Abstract
The outermost surfaces of plants are covered with an epicuticular wax layer that provides a primary waterproof barrier and protection against different environmental stresses. Glossy 1 (GL1) is one of the reported genes controlling wax synthesis. This study analyzed GL1-homologous genes in Oryza sativa and characterized the key members of this family involved in wax synthesis and stress resistance. Sequence analysis revealed 11 homologous genes of GL1 in rice, designated OsGL1-1 to OsGL1-11. OsGL1-1, -2 and -3 are closely related to GL1. OsGL1-4, -5, -6, and -7 are closely related to Arabidopsis CER1 that is involved in cuticular wax biosynthesis. OsGL1-8, -9, -10 and -11 are closely related to SUR2 encoding a putative sterol desaturase also involved in epicuticular wax biosynthesis. These genes showed variable expression levels in different tissues and organs of rice, and most of them were induced by abiotic stresses. Compared to the wild type, the OsGL1-2-over-expression rice exhibited more wax crystallization and a thicker epicuticular layer; while the mutant of this gene showed less wax crystallization and a thinner cuticular layer. Chlorophyll leaching experiment suggested that the cuticular permeability was decreased and increased in the over-expression lines and the mutant, respectively. Quantification analysis of wax composition by GC–MS revealed a significant reduction of total cuticular wax in the mutant and increase of total cuticular wax in the over-expression plants. Compared to the over-expression and wild type plants, the osgl1-2 mutant was more sensitive to drought stress at reproductive stage, suggesting an important role of this gene in drought resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Many characteristics of the leaf, such as water potential, osmotic adjustment, cell membrane stability, cuticular wax characteristics, and epidermal conductance, can affect drought resistance of plants. Among these, increasing attention has focused on the importance of cuticular wax on the aerial surfaces of plants because it is the outermost waterproof barrier and provides primary protection against environmental stresses (Post-Beittenmiller 1996). A cuticle layer covers most aerial organs of vascular plants and forms the outermost contacting zone between the plant and the environment (Kerstiens 1996a). Among the multiple functions of the cuticle (Barnes et al. 1996; Barthlott and Neinhuis 1997; Eigenbrode 1996; Jenks et al. 1994), the pivotal one is to provide a diffusion barrier against the uncontrolled loss or uptake of water and gases (Kerstiens 1996b, 2006).
A polyester matrix (cutin) is the major constituent of the cuticle that is overlaid or embedded with a long-chain hydrocarbon (cuticular wax) (Kunst and Samuels 2003; Riederer and Schreiber 2001; Vogg et al. 2004). Cuticular waxes are complex mixtures of primarily very-long-chain (VLC, >C18) fatty acids, hydrocarbons, alcohols, aldehydes, ketones, esters, triterpenes, sterols, and flavonoids (Kolattukudy 1980; Walton 1990). The proportions of the major constituents vary among plant species. Cutin, suberin, and wax are all derived from fatty acid precursors and their physical, functional, and biosynthetic relationships strongly suggest that wax production may be co-regulated with cutin and suberin production (James et al. 1999). An increase in cuticular permeability may be associated with a decrease (Jenks et al. 1994; Sturaro et al. 2005) or an increase (Chen et al. 2003) in cuticular thickness or coverage, or with a decrease (Chen et al. 2003; Jenks et al. 1994; Sturaro et al. 2005; Vogg et al. 2004) or increase (Aharoni et al. 2004) in wax coverage, or with a change in cuticle polymer (Xiao et al. 2004) or wax (Aharoni et al. 2004; Vogg et al. 2004) composition.
Plant cuticular wax biosynthesis and its loading to the plant surface are complicated but actively regulated processes (Broun et al. 2004; Jenks et al. 2002). Mutants with reduced wax accumulation or altered wax composition are commonly characterized by a bright green phenotype that can be detected visually (Aarts et al. 1995). In Arabidopsis, 120 cuticular wax mutants representing a total of 31 recessive mutant loci have been identified, although the dominant wax gene mutations have not been reported (Jenks et al. 2002). Wax-deficient mutants have been identified in other plants, including maize, sorghum, barley, and rape (Kunst and Samuels 2003). Studies of the eceriferum (cer) mutants in Arabidopsis and glossy (gl) mutants in maize led to the identification and isolation of a number of wax-related genes. To date, many genes involved in wax biosynthesis or its regulation have been identified in plants by molecular genetic approaches. Among these genes, CER1, CER2, CER6/CUT1, KCS1 (3-ketoacyl-CoA synthase), FIDDLEHEAD (FDH), and WAX2 from Arabidopsis, GL1 and GL8 from maize, and WXP1 from Medicago encode wax synthesis-related enzymes or proteins involved in the transport of wax compounds (Aarts et al. 1995; Chen et al. 2003; Fiebig et al. 2000; Hansen et al. 1997; Millar et al. 1999; Negruk et al. 1996; Pruitt et al. 2000; St-Pierre et al. 1998; Todd et al. 1999; Xia et al. 1996, 1997; Xu et al. 1997; Zhang et al. 2005), whereas CER3, GL2, GL15, and WIN1/SHN1 encode regulatory proteins (Aharoni et al. 2004; Broun et al. 2004; Hannoufa et al. 1996; Moose and Sisco 1996; Tacke et al. 1995). Mutation in most of these genes showed altered wax accumulation (Jenks et al. 2002). Co-suppression of some of the genes in Arabidopsis resulted in reduced wax on stems (Millar et al. 1999; Todd et al. 1999), and over-expression of some of these genes in the Arabidopsis mutants complemented the mutant phenotypes (Fiebig et al. 2000; Hannoufa et al. 1996). However, only a few reports discuss the effect of over-expression of these genes in the wild-type background. Over-expression of the condensing enzyme gene CER6/CUT1 under the control of the CaMV35S promoter failed to promote wax deposition (Millar et al. 1999), whereas under the control of the epidermis-specific CER6 promoter, CER6/CUT1 over-expression led to increased wax load in stems of Arabidopsis (Hooker et al. 2002). The only report on increased wax accumulation in leaf tissues of Arabidopsis was on the over-expression of AP2/EREBP transcriptional activator (Aharoni et al. 2004; Broun et al. 2004). Over-expression of WXP1 under the control of the CaMV35S promoter led to increased cuticular wax loading on the leaf surfaces, reduced water loss, and enhanced drought tolerance of transgenic alfalfa (Zhang et al. 2005). Transgenic expression of WXP1 or of its paralog WXP2 in Arabidopsis also leads to increased wax deposition and enhanced drought tolerance (Zhang et al. 2007).
Rice is one of the most important cereals and has become a model plant of monocot species for functional genomics studies. However, very few genes involved in leaf wax accumulation and drought resistance have been identified in rice. Identification of a T-DNA insertion mutant of the Wax-deficient anther1 (Wda1) gene in rice revealed a significant defect in the biosynthesis of VLC fatty acids in both layers (Jung et al. 2006). Recently, a cuticle-deficient rice mutant with T-DNA inserted in the Wax Crystal-sparse Leaf1 (WSL1) gene was identified (Yu et al. 2008). This mutant displayed pleiotropic phenotypes including reduced growth, leaf fusion, sparse crystals, enhanced sensitivity to drought, and low fertility. In this study, we report a systematic analysis of the Glossy 1-like gene family in rice (OsGL1) for sequence variation and expression profiles under normal and stress conditions. Further, we provide evidence for the function of this gene family in cuticular wax accumulation and drought resistance by checking the phenotypes of the over-expression and mutant plants of the OsGL1-2 gene.
Materials and methods
Identification and sequence analysis of GL1-homologous genes in rice
The reported GL1 protein sequence was used as a query to do BLAST search against the TIGR database (http://tigrblast.tigr.org/euk-blast/index.cgi?project=osa1) to identify putative OsGL1-like genes in the rice genome. The reported GL1 protein also was used to search as queries against the TAIR database (http://www.arabidopsis.org/index.jsp) and the NCBI (http://www.ncbi.nlm.nih.gov/blast/) to find a putative GL1 homolog in Arabidopsis and other species, respectively. All putative protein sequences were manually checked for a common fatty acid hydroxylase (FAH) domain (accession no. PF04116) using the PFAM program (http://www.sanger.ac.uk/Software/Pfam/) to confirm these sequences as members of GL1-homologous gene family.
The basic information of the protein sequences such as molecular weight and theoretical isoelectric point (PI) were obtained by the EXPASY PROTOPARAM tool (http://www.expasy.org/tools/protoparam.html). Protein sequences of all species were analyzed by the MEME program (http://www.meme.sdsc.edu/meme/meme.html) to predict the potential motifs. The full-length cDNAs or predicted coding sequences and the genomic sequences of GL1-like genes in rice were compared to predict the position of exons and introns by using Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/). The positions of domains in different GL1-like genes were also detected using this program.
Multiple sequence alignment of the GL1-like gene family was performed by ClustalX 1.81 with default settings (Thompson et al. 1997). The phylogenetic tree was constructed using PHYLIP software. The neighbor joining method was used to calculate the phylogenetic tree with 1,000 bootstrap replicates. The GENEDOC (Nicholas et al. 1997) and TreeView (Page 1996) programs were used for editing alignment and drawing the phylogenetic tree, respectively.
Plant growth and treatments
For detecting the transcript levels of OsGL1 genes, rice plants of Minghui 63 (Oryza sativa L. spp. indica) were grown in the greenhouse with a 14-h light/10-h dark cycle. Two-week-old seedlings were treated with abiotic and chemical stresses. Abiotic treatments were conducted according to Saijo et al. (2000). Drought stress was applied by exposing intact plants in the air without water supply and plant leaves were sampled at 0, 6, 24, and 48 h after treatment. For cold stress, seedlings were transferred into a growth chamber at 4°C and sampled at 0, 2, 4, 8, 12, and 24 h. The seedlings were submersed in a 200-mM NaCl solution for salt stress and sampled at 0, 6, 24, and 48 h after treatment. Chemical treatment was conducted by spraying leaves with 0.1 mM abscisic acid (ABA) and sampled at 0, 1, 3, and 12 h.
Transcript level analysis of GL1-like gene family in rice
The expression profile of the OsGL1-like gene family was pre-checked by looking up the gene chip expression profiling data of rice for more than 30 different tissues and organs of an elite hybrid rice parent Minghui 63 (Wang et al., unpublished data). In this study, 13 representative tissues and organs of Minghui 63 were selected for reverse transcriptase-polymerase chain reaction (RT-PCR) analysis to confirm the expression profiles. Total RNA was extracted by using TRIzol reagent (Invitrogen), according to the manufacturer’s instructions. Before reverse transcription, total RNA was treated with amplification-grade DNase I (Invitrogen) for 15 min to degrade possibly contaminated residual genomic DNA. The first-strand cDNA was synthesized by Super Script II reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. The PCR was performed in a volume of 50 μl with rTaq DNA polymerase (Takara) and about 1/20 of the first-strand cDNA template generated from 1 μg total RNA. PCR was performed with the following cycling profile: 94°C for 3 min, 25–30 cycles at 94°C for 40 s, 55–60°C for 40 s, and 72°C for 1 min. Rice Actin1 gene (accession no. X16280) was used as an internal control for RT-PCR. For each OsGL1-like gene, a pair of primers with an amplicon of 350–600 bp was used for RT-PCR (Supplementary Table 1). All RT-PCRs were repeated three times with independently reverse-transcribed templates.
Real-time PCR was performed in an optical 96-well plate with an ABI PRISM 7500 instrument (Applied Biosystems). The primers for real-time PCR were designed by Primer Expression Version 2.0 (Applied Biosystems; Supplementary Table 1). Each reaction contained 12.5 μl of 2× SYBR® Premix Ex Tag™ (Takara), 0.5 μl of 50× Rox Reference Dye II (Takara), 4 μl of cDNA samples and 200 nM gene-specific primers in a final volume of 25 μl. The thermal cycles used were as follows: 95°C for 10 s, 45 cycles of 95°C for 5 s, and 60°C for 34 s. Dissociation curve analysis was performed as follows: 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s. The rice Actin1 gene was used as an endogenous control. The relative expression levels were determined as described previously (Liang et al. 2006).
Generation of over-expression and mutant plants of OsGL1-2 gene
Based on the full-length cDNA sequences of GL1-like genes of rice in Knowledge-based Oryza Molecular biological Encyclopedia (KOME) database (http://cdna01.dna.affrc.go.jp/cDNA/), the full-length cDNA of OsGL1-2 was isolated from Minghui 63 by gene-specific primers (Supplementary Table 1) and cloned into pCAMBIA1301S under the control of Cauliflower mosaic virus 35S promoter. The construct was introduced into Agrobacterium tumefaciens strain EHA105 by electroporation and transformed into rice Zhonghua11 (Oryza sativa L. subsp. japonica) following the standard rice transformation protocol (Hiei et al. 1994). Expression of the transgene was checked by northern blot analysis. Total RNA (15 μg) was loaded in each lane of 1.2% agarose gels with formaldehyde for the northern blot analysis. Hybridization was performed with 32P-labeled gene-specific cDNA and results were detected by autoradiography.
To obtain rice mutants of GL1-like genes, the genomic sequences of OsGL1 genes were used to search our rice mutant database (RMD, http://RMD.ncpgr.cn/). Currently, one mutant is available for the OsGL1-2 gene. Genomic DNA was extracted from the young leaves of the osgl1-2 mutant plants, and PCR was performed to genotype the mutant plants by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.30 min with the primers A (gene-specific forward primer), B (gene-specific reverse primer), and C (NTLB5, T-DNA border specific primer) (Supplementary Table 1).
Phenotyping and measurements of transgenic and mutant plants
Transgenic seeds of T1 families were germinated on MS medium containing 50 mg/l hygromycin for stress testing. At the same time mutant and wild-type (WT) seeds were also germinated. Drought resistance at the reproductive stage was evaluated under drought stress conditions in the refined paddy field (sand:paddy soil = 1:3) facilitated with a movable rain-off shelter. For this experiment, positive transgenic and mutant lines (16 plants each in two rows) were planted along with the WT as a control to follow the randomized complete block design with three replications.
For chlorophyll leaching assay, the third leaf from the top was sampled from each tiller at heading stage plants and the leaf was cut into segments (about 3 cm) and immersed in 30 ml of 80% ethanol at room temperature (gently agitating in the dark). At 1, 3, 6, and 9 h, 3 ml of aliquot were taken out for chlorophyll quantification and the aliquot was poured back to the same tube after measurement. The chlorophyll concentration was quantified using a UNICO 7202B spectrophotometer at wavelengths of 663 and 645 nm by using the standard method (Lolle et al. 1997).
Scanning electron microscopy (SEM) was used to study both abaxial and adaxial surfaces of matured leaves of OsGL1-2 over-expression, mutant, and WT plants. Fragments of flag leaves were fixed in 5% glutaraldehyde and mounted on stubs. Samples were coated with 15–20 Ǻ grain-size gold particles for 15 min by using an EICO IB.5 ION coater. Coated samples were transferred to a JEOL JSM-3690LV scanning electron microscope for examination. For transmission electron microscopy (TEM) analysis, leaf segments (1 × 5 mm) were cut from the leaf (at 3-leaf stage) of OsGL1-2 over-expression, mutant, and WT plants. Samples were processed as described previously (Sturaro et al. 2005). Ultrathin sections were prepared using a Leica UC6 ultramicrotome. Sections were viewed in an H-7650 transmission electron microscope (Hitachi, Japan) operated at 80.0 kV in the public laboratory of Huazhong Agricultural University, PR China.
Wax extraction and quantification
Six leaf blades (each about 10 cm in length) from each plant were immersed in 30 ml chloroform for 30 s at room temperature. The same leaf blades were then re-extracted with chloroform at 60°C for 20 s, and the two chloroform extracts containing wax were pooled. Chloroform was evaporated by a nitrogen evaporator (Dry N-EVAP™111, Organomation Associates Inc. USA). Five microgram of n-tetracosane (C24) was added to each sample as an internal standard. After the samples were transferred to GC vials, 10 μl N,O-bis(trimethylsilyl)-trifluoroacetamide (BSTFA) and 10 μl pyridine were added and the samples were heated at 70°C for 1 h. BSTFA and pyridine were then removed under the stream of nitrogen. With 100 μl chloroform added, the samples were loaded for gas chromatography–mass spectrometry (GC–MS) analysis.
Wax components were separated using a 30 m × 0.32 mm HP-1 capillary column and helium as the carrying gas with constant flow of 2 ml/m in a GC–MS-QP2010 (Plus) gas chromatograph (SHIMADZU, Japan). GC was carried out with on-column injection temperature at 250°C, oven temperature of 2 min at 50°C, increasing at 40°C/min to 200°C, 2 min at 200°C, increasing at 3°C/min to 280°C, 20 min at 280°C. Wax composition was determined by comparing peak retention times with those of reference standards, and by a GC–MS analysis of representative samples. Wax loads were estimated by quantifying the areas of major peaks in comparison with the internal standard. Wax load per unit leaf area was calculated based on the area of leaves used for wax extraction.
Results
Identification of GL1 homologous genes in rice
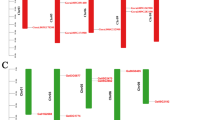
Using the reported GL1 protein sequence as a query to search against the annotated rice database, 11 putative GL1-like genes were found in the rice genome. These genes, designed OsGL1-1 ~ OsGL1-11, are distributed on chromosomes 2, 4, 6, 7, 10, and 11 (Table 1). In the KOME database, full-length cDNAs are available for all these genes except OsGL1-7. Almost all the rice GL1-like protein sequences contain the FAH domain (PF04116) except OsGL1-7 (Fig. 1a). Comparison of the full-length cDNAs or predicted coding sequences with the genomic sequences of OsGL1 genes revealed large variation in the number of exons among these genes (Fig. 1a; Table 1). We also noticed that the coding region of the FAH domain is interrupted by introns for the all genes except OsGL1-9, in which the entire FAH domain is located in the second exon. According to the PFAM database (http://pfam.sanger.ac.uk/family?acc=PF04116), the members of the FAH superfamily contain two copies of the HXXHH motif. Alignment of the FAH domain sequences suggested all the OsGL1 proteins have two copies of HXXHH motif and an additional His-rich motif (HXALH) (Supplementary Figure 1). By using the multiple EM for motifs elicitation (MEME) software, six putative motifs were identified in the GL1-like protein sequences (Fig. 1b; Table 2), and three of them (motif 1, 2, and 5) contain the His-rich residues. Although the putative functions of these motifs remain unknown, the motif composition patterns are useful clues for deducing the functions of the GL-like proteins. For example, a typical GL-like protein contains all six motifs and its FAH domain contains motifs 1, 2, 4, and 5. Four rice GL-like proteins (OsGL1-8, OsGL1-9, OsGL1-10, and OsGL1-11) only have a nontypical FAH domain with only two His-rich motifs. The sequence of OsGL1-7 is different because it has only two motifs (motif 3 and 6), which constitute the C-terminal region of a typical GL-like protein.
Genomic and structural organization GL1-like genes in rice. a Exon–intron structure of OsGL1-like genes. The line indicates introns, and rectangles indicate exons (untranslated regions [UTR] in gray, open reading frames [ORF] in white or black, and the FAH domains in black). b Six putative motifs in GL1-like gene family identified by MEME. Numbered boxes represent different putative motifs (annotation are listed in Table 2)
Phylogenetic analysis of GL1-like gene family
For phylogenetic analysis of the GL1-like gene family in plants, all available GL1-like sequences in rice, Arabidopsis and other plant species (such as maize, Medicago, Vitis vinifera, and Senecio odorus) were picked up based on BLASTP search using GL1 as the query. A total of 28 GL1 homologs were used to construct the phylogenetic tree. The results showed that these sequences can be classified into three groups: group I containing the GL1 protein, group II containing CER1 protein, and group III containing SUR2 and hydroxylase/desaturase protein (Fig. 2). Group I and group II each contain GL-like sequences from different species, but group III consists of genes only from rice. OsGL1-1, -2, and -3 were classified into group I and they have high sequence similarity (>60%) to the GL1 protein. OsGL1-4, -5, -6, and -7 were classified into group II and they have low sequence similarity (30% ~ 40%). OsGL1-8, -9, -10, and -11 were classified into group III and had very low sequence similarity to GL1. We also noticed that most of the GL-like sequences from monocot species are closely related in the tree. The phylogenetic result may provide clues for further deducing the putative functions of unknown GL-like sequences of rice.
Phylogenetic relationship of GL1-like gene family from diverse species based on protein sequences. The number at branching sites indicated the posterior probability values for nodal support. The unrooted tree was constructed using PHYLIP software to follow the neighbor joining method. The abbreviations of diverse species: AT, Arabidopsis thaliana; Mt, Medicago truncatula; Os, Oryza sativa; S. odorus, Senecio odorus; V. vinifera, Vitis vinifera; Z. mays, Zea mays
Expression levels of OsGL1-like family genes in different tissues and organs
RT-PCR was performed to detect the expression levels of OsGL1 genes in 13 tissues or organs (Fig. 3). OsGL1-1 had a relatively high expression level in germinating seeds (72 h after imbibitions), embryo and radical (after germination), and leaf (4–5 cm young panicle). For the OsGL1-2 gene, a relatively high expression was detected in germinating seeds (72 h after imbibitions), radical (48 h after emergence in dark), and leaf (4–5 cm young panicle). Relatively high expression was detected in germinating seeds (72 h after imbibitions) and stamens (1 day before flowering) for the OsGL1-3 gene. The expression of OsGL1-5 was detected in panicle (young panicle at stage 3), stamen (1 day before flowering), and hull (1 day before flowering). The OsGL1-6 gene showed high expression in germinating seeds (72 h after imbibitions) and shoot (seedling with two tillers). The genes OsGL1-8, -9, -10, and -11 were expressed in most of the tissues and organs investigated. On the other hand, the genes OsGL1-4 and OsGL1-7 had no or very faint expression in all the samples. According to the expression profiles, the genes in the OsGL1 family can be classified into two groups: tissue- or organ-specific expressed genes (OsGL1-1–7) that were expressed in very few tissue/organs and constitutively expressed genes (OsGL1-8–11) that were expressed in most of the tissue/organs. We searched the center of rice expression profiling (CREP) database that was established based on gene chip expression profiles of more than 30 tissues and organs (Wang et al., unpublished data) to check the relative expression levels of the OsGL1 gene family (Supplementary Figure 2). In general, the results from the gene chip analysis matched well with the RT-PCR results.
Semi-quantitative RT-PCR of OsGL1 genes in 13 different tissues and organs Tissues or organs: 1, seed (72 h after imbibition); 2, embryo and radicle (after germination); 3, radicle (48 h after emergence, dark); 4, callus (screening stage); 5, sheath (4–5-cm young panicle); 6, stem (heading stage); 7, panicle (young panicle at stage); 8, flag leaf (5 day before heading); 9, leaf (4–5-cm young panicle); 10, stamen (1 day before flowering); 11, root (seedling with two tillers); 12, shoot (seedling with two tillers); 13, hull (1 day before flowering). Numbers in brackets indicate the number of PCR cycles
Responsiveness of OsGL1 genes to abiotic stresses
To elucidate the roles of OsGL1 gene family in stress resistance, we first investigated the transcript levels of OsGL1 genes in rice plants under drought, cold, and salt stresses. Preliminary analysis suggested that six OsGL1 genes were responsive to drought or salt stress in microarray analysis (Zhou et al. 2007). These six genes were selected to test their responsiveness to drought, salt, cold, and ABA (see “Materials and methods” for details) in rice Minghui 63 by real-time PCR analysis. All six OsGL1 genes were responsive to more than one stresses (Fig. 4). Among them, three genes (OsGL1-1, -2 and -6) were induced by drought. The expression levels of OsGL1-1 and OsGL1-2 were gradually increased throughout the time course, whereas the OsGL1-6 showed strong induction only at 2 day after treatment. OsGL1-1, -3, -5, and -6 were induced by salt stress, and the expression levels of these genes except OsGL1-6 continued to increase throughout the time course. The expression level of OsGL1-6 peaked at 6 h after treatment and then decreased. With cold treatment, OsGL1-1 and OsGL1-4 showed strong induction at both 12 and 24 h after treatment, whereas OsGL1-6 showed high expression at 12 h but dramatically decreased expression at 24 h. Four genes (OsGL1-2, -3, -4, and -5) were induced by ABA treatment, and the highest induction was at 12 h after the treatment for all four genes.
Altered drought resistance and cuticular permeability in OsGL1-2 over-expression and mutant rice plants
For functional analysis of GL1-like genes in rice, we searched the T-DNA insertion mutant database (http://rmd.ncpgr.cn/) and three mutants (04Z11ML82, 04Z11ML83, and 05Z11AS53) were found only for the gene OsGL1-2 based on the flanking sequence tags of the mutants. Based on PCR analysis with two genomic primers flanking the insertion sites and a T-DNA vector primer (Fig. 5a), the insertion site was confirmed to be right for the mutant 04Z11ML82 (Fig. 5b) but not for 04Z11ML83 and 05Z11AS53. RT-PCR analysis showed that the transcript of OsGL1-2 gene was broken down in the 04Z11ML82 mutant (Fig. 5c). To study the function of the OsGL1-2 gene, the full-length cDNA of OsGL1-2 under the control of CaMV 35S promoter was transformed into japonica cultivar Zhonghua 11 for examining the phenotypes of wax accumulation and drought resistance along with the T-DNA mutant. Among 23 independent transgenic plants generated, nine transgenic plants showed high expression level of the transgene (Fig. 5d).
Altered drought resistance of the OsGL1-2 mutant. a Schematic gene structure of OsGL1-2 with a T-DNA insertion. b PCR for ten mutant plants. A, forward primer generated from the upstream region of OsGL1-2 gene; B, reverse primer generated from the coding region of the OsGL1-2 gene; C, reverse primer from the T-DNA region. H, homozygous; h, heterozygous; WT, wild-type; CK, control. Lanes 1, 2, and 7 are heterozygous; lanes 3, 6, 8, 9, and 10 are homozygous; lanes 4 and 5 are WT; lane 11 is a control. c Semi-quantitative RT-PCR of the OsGL1-2 transcript level in the osgl1-2 mutant compared to the WT. The expression level of Actin1 was used as an internal control. d Northern blot analysis to check the positive OsGL1-2 over-expression plants. CK, WT. e Appearance of mutant plants (two rows in middle side) under drought stress at reproductive stage compared to WT (both sides). f Mutant leaves became rolled (right side) compared to the non-rolled leaves of WT (left side) after drought stress
The most important physiologic function of the cuticle is to protect the plant from water loss. Therefore, we first carried out drought resistance testing for over-expression and mutant plants. Three independent over-expression transgenic families (S8, S9, and S15) and homozygous mutant plants were selected for drought resistance testing in the field (see “Materials and methods” for details). During the process of stress at the reproductive stage, the mutant showed obviously earlier leaf-rolling than WT (Fig. 5e–f). However, we did not observe obvious difference in leaf-rolling time between the over-expression and WT plants. We then checked the cuticular permeability of the over-expression and mutant plants by using the chlorophyll leaching method. Results showed that chlorophyll leaching from the over-expression plant leaves was significantly (t test, P < 0.01) slower than that from the WT leaves, whereas chlorophyll leaching from the mutant leaves was significantly (t test, P < 0.01) faster (Fig. 6a), indicating a decrease and increase of cuticular permeability in the over-expression and mutant plants, respectively. The drought resistance phenotype was also evaluated by measuring water loss in detached leaves over a period of time. Compared to WT, the detached leaves of the OsGL1-2 over-expression plants lost water slowly and the mutant lost water rapidly (Fig. 6b). Although the difference between the over-expression plants and WT or between the mutant and WT was not statistically significant, the difference between the over-expression plants and the mutant reached very significant level (t test, P < 0.01). These results suggest that the OsGL1-2 gene has a significant role in controlling the wax layer and thus protecting leaves from water loss in rice.
Altered cuticular permeability of the OsGL1-2 mutant and over-expression plants. a Chlorophyll leaching assays with matured leaves of OsGL1-2, mutant, and WT, immersed in 80% ethanol for different time intervals. The results are derived from three independent experiments and depicted with standard error of the mean from each time point. One asterisk indicates significant at P < 0.05 level and two asterisks indicate significant at P < 0.01 level by the t test. b Water loss rate of detached leaves of the over-expression, mutant and WT plants. The x-axis is the scale for different time points and the y-axis is percent of free water loss from leaves. Data is shown by mean ± SE with three replicates
Altered wax accumulation in the OsGL1-2 over-expression and mutant rice plants
We further checked the wax accumulation on the leaf surfaces of the over-expression and homozygous mutant plants along with the WT by SEM and TEM. Both adaxial and abaxial surfaces of the matured leaves of the over-expression plants were covered with a dense layer of wax crystals under SEM (Fig. 7a, b). Compared to the over-expression plant, fewer wax crystals were observed on both of the surfaces in the mutant (Fig. 7c, d) and WT (Fig. 7e, f). Ultrastructural analysis of the leaf cuticle by TEM indicated that the leaf cuticle membrane of the over-expression plant appears to be divided into an outermost and translucent layer (cuticle layer) and an innermost and opaque layer (reticulated cuticle layer) (Fig. 7g). Compared to WT (Fig. 7i), the cuticle membrane thickness of cuticle layer was increased the over-expression plants (Fig. 7g) and reduced in the mutant (Fig. 7h).
Electron microscopic analysis of the OsGL1-2-overexpression plant, mutant, and WT. a adaxial surface of the OsGL1-2-overexpression plant (OX), b abaxial surface of OX c adaxial surface of the mutant, d abaxial surface of the mutant, e adaxial surface of WT, f abaxial surface of WT. The scale of SEM images in a-f is 5 μm with images taken at ×5,000. g–i images of leaf epidermal cell sections for the OX (g), mutant (h), and WT (i) plants (images were taken at ×21,000). CUT, cuticle; CW, cell wall
Wax quantification analysis revealed significant difference in cuticular wax content among the over-expression, mutant and WT plants (Fig. 8). The total proportions of alkanes and fatty acids were significantly higher (t test, P < 0.01) in the over-expression plant leaves than in WT or mutant leaves (Fig. 8a). The total proportions of aldehydes and fatty acids in the mutant leaves were significantly less (P < 0.05) than that in WT (Fig. 8a). Contents of other wax constituents (alkanes and alcohol) were also reduced, but not statistically significant compared to WT. Significant decrease of individual wax constituent was detected for aldehyde C22 (P < 0.05) and fatty acid precursors C18 (P < 0.05) and C20 (P < 0.01) in the mutant compared to WT (Fig. 8b). In general, all the wax constituents detected showed increased levels in the over-expression plants and decreased levels in the mutant compared to WT, though some of them did not reach significant levels.
Quantification of cuticular wax composition in leaves of the OX, mutant and WT plants. a The coverage of total wax constituents including alkane, aldehyde, alcohol, and fatty acid. b The coverage of representative individual precursors of cuticular wax. Values are given as mean ± SE with three replicates. One and two asterisks indicate significance at P < 0.05 and P < 0.01 level, respectively, by t test between the mutant (or overexpression line) and WT
Discussion
A common feature of GL1-like proteins is the presence of eight conserved His-rich motifs in the tripartite domain H–X2–4–H, H–X2–3–H–H, (H/Q)–X2–3–H–H (where X stands for any amino acid), which form a di-iron-binding site essential for catalytic activity in a large family of integral membrane enzymes, such as acyl desaturases, alkyl hydroxylases, epoxydases, acetylenases, methyl oxidases, ketolases, and decarbonylases, activities found in prokaryotes and eukaryotes (Shanklin and Cahoon 1998). The positions of the first two His-rich motifs are close to the third motif. One reason for some variation in the spacing between the first two and the third His-rich motifs among protein families is that the structure of this protein part, rather than the spacing between the motifs, is important for the function of the proteins (Aarts et al. 1995). All the proteins with His-rich motifs described so far were found to contain long hydrophobic domains between the first two and the third His-rich motifs (Shanklin et al. 1994). These domains are able to span a membrane twice (Aarts et al. 1995). Our analysis (Supplementary Figure 1) suggested that all the members from different species were from the same ancestor with His-rich motifs (Chen et al. 2003; Kurata et al. 2003).
GL1 shows high sequence similarity with OsGL1-1 (76%), OsGL1-2 (62%), and OsGL1-3 (62%), suggesting that the OsGL1-1 ~ 3 may be homologs of GL1. OsGL1-4, OsGL1-5, and OsGL1-7 are very similar to CER1 (AT1g02205) (Fig. 2) indicating that they may be CER1 homologues in rice. The best-characterized cuticle-related gene in grasses that shows considerable homology (34–35%) to the barley CER1 is maize Glossy 1 (GL1; Hansen et al. 1997; Sturaro et al. 2005). Glossy 1 is the maize homologue of Arabidopsis CER1. The degree of similarity between the putative GL1 and Arabidopsis (AT5G57800) WAX2 proteins is 62%. Our result also shows that WAX2 proteins have close relation to OsGL1-1 ~ 3 proteins (Fig. 2). The WAX2 proteins have a 32% similarity to CER1 and contain certain regions with the homology to sterol desaturases and short-chain dehydrogenases/reductases (Aharoni et al. 2004). It was suggested that these enzymes play a metabolic role in both cutin and wax synthesis and therefore point to a link between cutin and wax metabolism.
We investigated the transcription levels of all OsGL1 genes in 13 different tissues or organs. The results revealed diverse expression patterns of the GL1-like genes in rice. Most of the gene expressed in leaf (4–5-cm panicle stage) but not in root (Fig. 3). Sturaro et al. (2005) also detected a similar expression pattern of the GL1-like genes in maize. OsGL1-1 ~ 3 showed high expression levels in seeds. However, Xu et al. (1997) reported a broad expression pattern of Glossy 8 in maize roots and seed. Kunst and Samuels (2003) reported the close relationship between Glossy 1 and Glossy 8. Most of the OsGL1-like genes were expressed in different panicle stages in rice, suggesting that some of these genes might be involved in the wax biosynthesis in panicle.
As pointed out by Jenks et al. (2002), alternation of wax accumulation in crop leaf tissues is an important physiologic process because leaves are the primary photosynthetic organs, comprise the primary biomass of most agronomical crops, and are often severely affected by environmental stress. To start elucidating the molecular events underlying stress-induced wax biosynthesis, mRNA steady-state levels of wax-related genes were assessed under drought, cold, and salt conditions. These analyses pointed out that in maize and Arabidopsis only a few structural genes involved in key steps of the wax biosynthetic pathway are regulated under stress (Gattuso et al. 2007). Transcript level changes of six OsGL1 genes in different stress treatments clearly suggest that different OsGL1 genes have different responses to stresses. All six genes were induced by more than one stress. In this study most of the drought- or salt stress-inducible OsGL1 genes were also induced by ABA (Fig. 4). The application of ABA to plants partially mimics the effect of stress conditions (Nambara and Marion-Poll 2005). These results indicate that OsGL1 genes may be regulated by substantially diverse regulatory systems under different stresses.
Genetic and mutant studies have suggested that wax accumulation is a potential drought adaptation trait (Jefferson 1994). The osgl1-2 mutant showed rapid water loss and decreased epidermal permeability compared to the OsGL1-2 over-expression and WT plants, suggesting that the mutant plants may be susceptible to drought. The faster leaf-rolling phenotype of the mutant compared to WT under drought stress in the field conditions further confirmed this speculation. In this drought testing, no obvious difference in leaf-rolling time was observed between the over-expression and WT plants, which may due to the acute drought stress (accompanied with high temperature) in that season that had caused a very narrow window of time to observe the difference in the field. Another piece of evidence is that OsGL1-2 over-expression and mutant plants were altered in the cuticular membrane as indicated by the chlorophyll leaching experiment. The OsGL1-2-overexpresion plants showed a significant decrease of chlorophyll leaching and the osgl1-2 mutant plants showed a significant increase of chlorophyll leaching. This result indicates that over-expression of OsGL1-2 genes decreased the cuticular permeability and perhaps can cause drought tolerance of the plants. Chen et al. (2003) reported similar results in the wax2 mutant of Arabidopsis.
Mutation of GL1 causes dramatic alternations in the crystallization patterns of cuticular waxes (Bianchi et al. 1985; Chen et al. 2003; Jenks et al. 1995; Lorenzoni and Salamini 1975). The crystallization patterns of cuticular waxes on matured leaves of the osgl1-2 mutant were significantly different and reduced compared to OsGL1-2 over-expression and WT plants (Fig. 7). The content of total wax and some of the wax constituents were significantly decreased in the mutant leaves compared to WT, indicating that mutation of this gene may block the elongation–decarboxylation pathway I (ED-I) or reduce accumulation of alkanes, aldehydes, and alcohols. Maddaloni et al. (1990) reported that gl1 mutation either blocks an early step in ED-I or interferes with the supply of precursors to ED-I in maize. This mutant has a dramatic reduction of alkanes, aldehydes, and alcohols, but the level of esters is similar to WT. The gl1 mutant, however, does not show an accumulation of metabolic intermediates due to a specific block in the pathway. Instead, there seems to be an increased flux of precursors into an alternative branch pathway, resulting in more complex changes in wax composition (Lemieux et al. 1994; von Wettstein-Knowles 1979).
According to the biosynthesis model of cuticular wax, the early precursors are short acyl chains activated by a soluble plastidic acyl carrier protein (ACP). The acyl chains are elongated by plastidic fatty acid synthetase (FAS) complex which condenses acetal groups from malonyl-ACP onto a growing chain (Ohlrogge and Jaworski 1997). Once C16 and C18 acyl-ACPs are synthesized in the plastids, acyl-ACP thioesterase cleaves the ACP and releases free C16 and C18 fatty acids (palmitic and stearic acids) into the cytoplasm, where they are activated by acyl-coenzyme A (CoA) synthetase via condensation with CoA. The C16 and C18 acyl-CoAs are recruited for conversion into waxes mainly via a membrane-associated pathway where they are modified by a series of cytoplasmic enzyme complexes. Our data showed that the fatty acid precursors C18 and C20 were significantly decreased in the osgl1-2 mutant compared to WT and the OsGL1-2-overexpression plants. This result suggests that OsGL1-2 may play a critical role in the biosynthesis of cuticular wax of leaf in rice.
In conclusion, the GL1-like genes in rice were systematically analyzed for their sequence and expression diversification in this study. Over-expression of one of these genes, OsGL1-2, resulted in an increased cuticular wax accumulation on the leaf surface of the transgenic rice, and the mutant of this gene showed reduced cuticular wax synthesis and significantly increased sensitivity to drought stress. Genetic modification of this gene may have great potential for improving drought resistance of rice.
Abbreviations
- FA:
-
Fatty acid
- GC–MS:
-
Gas chromatography–mass spectrometry
- GL1:
-
Glossy 1
- PCR:
-
Polymerase chain reaction
- RT:
-
Reverse transcriptase
- SEM:
-
Scanning electron microscopy
- TEM:
-
Transmission electron microscopy
- VLCFA:
-
Very long chain fatty acid
References
Aarts MGM, Keijzer CJ, Stiekema WJ et al (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7:2115–2127
Aharoni A, Dixit S, Jetter R et al (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16:2463–2480. doi:10.1105/tpc.104.022897
Barnes J, Percy K, Paul N et al (1996) The influence of UV-B radiation on the physiochemical nature of tobacco (Nicotiana tabacum L.) leaf surface. J Exp Bot 47:99–109. doi:10.1093/jxb/47.1.99
Barthlott W, Neinhuis C (1997) Purity of the sacred lotus, or escape from the contamination in biological science. Planta 202:1–8. doi:10.1007/s004250050096
Bianchi A, Bianchi G, Avato P et al (1985) Biosynthetic pathways of epicuticular wax of maize as assessed by mutation, light, plant age and inhibitor studies. Maydica 30:179–198
Broun P, Poindexter P, Osborne E et al (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 10:4706–4711. doi:10.1073/pnas.0305574101
Chen XB, Goodwin SM, Boroff VL et al (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15:1170–1185. doi:10.1105/tpc.010926
Eigenbrode SD (1996) Plant surface waxes and insect behaviour. In: Kerstiens G (ed) Plant cuticles-an integrated functional approach. BIOS Scientific Publishers Limited, Oxford, pp 201–222
Fiebig A, Mayfield JA, Miley NL et al (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12:2001–2008
Gattuso M, Bonomi M, Tateo F, et al (2007) Stress induced modulation of wax biosynthesis in maize and Arabidopsis. Proceedings of the 51st Italian Society of Agricultural Genetics Annual Congress. Poster Abstract C.02. Riva del Garda, Italy
Hannoufa A, Negruk V, Eisner G et al (1996) The CER3 gene of Arabidopsis thaliana is expressed in leaves, stems, roots, flowers and apical meristems. Plant J 10:459–467. doi:10.1046/j.1365-313X.1996.10030459.x
Hansen JD, Pyee J, Xia Y et al (1997) The glossy1 locus of maize and an epidermis-specific cDNA from Kleinia odora define a class of receptor-like proteins required for the normal accumulation of cuticular waxes. Plant Physiol 113:1091–1100. doi:10.1104/pp.113.4.1091
Hiei Y, Ohta S, Komari T et al (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282. doi:10.1046/j.1365-313X.1994.6020271.x
Hooker TS, Millar AA, Kunst L (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129:1568–1580. doi:10.1104/pp.003707
James T, Post-Beittenmiller D, Jaworski JG (1999) KCS1 encodes and fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17:119–130. doi:10.1046/j.1365-313X.1999.00352.x
Jefferson PG (1994) Genetic variation for epicuticular wax production in Altai wild rye populations that differ in glaucousness. Crop Sci 34:367–371
Jenks MA, Joly RJ, Peters PJ et al (1994) Chemically induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol 105:1239–1245
Jenks MA, Tuttle HA, Eigenbrode SD et al (1995) Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol 108:369–377
Jenks MA, Eigenbrode SD, Lemieux B (2002) Cuticular waxes of Arabidopsis. In: Somerville CR, Meyerowitz EM (eds) The Arabidopsis book. American Society of Plant Biologists, Rockville, MD. doi:10.1199/tab.0016, http://www.aspb.org/publications/arabidopsis/
Jung KH, Han MJ, Lee DY et al (2006) Wax deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18:3015–3032. doi:10.1105/tpc.106.042044
Kerstiens G (1996a) Signaling across the divide: a wider perspective of cuticular structure–function relationships. Trends Plant Sci 1:125–129. doi:10.1016/S1360-1385(96)90007-2
Kerstiens G (1996b) Cuticular water permeability and its physiological significance. J Exp Bot 47:1813–1832. doi:10.1093/jxb/47.12.1813
Kerstiens G (2006) Water transport in plant cuticles: an update. J Exp Bot 57:2493–2499. doi:10.1093/jxb/erl017
Kolattukudy PE (1980) Cutin, suberin, and waxes. In: Stumpf PK, Conn EE (eds) The biochemistry of plants. Academic, New York, pp 571–645
Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42:51–80. doi:10.1016/S0163-7827(02)00045-0
Kurata T, Kawabata-Awai C, Sakuradani S et al (2003) The YORE–YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J 36:55–66. doi:10.1046/j.1365-313X.2003.01854.x
Lemieux B, Koornneef M, Feldmann KA (1994) Epicuticular waxes and eceriferum mutants. In: Meyerowitz EM, Somerville CR (eds) Arabidopsis. Cold Spring Harbor Press, New York, pp 1031–1047
Liang D, Wu C, Li C et al (2006) Establishment of a patterned GAL4-VP16 transactivation system for discovering gene function in rice. Plant J 46:1059–1072. doi:10.1111/j.1365-313X.2006.02747.x
Lolle SJ, Berlyn GP, Engstrom EM et al (1997) Developmental regulation of cell interactions in the Arabidopsis fiddlehead-1 mutant: a role for the epidermal cell wall and cuticle. Dev Biol 189:311–321. doi:10.1006/dbio.1997.8671
Lorenzoni C, Salamini F (1975) Glossy mutant of maize. V. morphology of the epicuticular waxes. Maydica 20:5–19
Maddaloni M, Bossinger G, DiFonzo N et al (1990) Unstable alleles of the GLOSSY-1 locus of maize show a light-dependent variation in the pattern of somatic reversion. Maydica 35:409–420
Millar AA, Clemens S, Zachgo S et al (1999) CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11:825–838
Moose S, Sisco P (1996) Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev 10:3018–3027. doi:10.1101/gad.10.23.3018
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185. doi:10.1146/annurev.arplant.56.032604.144046
Negruk V, Yang P, Subramanian M et al (1996) Molecular cloning and characterization of the CER2 gene of Arabidopsis thaliana. Plant J 9:137–145. doi:10.1046/j.1365-313X.1996.09020137.x
Nicholas KB, Nicholas HB Jr, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNET News 4:1–4
Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Ann Rev Plant Physiol Mol Biol 48:109–136. doi:10.1146/annurev.arplant.48.1.109
Page RDM (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Post-Beittenmiller D (1996) Biochemistry and molecular biology of wax production in plants. Annu Rev Plant Physiol Plant Mol Biol 47:405–430. doi:10.1146/annurev.arplant.47.1.405
Pruitt RE, Vielle-Calzada JP, Ploense SE et al (2000) FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc Natl Acad Sci USA 97:1311–1316. doi:10.1073/pnas.97.3.1311
Riederer M, Schreiber L (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52:2023–2032. doi:10.1093/jexbot/52.363.2023
Saijo Y, Hata S, Kyozuka J et al (2000) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327. doi:10.1046/j.1365-313x.2000.00787.x
Shanklin J, Cahoon EB (1998) Desaturation and related modifications of fatty acids. Annu Rev Plant Physiol Plant Mol Biol 49:611–641. doi:10.1146/annurev.arplant.49.1.611
Shanklin J, Whittle E, Fox BG (1994) Eight histidine-residues are catalytically essential in a membrane-associated iron enzyme, stearoyl-CoA desaturase, and are conserved in alkane hydroxylase and xylene monooxygenase. Biochemistry 33:12787–12794. doi:10.1021/bi00209a009
St-Pierre B, Laflamme P, Alarco AM et al (1998) The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J 14:703–713. doi:10.1046/j.1365-313x.1998.00174.x
Sturaro M, Hartings H, Schmelzer E et al (2005) Cloning and characterization of GLOSSY1, a maize gene involved in cuticle membrane and wax production. Plant Physiol 138:478–489. doi:10.1104/pp.104.058164
Tacke E, Korfhage C, Michel D et al (1995) Transposon tagging of the maize Glossy2 locus with the transposable element En/Spm. Plant J 8:907–917
Thompson JD, Gibson TJ, Plewniak F et al (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality aided analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Todd J, Post BD, Jaworski JG (1999) KCS1 encodes a fatty acid elongase 3-ketoacyl-CoA synthase affecting wax biosynthesis in Arabidopsis thaliana. Plant J 17:119–130. doi:10.1046/j.1365-313X.1999.00352.x
Vogg G, Fischer S, Leide J et al (2004) Tomato fruit cuticular waxes and the effects on transpiration barrier properties: functional characterization of a mutant deficient in a very-long-chain fatty acid b-ketoacyl- CoA synthase. J Exp Bot 55:1401–1410. doi:10.1093/jxb/erh149
von Wettstein-Knowles P (1979) Genetics and biosynthesis of plant epicuticular waxes. In: Appelqvist L-A, Liljenberg C (eds) Advances in the biochemistry and physiology of plant lipids. Elsevier North-Holland Biomedical Press, Amsterdam, pp 1–26
Walton TJ (1990) Waxes, cutin and suberin. In: Harwood JL, Bowyer JR (eds) Methods in plant biochemistry lipids, membranes and aspects of photobiology. Academic, San Diego, pp 105–158
Xia Y, Nikolau BJ, Schnable PS (1996) Cloning and characterization of CER2, an Arabidopsis gene that affects cuticular wax accumulation. Plant Cell 8:1291–1304
Xia Y, Nikolau BJ, Schnable PS (1997) Developmental and hormonal regulation of the Arabidopsis CER2 gene that codes for a nuclear-localized protein required for the normal accumulation of cuticular waxes. Plant Physiol 115:925–937. doi:10.1104/pp.115.3.925
Xiao FM, Goodwin SM, Xiao YM et al (2004) Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J 23:2903–2913. doi:10.1038/sj.emboj.7600290
Xu X, Dietrich CR, Delledonne M et al (1997) Sequence analysis of the cloned glossy8 gene of maize suggests that it may code for a [beta]-ketoacyl reductase required for the biosynthesis of cuticular waxes. Plant Physiol 115:501–510. doi:10.1104/pp.115.2.501
Yu D, Ranathunge K, Huang H et al (2008) Wax crystal-sparse leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228:675–685. doi:10.1007/s00425-008-0770-9
Zhang JY, Broeckling CD, Blancaflor EB et al (2005) Over-expression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42:689–707. doi:10.1111/j.1365-313X.2005.02405.x
Zhang JY, Broeckling CD, Sumner LW et al (2007) Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol 64:265–278. doi:10.1007/s11103-007-9150-2
Zhou J, Wang X, Jiao Y et al (2007) Global genome expression analysis of rice in response to drought and high salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63:591–608. doi:10.1007/s11103-006-9111-1
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China, the National Special Key Project of China on Functional Genomics of Major Plants and Animals, the National Program on the Development of Basic Research, and the Ministry of Education of China (No 707045).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Islam, M.A., Du, H., Ning, J. et al. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70, 443–456 (2009). https://doi.org/10.1007/s11103-009-9483-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9483-0