiol Plant 84:55–60
Islam MA, Du H, Ning J, Ye H, Xiong L (2009) Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70:443–456. https://doi.org/10.1007/s11103-009-9483-0
Kamthan A, Kamthan M, Azam M, Chakraborty N, Chakraborty S, Datta A (2012) Expression of a fungal sterol desaturase improves tomato drought tolerance, pathogen resistance and nutritional quality. Sci Rep 2:951. https://doi.org/10.1038/srep00951
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2016) Genetically modified (GM) crops: milestones and new advances in crop improvement. Theor Appl Genet 129:1639–1655. https://doi.org/10.1007/s00122-016-2747-6
Kamthan A, Kamthan M, Datta A (2017) Expression of C-5 sterol desaturase from an edible mushroom in fisson yeast enhances its ethanol and thermotolerance. PLoS One 12:e0173381. https://doi.org/10.1371/journal.pone.0173381
Kim HJ et al (2018a) Confirmation of drought tolerance of ectopically expressed AtABF3 gene in Soybean. Mol Cells 41:413–422. https://doi.org/10.14348/molcells.2018.2254
Kim HJ et al (2018b) Erratum to: confirmation of drought tolerance of ectopically expressed AtABF3 gene in Soybean. Mol Cells 41:703. https://doi.org/10.14348/molcells.2018.1254
Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C (2015) Cell wall metabolism in response to abiotic stress. Plants (Basel) 4:112–166. https://doi.org/10.3390/plants4010112
Lee HJ et al (2013) Overexpression of the glutamine synthetase gene modulates oxidative stress response in rice after exposure to cadmium stress. Plant Cell Rep 32:1521–1529. https://doi.org/10.1007/s00299-013-1464-8
Leite JP et al (2014) Overexpression of the activated form of the AtAREB1 gene (AtAREB1DeltaQT) improves soybean responses to water deficit. Genet Mol Res 13:6272–6286. https://doi.org/10.4238/2014.august.15.10
Li Y et al (2013) Expression of an Arabidopsis molybdenum cofactor sulphurase gene in soybean enhances drought tolerance and increases yield under field conditions. Plant Biotechnol J 11:747–758. https://doi.org/10.1111/pbi.12066
Li Y et al (2017) Overexpression of GmFDL19 enhances tolerance to drought and salt stresses in soybean. PLoS One 12:e0179554. https://doi.org/10.1371/journal.pone.0179554
Manavalan LP, Guttikonda SK, Tran LS, Nguyen HT (2009) Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol 50:1260–1276. https://doi.org/10.1093/pcp/pcp082
Mansour MMF, Ali EF (2017) Evaluation of proline functions in saline conditions. Phytochemistry 140:52–68. https://doi.org/10.1016/j.phytochem.2017.04.016
Miflin BJ, Habash DZ (2002) The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J Exp Bot 53:979–987
Miyazaki Y et al (1999) Cloning, sequencing, expression and allelic sequence diversity of ERG3 (C-5 sterol desaturase gene) in Candida albicans. Gene 236:43–51
Ogata T, Nagatoshi Y, Yamagishi N, Yoshikawa N, Fujita Y (2017) Virus-induced down-regulation of GmERA1A and GmERA1B genes enhances the stomatal response to abscisic acid and drought resistance in soybean. PLoS One 12:e0175650. https://doi.org/10.1371/journal.pone.0175650
Paz MM, Martinez JC, Kalvig AB, Fonger TM, Wang K (2006) Improved cotyledonary node method using an alternative explant derived from mature seed for efficient Agrobacterium-mediated soybean transformation. Plant Cell Rep 25:206–213. https://doi.org/10.1007/s00299-005-0048-7
Rodrigues NF, Fonseca GC, Kulcheski FR, Margis R (2017) Salt stress affects mRNA editing in soybean chloroplasts. Genet Mol Biol 40:200–208. https://doi.org/10.1590/1678-4685-GMB-2016-0055
Suutari M, Liukkonen K, Laakso S (1990) Temperature adaptation in yeasts: the role of fatty acids. J Gen Microbiol 136:1469–1474. https://doi.org/10.1099/00221287-136-8-1469
Taton M, Rahier A (1996) Plant sterol biosynthesis: identification and characterization of higher plant delta 7-sterol C5(6)-desaturase. Arch Biochem Biophys 325:279–288. https://doi.org/10.1006/abbi.1996.0035
Thao NP, Tran LS (2012) Potentials toward genetic engineering of drought-tolerant soybean. Crit Rev Biotechnol 32:349–362. https://doi.org/10.3109/07388551.2011.643463
Valente MA et al (2009) The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J Exp Bot 60:533–546. https://doi.org/10.1093/jxb/ern296
Valliyodan B, Ye H, Song L, Murphy M, Shannon JG, Nguyen HT (2017) Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J Exp Bot 68:1835–1849. https://doi.org/10.1093/jxb/erw433
van Leeuwen M, Kremens RL, van Aardt J (2015) Tracking diurnal variation in photosynthetic down-regulation using low cost spectroscopic instrumentation. Sensors (Basel) 15:10616–10630. https://doi.org/10.3390/s150510616
Weber RL et al (2014) Expression of an osmotin-like protein from Solanum nigrum confers drought tolerance in transgenic soybean. BMC Plant Biol 14:343. https://doi.org/10.1186/s12870-014-0343-y
Yamada T, Takagi K, Ishimoto M (2012) Recent advances in soybean transformation and their application to molecular breeding and genomic analysis. Breed Sci 61:480–494. https://doi.org/10.1270/jsbbs.61.480
Yang L, Tang R, Zhu J, Liu H, Mueller-Roeber B, Xia H, Zhang H (2008) Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2beta, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol Biol 66:329–343. https://doi.org/10.1007/s11103-007-9267-3
Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42:689–707. https://doi.org/10.1111/j.1365-313x.2005.02405.x
Zhang XX et al (2013) OsDREB2A, a rice transcription factor, significantly affects salt tolerance in transgenic soybean. PLoS One 8:e83011. https://doi.org/10.1371/journal.pone.0083011
Zhao C, Haigh AM, Holford P, Chen ZH (2018) Roles of chloroplast retrograde signals and ion transport in plant drought tolerance. Int J Mol Sci. https://doi.org/10.3390/ijms19040963
Acknowledgements
This research work was supported by the Agricultural Science and Technology Innovation Project of Jilin Province (CXGC2017ZY02), the National Natural Science Foundation of China (31501327), and the Ministry of Agriculture of China for Transgenic Research (2016ZX08004-004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Additional information
Communicated by Zheng-Yi Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, L., Li, T., Wang, Y. et al. FvC5SD overexpression enhances drought tolerance in soybean by reactive oxygen species scavenging and modulating stress-responsive gene expression. Plant Cell Rep 38, 1039–1051 (2019). https://doi.org/10.1007/s00299-019-02424-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-019-02424-y