Abstract
Key message
CmWRKY17 was induced by salinity in chrysanthemum, and it might negatively regulate salt stress in transgenic plants as a transcriptional repressor.
Abstract
WRKY transcription factors play roles as positive or negative regulators in response to various stresses in plants. In this study, CmWRKY17 was isolated from chrysanthemum (Chrysanthemum morifolium). The gene encodes a 227-amino acid protein and belongs to the group II WRKY family, but has an atypical WRKY domain with the sequence WKKYGEK. Our data indicated that CmWRKY17 was localized to the nucleus in onion epidermal cells. CmWRKY17 showed no transcriptional activation in yeast; furthermore, luminescence assay clearly suggested that CmWRKY17 functions as a transcriptional repressor. DNA-binding assay showed that CmWRKY17 can bind to W-box. The expression of CmWRKY17 was induced by salinity in chrysanthemum, and a higher expression level was observed in the stem and leaf compared with that in the root, disk florets, and ray florets. Overexpression of CmWRKY17 in chrysanthemum and Arabidopsis increased the sensitivity to salinity stress. The activities of superoxide dismutase and peroxidase and proline content in the leaf were significantly lower in transgenic chrysanthemum than those in the wild type under salinity stress, whereas electrical conductivity was increased in transgenic plants. Expression of the stress-related genes AtRD29, AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 was reduced in the CmWRKY17 transgenic Arabidopsis compared with that in the wild-type Col-0. Collectively, these data suggest that CmWRKY17 may increase the salinity sensitivity in plants as a transcriptional repressor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The WRKY family is among the largest families of transcription factors in plants and is named after the highly conserved sequence motif WRKYGQK (Eulgem et al. 2000). Since Ishiguro and Nakamura isolated the first WRKY gene, SPF1, from Ipomoea batatas in 1994 (Ishiguro and Nakamura 1994), an increasing number of WRKY genes have been identified from a diverse variety of plants (Abbruscato et al. 2012; Atamian et al. 2012; Borrone et al. 2004; Guo et al. 2011; Zheng et al. 2011; Lagacé and Matton 2004; Li et al. 2013; Liu et al. 2012; Marè et al. 2004; Niu et al. 2012; Pan et al. 2009; Park et al. 2006). Over 70 and 100 members of the family are known in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively (Agarwal et al. 2011; Kalde et al. 2003; Wang et al. 2011). On the basis of the primary structure, WRKY proteins have been classified into three groups (I, II, and II) and various subgroups (Eulgem et al. 2000). Group II members contain a single C2H2 zinc finger with one conserved WRKY (WRKYGQK) domain, group III proteins contain one C2HC zinc finger and the conserved WRKY domain as in group II, whereas group I proteins contain two C2H2 zinc fingers and two WRKY domains. The WRKY proteins can specifically bind to the W-box [TTGAC(C/T)], a common cis-element in the promoters of target genes (Maeo et al. 2001).
The WRKY proteins play various roles as positive or negative regulators in response to biotic and abiotic stresses (Agarwal et al. 2011; Rushton et al. 2010). In Arabidopsis, overexpression of WRKY25 can improve the salinity tolerance of the transgenic plants (Jiang and Deyholos 2006), constitutive expression of WRKY33 enhances resistance to heat stress (Li et al. 2011), while WRKY8 plays a role in response to salt stress (Hu et al. 2013). GmWKRY21-transgenic Arabidopsis plants are tolerant to cold stress, and moreover GmWRKY54 confers salt and drought tolerance. However, overexpression of GmWRKY13 causes increased sensitivity to salt stress (Zhou et al. 2008). ZmWRKY33 (Li et al. 2013) and TaWRKY2/TaWRKY19 (Niu et al. 2012) are similarly induced by abiotic stresses, such as drought, salt, and abscisic acid (ABA) treatments. Transgenic Arabidopsis plants overexpressing TaWRKY2 exhibit enhanced salt and drought tolerance, whereas overexpression of TaWRKY19 confers increased tolerance to salt, drought, and freezing stresses. AtWRKY17 acts as a negative regulator of basal resistance to Pseudomonas syringae pv. Tomato (Journot-Catalino et al. 2006). OsWRKY28 (Chujo et al. 2013) from rice and CaWRKY58 (Wang et al. 2013) from Capsicum annuum negatively regulate resistance to the rice blast fungus Magnaporthe oryzae and Ralstonia solanacearum, respectively. NaWRKY3 and NaWRKY6 from the native tobacco Nicotiana attenuata can be significantly up-regulated in plants attacked by Manduca sexta (Hui et al. 2003). Besides abiotic and biotic stress response, WRKY proteins also play important roles in plant growth and development processes, such as seed coat development (Johnson et al. 2002), embryonic development (Lagacé and Matton 2004), lateral root growth (Zhang et al. 2008; Zhou et al. 2008), fruit ripening (Devaiah et al. 2007), and senescence (Hinderhofer and Zentgraf 2001; Ülker et al. 2007).
Chrysanthemum (Chrysanthemum morifolium), a widely grown ornamental plant, is sensitive to salinity, which induces extensive leaf chlorosis, retards growth, and in some cases even kills the plant. To our knowledge, little information on the isolation and functional identification of WRKY transcription factors in chrysanthemum is available. We have previously isolated 15 WRKY genes, which are induced by various stresses (Song et al. 2014a). Overexpression of DgWRKY3, which was isolated from chrysanthemum, increased salt stress tolerance in tobacco (Liu et al. 2013). Here, we identified a novel group II WRKY transcription factor, CmWRKY17, from chrysanthemum ‘Zhongshanzigui’, and showed that it was up-regulated by salinity stress. Overexpression of CmWRKY17 in chrysanthemum and Arabidopsis caused increased sensitivity to salinity stress compared with the wild-type (WT) plants.
Materials and methods
Plants materials and growth conditions
The chrysanthemum cultivars ‘Zhongshanzigui’ and ‘Jinba’ were obtained from the Chrysanthemum Germplasm Resource Conservation Centre, Nanjing Agricultural University, China. The plants were potted in a 1:1 (v/v) mixture of soil and vermiculite, and grown in a greenhouse. A. thaliana ecotype Columbia (Col-0) plants were grown in a 3:1 (v/v) mixture of vermiculite and soilrite in a growth chamber (SANYO, MLR-359H, Japan) with a 16-h light period (80–100 µmol/m2/s illumination) at 23 °C, followed by an 8-h dark period at 18 °C, with a relative humidity of 70 %.
Cloning of CmWRKY17 and sequence analyses
Total RNA was isolated from ‘Zhongshanzigui’ leaves using the RNAiso reagent (TaKaRa, Japan), following the manufacturer’s instructions, and then treated with RNase-free DNase I (TaKaRa, Japan) to remove potential genomic DNA contamination. The first-strand cDNA was synthesized with the SuperScript III reverse transcriptase (Invitrogen). On the basis of chrysanthemum expressed sequence tags (Chen et al. 2009), a primer pair (CmWRKY17-F/R) was designed to amplify a fragment of CmWRKY17, then random amplification of cDNA ends (RACE) PCR was used to obtain the full-length cDNA. For 3′ RACE, the first-strand cDNA was synthesized using an oligo (dT) primer incorporating the sequence of the adaptor primer, followed by a nested PCR using the primers CmWRKY17-3-F1/F2 and the adaptor primer dT-R (Table 1). For 5′ RACE, the primers AAP and AUAP, and specific primers (CmWRKY17-5-R1/R2) were used in the nested PCR with the 5′ RACE System Kit v2.0 (Invitrogen). The PCR products were purified with the Biospin Gel Extraction Kit (BioFlux, China) and cloned into the pMD19-T easy vector (TaKaRa) for sequencing. Consequently, a pair of primers (CmWRKY17-ORF-F/R) was designed to amplify the complete CmWRKY17 open reading frame (ORF) with the first-strand cDNA as the template.
The CmWRKY17 amino acid sequence was aligned with its homologs using the DNAman 5.2.2 software and BLAST software online (http://www.ncbi.nlm.gov/blast). A phylogenetic tree was constructed using the neighbor-joining method with BioEdit and MEGA 5.2.2.
Assay of CmWRKY17 expression in different tissues and under salinity stress
For salinity treatment, ‘Zhongshanzigui’ plants at the six- to eight-leaf stage were used. The plants were watered with 200 mM NaCl. Each experiment contained three biological replicates. Leaves of NaCl-treated plants were harvested after 0, 1, 2, 4, 8, 12, and 24 h, frozen in liquid nitrogen, and stored at −80 °C for RNA extraction.
Total RNA was extracted from different tissues (root, stem, leaf, disk florets, and ray florets) in chrysanthemum and salinity-stressed plants at the above-mentioned time points using the RNAiso reagent (TaKaRa, Japan) following the manufacturer’s instructions. The RNA was treated with RNase-free DNase I (TaKaRa) to remove genomic DNA contamination. The first cDNA strand was synthesized from 1 μg total RNA using M-MLV reverse transcriptase (TaKaRa) in accordance with the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) using SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa) was used to detect the expression of CmWRKY17. Each 20 μL qRT-PCR contained 10 μL SYBR Green PCR master mix, 0.4 μL of each primer (10 μM), 4.2 μL H2O, and 5 μL cDNA template. The PCR cycling regime comprised an initial denaturation (95 °C for 2 min), followed by 40 cycles of 95 °C for 10 s, 55 °C for 15 s, and 72 °C for 20 s. The resulting data were represented by the mean ± SD of three biological replicates. The primer pair CmWRKY17-RT-F/R was used and the CmEF1α gene (KF305681) acted as a reference sequence after sequencing of the amplified fragment by primer pairs. Relative transcript abundances were calculated using the 2−ΔΔCT method. The data were scaled by setting the expression of CmWRKY17 in untreated leaves at 0 h as 1.
Transcriptional activity analysis of CmWRKY17
CmWRKY17 was examined for transactivation activity firstly using a yeast assay system. The ORF of CmWRKY17 without the termination codon was amplified using the Phusion HighFidelity PCR Kit (New England Biolabs) with the primer pair CmWRKY17-BD-F/R (Table 1). The PCR products were subcloned into the pMD19-T vector (TaKaRa) and confirmed by sequencing. Both pMD19-T harboring CmWRKY17 and pGBKT7 were digested with EcoR I and BamH I, then the corresponding bands were recovered and ligated into pGBKT7 to yield the expression vector pGBKT7-CmWRKY17. The pGBKT7-CmWRKY17 construct, pCL1 (positive control), and pGBKT7 (negative control) were introduced into Saccharomyces cerevisiae strain Y2HGold (Clontech) following the manufacturer’s protocol. Selection for transformants carrying either pGBKT7-CmWRKY17 or pGBKT7 employed an SD/-Trp medium, whereas pCL1 was selected on SD/-Leu medium. As a positive control, Y2H cells carrying pCL1 can grow on SD/-His-Ade medium, whereas Y2H cells carrying pGBKT7, the negative control, cannot grow on the medium.
Then the luminescence assay of CmWRKY17 was taken to further understand its transcriptional activity. To generate 35S::GAL4DB-CmWRKY17 fusion construct, the CmWRKY17 ORF was amplified by PCR using the primer set CmWRKY17-GATE-F/R (Table 1) harboring the BamH I and Not I sites. The amplified DNA fragment was directly cloned into the pENTR™1A dual selection vector (Invitrogen) to obtain pENTR™1A-CmWRKY17 by sequencing. The plasmid was subjected to the vector 35S::GAL4DB to obtain the construction of 35S::GAL4DB-CmWRKY17 using the LR reaction (Invitrogen). Protoplasts of Arabidopsis were prepared and transfected based on the protocol as described by Yoo et al. (2007). For the transfection, 7.5 μg of 35S::GAL4DB-AtARF5, 35S::GAL4DB, or 35S::GAL4DB-CmWRKY17 was transfected, respectively, with additional 7.5 μg 5×GAL4-LUC as reporter plasmid which has a reporter gene for luciferase driven by five copies of GAL4-binding elements. The luciferase activities were measured as previously reported (Song et al. 2013). Three independent experiments were carried out.
DNA-binding assay of CmWRKY17 using a yeast one-hybrid system
To examine whether CmWRKY17 has DNA-binding ability to W-box, a yeast one-hybrid analysis was performed using a Clontech system (Clontech, Mountain View, CA, USA). A W-box-related cis-acting element from RD29A in A. thaliana and its corresponding W-box mutant (mW-box) element were synthesized. The nucleotide primer sequences of W-box were pHis-W–F/R (Table 1), which contains three TTGACT repeats. The primer sequences of mW-box were pHis-mW-F/R (Table 1), in which the TTGACT core sequence of each W-box is changed to TAATAT. The two elements were obtained following the manufacturer’s instruction (Clontech), and then inserted into the EcoR I and Xba I of pHis vector to generate pW-box-His and pmW-box-His constructs. The coding region of CmWRKY17 was cloned into the yeast expression vector pGADT7 by LR reaction from pENTR™1A-CmWRKY17 to obtain pGADT7-CmWRKY17. All the constructs were transferred into yeast strain Y1H (Clontech). The empty vector pGADT7 was transformed as a negative control. The growth status of transformed yeast cells was tested on SD/-His-Ura-Leu medium containing 50 mM 3-aminotrizole (3-AT).
Subcellular localization of CmWRKY17
To generate the green fluorescent protein (GFP)-CmWRKY17 fusion construct, the plasmid pENTR™1A-CmWRKY17 previously was subjected to the LR reaction to obtain a GFP-fused construct using the binary vector pMDC43, resulting in the plasmid 35S::GFP-CmWRKY17. Onion (Allium cepa) epidermal strips were placed on Murashige and Skoog (MS) medium with the inner cell surface oriented upward. Both the 35S::GFP-CmWRKY17 plasmid and the vector 35 s::GFP were transiently introduced into the onion epidermal cells by a helium-driven particle accelerator (PDS-1000; Bio-Rad). After bombardment, the cells were incubated in the same petri dishes for 16 h at 22 °C in the dark before observation. An AX80T microscope (Olympus, Japan) was used to monitor the expression of GFP.
Plasmid construction and transformation of chrysanthemum and Arabidopsis
To further investigate the function of CmWRKY17, CmWRKY17-overexpressing chrysanthemum and Arabidopsis transformants were obtained. The CmWRKY17 coding sequence was amplified with the primer pair CmWRKY17-1301-F/R harboring the BamH I and Sac I sites (Table 1). The amplicons digested by BamH I and Sac I were directly cloned into the pCAMBIA1301 cassette, containing the CaMV 35S promoter, to generate the plasmid 35S::CmWRKY17. This plasmid was transformed into Agrobacterium tumefaciens strain EHA105 using the freeze–thaw transformation method.
Transformation of chrysanthemum was performed as described previously (Cui et al. 2009). Leaf disks (5 mm diameter) taken from young leaves of ‘Jinba’ cultured in vitro were used as explants (Song et al. 2014b). The transformants were selected on culture medium containing 8 mg/L hygromycin. After regeneration, RNA was extracted from the putative transgenic chrysanthemum and WT plants, treated with RNase-free DNase I (TaKaRa), and reverse transcribed using Reverse Transcription M-MLV (TaKaRa). For qRT-PCR analysis, SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa) was employed to detect CmWRKY17 expression using the primer pair CmWRKY17-RT-F/R. The primer pair CmEF1α-F/R was used to amplify the reference gene CmEF1α. The transcription data (with three biological replicates) were calculated using the 2−ΔΔCt method.
Arabidopsis transformants were generated by introducing 35S::CmWRKY17 into the Col-0 ecotype using the floral dip method (Clough and Bent 1998). Transformants were selected by germination of seeds on MS medium containing 50 μg mL−1 hygromycin. The T3 generations were obtained using RT-PCR with the primer pair CmWRKY17-ORF-F/R.
Salinity treatment of transgenic chrysanthemum and Arabidopsis plants
For the salinity tolerance test, plants of the transgenic chrysanthemum (OX plants) and the non-transgenic chrysanthemum ‘Jinba’ (WT) at the six- to eight-leaf stage were irrigated with 200 mM NaCl for 2 weeks. After treatment, the roots were washed with distilled water and the plants were repotted in fresh soil [1:1 (v/v) mixture of soil and vermiculite] and allowed to recover for 2 weeks (An et al. 2014), after which the survival rate was calculated. The median leaves (fourth to fifth leaves counted from the base of the stem) harvested before the NaCl treatment (control) and at 48 h after NaCl treatment were used for physiological analysis.
For salinity tolerance evaluation, 3-week-old Col-0 and CmWRKY17-transgenic Arabidopsis plants with 90 of each lines were watered for 12 days at 4-day intervals, initially with 100 mM NaCl, then 200 mM NaCl, and finally with 300 mM NaCl before being scored and photographed following Gao et al. (2012).
Physiological changes in transgenic chrysanthemum
Physiological traits of OX-3, OX-7 and WT plants were measured at 0 h and 48 h of the salinity test. The activities of superoxide dismutase (SOD) and peroxidase (POD), leaf proline content, and the electrical conductivity (EC) were investigated. Activities of SOD and POD were assessed as described by Pan et al. (2006). One unit of SOD activity was defined as the amount of enzyme required to cause a 50 % inhibition and expressed as enzyme units per g of protein, while a unit of POD activity was expressed as the change in absorbance per minute at 470 nm and specific activity as enzyme units per g soluble protein, respectively. Protein concentration was measured based on the method of Bradford (1976), using bovine serum albumin as a standard. Leaf proline content was estimated following the method of Liu et al. (2005). The EC of the leaf tissue was determined using a Mode DDS-320 conductivity meter as described previously (Xu and Chen 2008).
Expression analysis of salinity stress-related genes in CmWRKY17-transformed Arabidopsis
To analyze the expression of salinity stress-responsive genes, cDNA was synthesized using RNA from the leaves of Col-0 and transgenic Arabidopsis subjected to salinity stress as template by qRT-PCR. The stress-related genes monitored were AtRD29A (AT5G52310.1), AtDREB2B (AT3G11020.1), AtSOS1 (AT2G01980.1), AtSOS2 (AT5G35410.1), AtSOS3 (AT5G24270.1), and AtNHX1 (AT5G27150.1). AtAct2 (AT3G18780.2) was used as the reference gene. The sequences of all relevant primers are listed in Table 1.
Statistical analysis
A one-way analysis of variance, followed using Tukey’s multiple range test (P = 0.05), was employed to identify treatment means that differed statistically. SPSS v17.0 software (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Cloning and sequence analysis of CmWRKY17
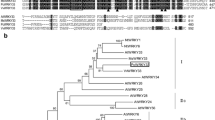
The WRKY transcription factor CmWRKY17 (KM359572) was isolated from chrysanthemum. The gene consisted of 904 bp with a 681 bp ORF encoding a 227-amino acid protein. CmWRKY17 contained one WRKY domain of WKKYGEK, which differed from the typical WRKYGQK domain, and one C2H2 zinc finger motif (C-X5-C-X23-H–X1-H) (Fig. 1a). Therefore, we assigned the cloned CmWRKY17 to group II of the WRKY family based on the characteristics of the conserved domains. Phylogenetic analysis showed that CmWRKY17 showed a higher similarity with AtWRKY17 and was most closely related to PqWRKY10 from Panax quinquefolius (Fig. 1b). Furthermore, except for the conserved WRKY domain, CmWRKY17 shared few similarities between different WRKYs from a variety of plant species (data not shown).
Deduced peptide sequences of CmWRKY17 and other WRKY proteins. a Alignment of the putative amino acid sequence of CmWRKY17 with homologous proteins. Features of the sequence include a WRKY domain (WRKYGQK) and C2H2 zinc finger (both highlighted by lines above the alignment). b Phylogenetic analysis of relationships between CmWRKY17 and WRKY proteins from other plant species. The amino acid sequences were aligned with Clustal W and the phylogenetic tree was constructed using the neighbor-joining method with MEGA 5.0. The sequence details are as follows: BnWRKY17 (ACQ76799.1), AtWRKY17 (AAL13049.1), CrWRKY11 (AAS66779.1), AtWRKY11 (AAK96194.1), MnWRKY11 (EXC32736.1), TcWRKY1 (XP_007011614.1), GhWRKY84 (AIE43859.1), MtWRKY (XP_003610020.1), PqWRKY10 (AEQ29023.1), NtWRKY3 (BAA77358.1), StWRKY3 (NP_001274938.1), VvWRKY11 (XP_002266188.1), HvWRKY7 (ABI13373.1), AsWRKY3 (AAD32676.1), AtWRKY15 (NP_179913.1), JcWRKY42 (AGQ04232.1), AtWRKY7 (AEE84877.1). CmWRKY17 is boxed
Expression profiles of CmWRKY17 in different tissues of chrysanthemum and in response to salinity stress
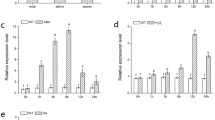
Expression patterns of CmWRKY17 in different tissues of chrysanthemum plants were investigated using qRT-PCR. Transcripts of CmWRKY17 were detected in all tissues analyzed. The highest expression level was observed in the stem and leaf, whereas the lowest level was observed in ray florets (Fig. 2a). Under treatment with 200 mM NaCl, CmWRKY17 transcript expression was induced at an early stage, increasing rapidly to be 82-fold higher than that of the control 4 h after treatment (Fig. 2b). However, thereafter the CmWRKY17 relative expression level decreased, but was maintained at a higher level compared with that of the untreated control at 24 h after treatment. It was inferred that CmWRKY17 might regulate the plant’s response to salinity stress.
Expression of CmWRKY17 in chrysanthemum plants in different organs and under salinity treatment as revealed by quantitative real-time PCR. a Expression patterns of CmWRKY17 in the root, stem, leaf, disk florets, and ray florets under non-stress conditions. b Expression of CmWRKY17 in response to 200 mM NaCl treatment
Transrepression assay of CmWRKY17
Yeast one-hybrid expression and luminescence assay were used to investigate the transcriptional activation of CmWRKY17. Yeast cells harboring the negative control pGBKT7 or pGBKT7-CmWRKY17 construct were unable to grow on SD/-His-Ade medium, whereas the positive control pCL1 grew extremely well (Fig. 3a). CmWRKY17 showed no transcriptional activation in yeast cells. To further analyze the transactivation function of CmWRKY17, the plasmid encoding CmWRKY17 was used to transfect Arabidopsis protoplasts together with reporter plasmid. The relative LUC units (RLU) of 35S::GAL4DB-CmWRKY17 were significantly lower than that of 35S::GAL4DB-AtARF5 in Arabidopsis protoplasts (P < 0.01), while the RLU of 35S::GAL4DB-CmWRKY17 were lower than that of 35S::GAL4DB (P < 0.05) (Fig. 3b), suggesting that CmWRKY17 was a repressor of transcription.
Transactivation analysis and subcellular localization of CmWRKY17. a The transcriptional activity analysis of CmWRKY17 in a yeast assay system, in which pCL1 acts as a positive control, pGBKT7 as a negative control. b Relative luciferase activities in Arabidopsis mesophyll protoplasts after transfection with 35S::GAL4DB-CmWRKY17. c Assay of CmWRKY17 binding to W-box using a yeast system. d Subcellular localization of the CmWRKY17. Bar 100 μm
DNA-binding assay of CmWRKY17
All the yeast cells harboring the two kinds of plasmids grew well on SD/-His-Ura-Leu medium (Fig. 3c). On SD/-His-Ura-Leu medium plus 50 mM 3-AT, the cells containing pGADT7-CmWRKY17 plus pW-box-His vectors grew well, and pGADT7-CmWRKY17 plus pmW-box-His grew slowly (Fig. 3c), suggesting that interaction between CmWRKY17 and the W-box element activates transcription of the reporter gene HIS3. In contrast, yeast cells with both pW-box-His or pmW-box-His and pGADT7 vectors did not grow on SD/-His-Ura-Leu medium supplemented with 50 mM 3-AT (Fig. 3c), as the negative control. These results suggested that CmWRKY17 binds to TTGACT strongly, and to the mutant TAATAT slightly.
Subcellular localization of CmWRKY17
The construct 35 s::GFP-CmWRKY17 and a positive vector harboring only 35S::GFP were introduced into onion epidermal cells via particle bombardment. Onion epidermal cells expressing 35S::GFP showed GFP fluorescence throughout the cells (Fig. 3d). In contrast, GFP fluorescence localized solely in nuclei of the onion epidermal cells transformed with the 35 s::GFP-CmWRKY17 fusion protein (Fig. 3d). These results indicated that CmWRKY17 was localized to the nucleus in vivo.
CmWRKY17 overexpression increased sensitivity of chrysanthemum to salinity stress
Transgenic chrysanthemum lines overexpressing CmWRKY17 were regenerated. The transcript levels of CmWRKY17 were measured by qRT-PCR. In the overexpressing (OX) plants, the CmWRKY17 transcript levels in OX-3 and OX-7 were significantly higher than that in WT plants (Fig. 4a). Therefore, the lines OX-3 and OX-7 were selected for further salinity tolerance evaluation.
Salinity tolerance of wild-type ‘Jinba’ and transgenic chrysanthemum plants overexpressing CmWRKY17. a Transcript abundance of CmWRKY17 in wild-type ‘Jinba’ and transgenic chrysanthemum plants overexpressing CmWRKY17. WT, Wild type; OX-3, OX-7, OX-8, and OX-9, transgenic plants overexpressing CmWRKY17. b Plant survival measured at the end of the recovery period. c Phenotypic effect of watering with 200 mM NaCl for 2 weeks, followed by a 2-week recovery period
With NaCl treatment for 2 weeks, the leaves of both OX-3 and OX-7 plants became severely wilted, withered, and some plants even died (Fig. 4c). In contrast, the WT plants were less affected by salinity stress and showed less severe withering of the leaves, and most plants survived the stress. The percentage survival of OX-3 and OX-7 plants was 28.6 % and 18.6 %, respectively, whereas that of WT plants was 50.0 % (Fig. 4b).
After 48 h of salinity stress, SOD and POD activities, proline content, and electrolyte leakage (EL) in leaves were measured in both WT and OX plants. Activities of SOD and POD and proline content were significantly higher in WT than in OX-3 or OX-7 plants (Fig. 5a–c). In contrast, EL was significantly higher in OX-3 and OX-7 relative to WT plants (Fig. 5d). These results indicated that the transgenic plants suffered oxidative damage much more severely than did WT plants. This suggested that CmWRKY17 overexpression increased the salinity sensitivity in chrysanthemum.
Physiological effects of salinity stress on wild-type ‘Jinba’ and transgenic chrysanthemum plants overexpressing CmWRKY17. a Leaf superoxide dismutase (SOD) activity, b leaf peroxidase (POD) activity, c Leaf proline content, d leaf electrical conductivity (EC). Data represent means and standard errors of three replicates
Overexpression of CmWRKY17 reduced salinity tolerance in transgenic Arabidopsis
Transgenic Arabidopsis plants overexpressing CmWRKY17 were successfully generated. Exogenous CmWRKY17 was expressed in transgenic Arabidopsis 17-5, 17-6, 17-8, and 17-9 lines but not in the Col-0 wild type (Fig. 6a). We selected the lines 17-5 and 17-6, which showed a high expression level of CmWRKY17, for further study. No difference in phenotypes was observed between transgenic and Col-0 plants under non-stress conditions. However, after salinity treatment, the transgenic lines were severely withered, whereas Col-0 plants were less affected by salinity (Fig. 6b). The percentage survival of the transgenic lines 17-5 and 17-6 was 8.9 and 7.8 %, respectively, compared with 24.4 % for Col-0 (Fig. 6c). Thus, CmWRKY17 was indicated to increase the salinity sensitivity of the transgenic Arabidopsis plants.
Salinity tolerance of Col-0 and transgenic Arabidopsis overexpressing CmWRKY17. a CmWRKY17 expression in Col-0 and transgenic Arabidopsis plants under non-stress conditions. b Salinity tolerance of Col-0 and the 35 s::CmWRKY17 transgenic lines. c Survival rate of Col-0 and transgenic Arabidopsis plants under salinity stress
To explain the possible regulatory mechanisms of CmWRKY17 in response to salinity stress, the expression levels of four genes involved in Na+ homeostasis, comprising AtSOS1, AtSOS2, AtSOS3, and AtNHX1, as well as AtRD29A and AtDREB2B were compared between transgenic and Col-0 plants (Fig. 7). Under non-stress conditions, the expression of AtRD29A, AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 was inhibited in CmWRKY17-overexpressing plants. Consistent with the finding that the background level of AtRD29A expression was inhibited in CmWRKY17-overexpressing plants, induction of AtRD29A in response to salinity treatment was reduced in CmWRKY17-overexpressing plants compared with that in Col-0 (Fig. 7a). More specifically, the early phase of induction (at 1 h) was reduced by 74 %. An approximately twofold reduction in relative expression level was observed at 2 and 4 h. Similarly, the expression levels of AtSOS1, AtSOS2, and AtSOS3 in the early phase of induction were reduced by 44 % (at 1 h), 60 % (at 2 h), and 48 % (at 1 h), respectively, compared with that of Col-0. Subsequently, a twofold, twofold, and threefold reduction were observed at 12 h for AtSOS1, 24 h for AtSOS2, and 4 h for AtSOS3, respectively (Fig. 7c–e). For clarity, we divided the stress gene mRNAs in CmWRKY17-overexpressing plants into two pools: “constitutive” and “induced”. To assess the change in the induced pool, we subtracted the mRNA level at 0 h from the mRNA level at the other time points. We found that AtDREB2B and AtNHX1 gene expression in the CmWRKY17-overexpressing plants remained at the “basal” level and did not increase during the first 2 h of induction, whereas induction in Col-0 was clearly detected within 1 h (Fig. 7b, f). Thus, CmWRKY17 may increase salinity sensitivity in Arabidopsis via inhibition of the expression of salinity-related genes.
Discussion
WRKY proteins positively or negatively regulate the response to various stresses and development in plants (Agarwal et al. 2011; Rushton et al. 2010). In the present study, we identified a group II WRKY gene, CmWRKY17, from chrysanthemum. Sequence analysis showed that it contains a special WRKY domain characterized by the sequence WKKYGEK, which differs from the classical conserved WRKY domain of WRKYGQK (Fig. 1a), and one C2H2 zinc finger motif (C-X5-C-X23-H-X-H). The protein containing the WRKYGKK motif is unable to bind to the W-box (Zhou et al. 2008), but binds to other elements (van Verk et al. 2008). Our result showed that CmWRKY17 can bind to W-box strongly, and slightly bind to the mutant W-box (Fig. 3c), suggesting that CmWRKY17 might bind to the multiple promoter elements of the target genes. The amino acid of CmWRKY17 was similar to AtWRKY17 in Arabidopsis, which negatively regulated the basal resistance in A. thaliana (Journot-Catalino et al. 2006).
CmWRKY17 was significantly induced by salinity stress, which was similar to that of AtWRKY25, AtWRKY33 in A. thaliana (Jiang and Deyholos 2006), TcWRKY53 in Thlaspi caerulescens (Wei et al. 2008), and GmWRKY13, 27, 54 in soybean (Zhou et al. 2008). In addition, CmWRKY17 did not show significant change under cold, heat, PEG, or ABA treatments (Fig. S1). CmWRK17 was supposed to be localized to the nucleus on the basis of the subcellular localization, which was in consistent with all the WRKY proteins ever reported before. The pGBKT7-CmWRKY17 cannot grow on the SD/His-Ade-, which suggested that CmWRKY17 might have no transcription activity in yeast cells (Fig. 3a). Further study of luminescence assay showed that it might act as a transcription repressor in Arabidopsis proroplasts (Fig. 3b). As we know, AtWRKY7 functions as a transcriptional repressor in plant cells (Kim et al. 2006), with higher similarity to the amino acid of CmWRKY17.
To further understand the function of CmWRKY17 in response to salinity stress, we generated several transgenic chrysanthemum (Fig. 4a) and Arabidopsis lines (Fig. 6a), respectively. There were two transgenic lines to exclude the effect of the insertion site on the characteristics of the offspring. Additionally, there was no significant difference between the empty vector lines and wild plants before or after the salt treatment as descried previously (Gao et al. 2012; Shan et al. 2012; Song et al. 2014 b).
Salinity stress causes the accumulation of reactive oxygen species (ROS), leading to a marked increase in oxidative damage to plants (Hasegawa et al. 2000). Enzymes such as SOD and POD are involved in oxidative protection; SOD is a major scavenger of O2 −, and it catalyzes the dismutation reaction of superoxide radical anions into O2 and H2O2. CAT dismutates H2O2 into water and molecular oxygen, whereas POD decomposes H2O2 by oxidation of co-substrates such as phenolic compounds and/or antioxidants (Pan et al. 2006). The proline content would strangely congest in response to a variety of stresses in plants, while electrolyte leakage reflects membrane injury caused by stresses. In the present study, no difference in the activities of ROS-scavenging enzymes (SOD and POD) was observed in either OX or WT plants under non-stress conditions, whereas the activities in OX plants were significantly lower than those in WT plants after salinity stress (Fig. 5a, b). There was no obvious difference of CAT before or after the salt treatment between WT and transgenic plants (Data not shown). Higher quantities of proline were accumulated in the leaves of WT plants compared with those in OX plants under salinity stress (Fig. 5c). However, leaf EC was much higher in OX plants than in WT plants (Fig. 5d). These results indicated that overexpression of CmWRKY17 in chrysanthemum increased the salinity sensitivity because of a decline in ROS scavenging and that salinity caused ROS damage in chrysanthemum. Low activity of ROS-scavenging enzymes and low proline contents in transgenic chrysanthemum might underlie the increased sensitivity to salinity in CmWRKY17-overexpressing plants, thereby indicating that CmWRKY17 alters the salinity sensitivity via regulation of ROS levels.
Similarly, AtWRKY17 (Journot-Catalino et al. 2006) and OsWRKY28 (Chujo et al. 2013) negatively regulate the basal resistance to Pseudomonas syringae pv. tomato and Magnaporthe oryzae (strain Ina86-137), respectively. Transgenic Arabidopsis overexpressing GmWRKY13, a group II WRKY, showAQ increased sensitivity to salt stress, while it was strongly induced by salinity (Zhou et al. 2008). In the current study, heterologous expression of CmWRKY17 increased the sensitivity to salinity stress in Arabidopsis (Figs. 6, 7), which was significantly induced by salt stress, showing similar function. As a well-known stress marker gene and a lea-type stress protein, RD29A has been proposed to function in the detoxification or alleviation of stress-related damage (Zhu 2001). The DREB transcription factor mediates the expression of some stress-related genes in response to secondary stresses caused by high salinity (Zhu 2001). Genes such as AtSOS1, AtSOS2, AtSOS3, and AtNHX1 are involved in a regulatory pathway for ionic homeostasis under salt stress. Recently, the SOS1 gene was shown to encode a putative plasma membrane Na+-H+ antiporter (Shi et al. 2000). SOS3 interacts physically with SOS2, which is a serine/threonine protein kinase (Halfter et al. 2000; Liu et al. 2000). One downstream target of the SOS3–SOS2 kinase complex is SOS1, which exports Na+ from the cell (Shi et al. 2000). Overexpression of AtNHX1 appears to improve plant salt tolerance substantially (Apse et al. 1999). The present data showed that salinity-induced expression of AtRD29A was inhibited by overexpression of CmWRKY17 (Fig. 7a). Similarly, under salinity stress, the induction of the genes AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 was reduced in CmWRKY17-overexpressing plants compared with that in the control (Col-0) (Fig. 7b–f). These results indicated that CmWRKY17 might reduce the expression of SOS pathway genes to increase the salinity sensitivity in Arabidopsis. The complemented line shows lower induction of all genes compared with that in Col-0, which is consistent with the earlier observation that higher levels of CmWRKY17 expression inhibited salt induction.
WRKY transcription factors are able to bind to the W-box in the promoter of the target genes. Although CmWRKY17 contained a WKKYGEK domain, which is quite different from the typical WRKYGQK motif, and a yeast one-hybrid system showed that CmWRKY17 can bind to the W-box, and it might bind to the mutant W-box, further regulation of expression of the genes in the salinity stress-response pathway remains unknown. Increased expression of CmWRKY17 reduced salt-induced expression of stress-responsive genes such as AtRD29A and AtDREB2B, as well as AtSOS1, AtSOS2, AtSOS3, and AtNHX1. In this study, the data showed that CmWRKY17 acted as a transcription repressor. Collectively, these results suggest that CmWRKY17 may serve as a negative regulator of the salinity response pathway through inhibiting the expression of stress-related gene.
In summary, a novel group II WRKY gene, CmWRKY17, a transcriptional repressor, was induced by salinity stress. CmWRKY17-overexpressing transgenic chrysanthemum and Arabidopsis plants showed increased salinity sensitivity compared with the wild-type plants. These results indicated that CmWRKY17 might directly or indirectly negatively regulate the stress-related genes, at least partially through regulation of the SOS pathway, to increase the sensitivity to salinity.
Author contribution statement
LPL, CFD, and CSM conceived and designed the experiments. LPL, SAP, and GCY performed the experiments. LPL, WLX, WYJ, and SJ analyzed the data. JJF, CFD, and CSM contribute reagents/materials/analysis tools. LPL, CFD, and CSM wrote the paper. All authors read and approved the manuscript.
References
Abbruscato P, Nepusz T, Mizzi L, Del Corvo M, Morandini P, Fumasoni I, Michel C, Paccanaro A, Guiderdoni E, Schaffrath U (2012) OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol Plant Pathol 13:828–841
Agarwal P, Reddy M, Chikara J (2011) WRKY: its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol Biol Rep 38:3883–3896
An J, Song A, Guan Z, Jiang J, Chen F, Lou W, Fang W, Liu Z, Chen S (2014) The over-expression of Chrysanthemum crassum CcSOS1 improves the salinity tolerance of chrysanthemum. Mol Biol Rep 41:4155–4162
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na +/H + antiport in Arabidopsis. Science 285:1256–1258
Atamian HS, Eulgem T, Kaloshian I (2012) SlWRKY70 is required for Mi-1-mediated resistance to aphids and nematodes in tomato. Planta 235:299–309
Borrone JW, Kuhn DN, Schnell RJ (2004) Isolation, characterization, and development of WRKY genes as useful genetic markers in Theobroma cacao. Theor Appl Genet 109:495–507
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chen S, Miao H, Chen F, Jiang B, Lu J, Fang W (2009) Analysis of expressed sequence tags (ESTs) collected from the inflorescence of chrysanthemum. Plant Mol Biol Rep 27:503–510
Chujo T, Miyamoto K, Shimogawa T, Shimizu T, Otake Y, Yokotani N, Nishizawa Y, Shibuya N, Nojiri H, Yamane H (2013) OsWRKY28, a PAMP-responsive transrepressor, negatively regulates innate immune responses in rice against rice blast fungus. Plant Mol Biol 82:23–37
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui X, Chen F, Chen S (2009) Establishment of regeneration and transformation system of ground-cover chrysanthemum Yuhuaxunzhang [J]. J Nanjing Agric Univ 32:40–46
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Gao H, Song A, Zhu X, Chen F, Jiang J, Chen Y, Sun Y, Shan H, Gu C, Li P (2012) The heterologous expression in Arabidopsis of a chrysanthemum Cys2/His2 zinc finger protein gene confers salinity and drought tolerance. Planta 235:979–993
Guo R, Yu F, Gao Z, An H, Cao X, Guo X (2011) GhWRKY3, a novel cotton (Gossypium hirsutum L.) WRKY gene, is involved in diverse stress responses. Mol Biol Rep 38:49–58
Halfter U, Ishitani M, Zhu J (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci 97:3735–3740
Hasegawa PM, Bressan RA, Zhu J, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Ann Rev Plant Biol 51:463–499
Hinderhofer K, Zentgraf U (2001) Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213:469–473
Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74:730–745
Hui D, Iqbal J, Lehmann K, Gase K, Saluz HP, Baldwin IT (2003) Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: V. Microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol 131:1877–1893
Ishiguro S, Nakamura K (1994) Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol Gen Genet 244:563–571
Jiang Y, Deyholos MK (2006) Comprehensive transcriptional profiling of NaCl-stressed Arabidopsis roots reveals novel classes of responsive genes. BMC Plant Biol 6:25
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14:1359–1375
Journot-Catalino N, Somssich IE, Roby D, Kroj T (2006) The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18:3289–3302
Kalde M, Barth M, Somssich IE, Lippok B (2003) Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Mol Plant-microbe Interact 16:295–305
Kim KC, Fan B, Chen Z (2006) Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol 142:1180–1192
Lagacé M, Matton DP (2004) Characterization of a WRKY transcription factor expressed in late torpedo-stage embryos of Solanum chacoense. Planta 219:185–189
Li S, Fu Q, Chen L, Huang W, Yu D (2011) Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233:1237–1252
Li H, Gao Y, Xu H, Dai Y, Deng D, Chen J (2013) ZmWRKY33, a WRKY maize transcription factor conferring enhanced salt stress tolerances in Arabidopsis. Plant Growth Regul 70:207–216
Liu J, Ishitani M, Halfter U, Kim C, Zhu J (2000) The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci 97:3730–3734
Liu F, Liu Q, Liang X, Huang H, Zhang S (2005) Morphological, anatomical, and physiological assessment of ramie [Boehmeria Nivea (L.) Gaud.] tolerance to soil drought. Genet Resour Crop Evols 52:497–506
Liu J, Que Y, Guo J, Xu L, Wu J, Chen R (2012) Molecular cloning and expression analysis of a WRKY transcription factor in sugarcane. Afr J Biotechnol 11:6434–6444
Liu Q, Zhong M, Li S, Pan Y, Jiang B, Jia Y, Zhang H (2013) Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol Biochem 69:27–33
Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K (2001) Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci Biotechnol Biochem 65:2428–2436
Marè C, Mazzucotelli E, Crosatti C, Francia E, Cattivelli L (2004) Hv-WRKY38: a new transcription factor involved in cold-and drought-response in barley. Plant Mol Biol 55:399–416
Niu C, Wei W, Zhou Q, Tian A, Hao Y, Zhang W, Ma B, Lin Q, Zhang Z, Zhang J (2012) Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant, Cell Environ 35:1156–1170
Pan Y, Wu L, Yu Z (2006) Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul 49:157–165
Pan Y, Cho C, Kao Y, Sun C (2009) A novel WRKY-like protein involved in transcriptional activation of cyst wall protein genes in Giardia lamblia. J Biol Chem 284:17975–17988
Park C, Shin Y, Lee B, Kim K, Kim J, Paek K (2006) A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris. Planta 223:168–179
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Shan H, Chen S, Jiang J, Chen F, Chen Y, Gu C, Li P, Song A, Zhu X, Gao H (2012) Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance, increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana. Mol Biotechnol 51:160–173
Shi H, Ishitani M, Kim C, Zhu J (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na +/H + antiporter. Proc Natl Acad Sci 97:6896–6901
Song A, Lou W, Jiang J, Chen S, Sun Z, Guan Z, Fang W, Teng N, Chen F (2013) An isoform of eukaryotic initiation factor 4E from Chrysanthemum morifolium Interacts with chrysanthemum virus B coat protein. PLoS ONE 8:e57229
Song A, Li P, Jiang J, Chen S, Li H, Zeng J, Shao Y, Zhu L, Zhang Z, Chen F (2014a) Phylogenetic and Transcription Analysis of Chrysanthemum WRKY Transcription Factors. Int J Mol Sci 15:14442–14455
Song A, Zhu X, Chen F, Gao H, Jiang J, Chen S (2014b) A chrysanthemum heat shock protein confers tolerance to abiotic stress. Int J Mol Sci 15:5063–5078
Ülker B, Mukhtar MS, Somssich IE (2007) The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 226:125–137
van Verk MC, Pappaioannou D, Neeleman L, Bol JF, Linthorst HJ (2008) A novel WRKY transcription factor is required for induction of PR-1a gene expression by salicylic acid and bacterial elicitors. Plant Physiol 146:1983–1995
Wang Q, Wang M, Zhang X, Hao B, Kaushik S, Pan Y (2011) WRKY gene family evolution in Arabidopsis thaliana. Genetica 139:973–983
Wang Y, Dang F, Liu Z, Wang X, Eulgem T, Lai Y, Yu L, She J, Shi Y, Lin J (2013) CaWRKY58, encoding a group I WRKY transcription factor of Capsicum annuum, negatively regulates resistance to Ralstonia solanacearum infection. Mol Plant Pathol 14:131–144
Wei W, Zhang Y, Han L, Guan Z, Chai T (2008) A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep 27:795–803
Xu Y, Chen F (2008) The LT_(50) and cold tolerance adaptability of chrysanthemum during a natural drop in temperature. Acta Horticulturae Sinica 35:559–564
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protoc 2:1565–1572
Zhang J, Peng Y, Guo Z (2008) Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res 18:508–521
Zheng J, Zou X, Mao Z, Xie B (2011) A novel pepper (Capsicum annuum L.) WRKY gene, CaWRKY30, is involved in pathogen stress responses. J Plant Biol 54:329–337
Zhou Q, Tian A, Zou H, Xie Z, Lei G, Huang J, Wang C, Wang H, Zhang J, Chen S (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503
Zhu J (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Acknowledgments
This work was supported by the Natural Science Fund of Jiangsu Province (BK2011641, BK2012773), the Program for New Century Excellent Talents in University of the Chinese Ministry of Education (Grant nos. NCET-10-0492, NCET-12-0890), the Fundamental Research Funds for the Central Universities (KYZ201112, KYZ201147), Fund for Independent Innovation of Agricultural Sciences in Jiangsu Province [CX(12)2020], and the Program for Hi-Tech Research, Jiangsu, China (Grant nos. BE2012350, BE2011325).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by L. Peña.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2015_1793_MOESM1_ESM.tif
Supplementary material 1 (TIFF 2042 kb). Expression of CmWRKY17 in chrysanthemum plants under different treatments. Cold: 4 °C; Heat: 40 °C; PEG: 20 % PEG6000; ABA: 50 μM ABA
Rights and permissions
About this article
Cite this article
Li, P., Song, A., Gao, C. et al. Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep 34, 1365–1378 (2015). https://doi.org/10.1007/s00299-015-1793-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1793-x