Abstract
Key message
PcWRKY33 is a transcription factor which can reduce salt tolerance by decreasing the expression of stress-related genes and increasing the cellular levels of reactive oxygen species (ROS).
Abstract
WRKY transcription factors play important roles in the regulation of biotic and abiotic stresses. Here, we report a group I WRKY gene from Polygonum cuspidatum, PcWRKY33, that encodes a nucleoprotein, which specifically binds to the W-box in the promoter of target genes to regulate their expression. The results from qPCR and promoter analysis show that expression of PcWRKY33 can be induced by various abiotic stresses, including NaCl and plant hormones. Overexpression of PcWRKY33 in Arabidopsis thaliana reduced tolerance to salt stress. More specifically, several physiological parameters (such as root length, seed germination rate, seedling survival rate, and chlorophyll concentration) of the transgenic lines were significantly lower than those of the wild type under salt stress. In addition, following exposure to salt stress, transgenic plants showed decreased expression of stress-related genes, a weakened ability to maintain Na+/K+ homeostasis, decreased activities of reactive oxygen species- (ROS-) scavenging enzymes, and increased accumulation of ROS. Taken together, these results suggest that PcWRKY33 negatively regulates the salt tolerance in at least two ways: by down-regulating the induction of stress-related genes and by increasing the level of cellular ROS. In sum, our results indicate that PcWRKY33 is a group I WRKY transcription factor involved in abiotic stress regulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saline water covers about 75% of the earth’s surface, and approximately 3% of land is affected by salt under normal circumstances (FAO 2005). In addition, human activities, such as intensive agriculture and irrigation, are increasing the degree of salinization of arable soils. Hence, soil salinity has become a primary environmental risk for plant cultivation and markedly reduces the yield and quality of crops. It is predicted that more than 50% of the productivity of the world’s cultivated land will be lost by 2050, as a result of soil salinity, exacerbated by global climate change (Mahajan and Tuteja 2005). Growing in such an environment, salt-adapted plant species have evolved multifaceted mechanisms to tolerate salt stress, including morphological, physiological, and biochemical changes. Although the molecular mechanisms underlying plant salt tolerance are still not fully clarified, there is no doubt that transcription factors play important roles. Over the past few decades, increasing attention has been paid to the regulation of plant abiotic stress tolerance by transcription factors, such as AP2/ERF, WRKY, MYB, NAC, and bHLH (Cui et al. 2013; Xu et al. 2015; Zhou et al. 2009).
The WRKY transcription factor family is one of the largest transcription factor families in higher plants (Ulker and Somssich 2004). WRKY is named after the conserved WRKYGQK sequence which contains about 60 amino acid residues at the N-terminal end. Members of this family contain one or two WRKY domains at the N-terminus and a zinc-finger motif at the C-terminus, and are divided into three groups, namely group I, II, and III. Group I contains two WRKY domains and a C2H2 (C–X4–5–C–X22–23–H–X–H) zinc-finger motif; group II includes one WRKY domain and a C2H2 zinc-finger motif and can be further split into five subgroups (IIa–e) (Rushton et al. 2008; Zhang and Wang 2005); and group III comprises one WRKY domain and a C2HC (C–X7–C–X23–H–X–C) zinc-finger motif (Eulgem et al. 2000).
WRKY transcription factors are reported to be involved in responses to biotic and abiotic stresses (Li et al. 2004; Zhou et al. 2008), and in metabolic and developmental regulation (Devaiah et al. 2007; Kato et al. 2007), through specifically binding to the W-box (TTGACC/T) in the promoter region of target genes (Rushton et al. 1996). Thus far, most studies on the function of group I WRKY transcription factors have focused on their roles in disease resistance and in response to abiotic stresses. In grape, VvWRKY33 induction and the increased expression of the VvPR10.1 pathogenesis-related gene in the leaves of the mildew-resistant cultivar ‘Regent’ after infection with Plasmopara viticola were correlated, whereas, in the susceptible cultivar ‘Lemberger’, VvWRKY33 and VvPR10.1 were not induced. The results of ectopic expression studies showed that VvWRKY33 could strongly enhance resistance to P. viticola in the susceptible cultivar ‘Shiraz’ and reduce pathogen sporulation by about 50–70% (Merz et al. 2015). In Arabidopsis thaliana, the overexpression of a poplar (Populus trichocarpa) salicylic acid- (SA-) inducible WRKY gene, PtrWRKY73, increased resistance to biotrophic pathogens but reduced resistance against necrotrophic pathogens (Duan et al. 2015). A series of experiments on AtWRKY34 overexpressing plants showed that this gene negatively regulated cold-sensitivity of mature Arabidopsis pollen by participating in the CBF signal cascade (Zou et al. 2010). In addition, CsWRKY2, a WRKY transcription factor gene from tea (Camellia sinensis), was reported to play an important role in cold stress response via the ABA signaling pathway (Wang et al. 2016).
A growing number of studies have demonstrated that many group I WRKY genes can play either positive or negative roles in response to salt stress. For instance, overexpression of GmWRKY54 from soybean in Arabidopsis improved salt and drought tolerance, possibly by regulating the expression of DREB2A and STZ/Zat10, whereas GmWRKY13-transgenic Arabidopsis plants showed an increase in salt and mannitol sensitivity, and a reduction in ABA sensitivity (Zhou et al. 2008). Both the Atwrky33 null mutants and the Atwrky25Atwrky33 double mutants showed a greater sensitivity to NaCl and ABA, and genetic complementary experiments confirmed that overexpression of either AtWRKY25 or AtWRKY33 was sufficient to increase Arabidopsis NaCl tolerance (Jiang and Deyholos 2009). Studies on GhWRKY25, a group I WRKY gene from cotton, indicated that its heterologous expression in Nicotiana benthamiana reduced the tolerance to drought stress but increased the tolerance to salt stress (Liu et al. 2016). Transformation with RtWRKY1, from the recretohalophyte Reaumuria trigyna, was also reported to confer salt tolerance to the transgenic Arabidopsis by regulating plant growth, osmotic balance, Na+/K+ homeostasis, and the antioxidant system (Chao et al. 2017). In chrysanthemum (Dendranthema grandiflorum), DgWRKY1, DgWRKY3, DgWRKY4, and DgWRKY5 were reported to increase tolerance to salt stress in transgenic tobacco or chrysanthemum (Liang et al. 2017; Liu et al. 2014, 2013; Wang et al. 2017). On the contrary, CmWRKY17 negatively regulated salt tolerance in transgenic chrysanthemum, Chrysanthemum morifolium (Li et al. 2015). The above findings indicated that WRKY genes regulate salt stress tolerance through multiple processes, mainly by regulating the cellular levels of reactive oxygen species (ROS), controlling the ABA signaling pathway, altering the expression of stress-related genes, maintaining the Na+/K+ homeostasis, and maintaining membrane stability.
Biotic and abiotic stresses can cause changes in gene expression within a short time. Overexpression of GhWRKY34 enhanced salt tolerance in Arabidopsis, largely by up-regulating the expression of the stress-related genes AtRD29A, AtRD29B, AtABF4, AtSOS2, AtSOS1, and AtCBL1 (Zhou et al. 2015). On the contrary, exposure to salt stress reduced the expression of the stress-related genes AtRD29A, AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 in CmWRKY17 transgenic Arabidopsis plants and enhanced the sensitivity to salt (Li et al. 2015). In addition, salt stress also causes the accumulation of ROS in plants. For example, overexpression of GhWRKY17 in Nicotiana benthamiana significantly reduced plant tolerance to salt stress through reduced expression of genes encoding ROS-scavenging enzymes, increased accumulation of ROS, increased malondialdehyde (MDA) content, caused by the interaction between ROS and the lipids of cell membranes, reduced proline content and antioxidant enzyme activities [superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and ascorbate peroxidase (APX)], and increased electrolyte leakage (Yan et al. 2014). WRKY genes were reported to affect salt tolerance by regulating the expression of stress-related genes and the cellular level of ROS simultaneously. For example, overexpression of a novel cotton gene GhWRKY6-like in Arabidopsis enhanced salt tolerance by reducing the accumulation of ROS, increasing the levels of ROS-scavenging enzymes, and increasing the expression of stress-related genes (AtSOS2, AtRD29a, and AtRD29b) (Ullah et al. 2017).
Polygonum cuspidatum Sieb. et Zucc. is a perennial plant widely used in herbal medicine in Asian countries, such as China and Japan, because of its pharmacological effects with respect to antifungal (Agarwal et al. 2000), antioxidant (Yen et al. 2000), antiviral (Barnard et al. 1995), and antineoplastic activities (Su et al. 2005), with the active organs being the roots and rhizomes. P. cuspidatum has also become a serious invasive alien plant in Europe and part of the US. In large-scale planting of P. cuspidatum, soil salinity is one of the important factors influencing yield and quality. Thus far, most Asian studies on P. cuspidatum have dealt with its pharmacodynamic effects, with apparently no reports on the regulation of abiotic stress by P. cuspidatum transcription factors. In the present study, we report the isolation and molecular characterization of a group I WRKY gene, PcWRKY33, from P. cuspidatum. Our results showed that PcWRKY33 overexpression in A. thaliana can negatively regulate salt stress through decreasing the expression of stress-related genes and regulating cellular levels of ROS.
Materials and methods
Plant materials, growth conditions, and treatments
Polygonum cuspidatum plants were maintained in the medicinal plant garden and in a greenhouse at the Institute of Botany, The Chinese Academy of Sciences, Beijing, China. Seeds were surface-sterilized and sown on Murashige and Skoog (MS) agar medium in a growth chamber at 24 °C with a 16/8 h light/dark cycle. One-month-old seedlings were used in the subsequent treatments. For plant hormone treatment, the seedlings were uniformly sprayed with 0.1 mM methyl jasmonate (MeJA), 0.1 mM ABA, or 500 µM gibberellic acid (GA3), and the solutions had been filter-sterilized before use. For salt treatment, the 1-month-old seedlings were transplanted to MS agar medium containing 100 mM NaCl. For drought treatment, seedlings, transplanted from agar to a 1:1 (v/v) mixture of soil and vermiculite in pots, and grown in greenhouse at 24 °C with a 16/8 h light/dark cycle, were used. All the samples were frozen in liquid nitrogen at the appropriate time and stored at -80 °C before RNA extraction. Each treatment was repeated at least twice and the results were expressed as the mean ± standard deviation (SD) of two replicate experiments.
Seeds of wild-type A. thaliana Columbia ecotype (Col-0) (and of transgenic lines derived from the wild type) were potted in a 1:1 (v/v) mixture of soil and vermiculite, and grown in a greenhouse at 22 ± 1 °C with a 16/8 h light/dark cycle and a relative humidity of 60%.
Gene cloning, promoter cloning, and sequence analyses
Total RNA was isolated from P. cuspidatum leaves using the Plant Total RNA Purification Kit (GeneMark). The first-strand cDNA was synthesized using 5 µg total RNA, and TransScript®One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) in a reaction volume of 20 µL. The primers used are listed in Table S1. The full-length sequence of PcWRKY33 was cloned into the pCloneEZ-TA-Amp/HC (ThBio, Beijing, China) for sequencing.
Genomic DNA was isolated from seedling leaves using Plant Genomic DNA Kit (TIANGEN, Beijing, China), and was then digested by EcoRV, StuI, DraI, and PvuII, respectively. After purification, a 1:1 (v/v) mixture of GW-AP1 and GW-AP2 was used as an adapter to connect with the treated DNA (Siebert et al. 1995). Two rounds of PCR were carried out and the products were cloned into the pCloneEZ-TA-Amp/HC (ThBio, Beijing, China) for sequencing.
The PcWRKY33 amino acid sequence was aligned with its homologs using the ClustalX program (http://www.clustal.org) and BLAST online software (http://www.ncbi.nlm.gov/blast). The phylogenetic relationships of the WRKY proteins were analyzed with the neighbor-joining method using MAGE 7.1 (http://www.megasoftware.net/). Physicochemical property analysis was carried out using the ProtParam program (http://au.expasy.org/tools/protparam.html). Identification of protein domains and significant sites was performed with the ScanProsite tool (http://prosite.expasy.org/scanprosite/). Analysis of the promoter sequence of PcWRKY33 was performed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The localization of the PcWRKY33 protein was predicted by the ProtComp program (http://linux1.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc).
Quantitative real-time PCR
Total RNA was isolated by the Plant Total RNA Purification Kit (GeneMark). The first-strand cDNA was synthesized using 3 µg total RNA and HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) (Vazyme, Nanjing, China) in a reaction volume of 20 µL. Quantitative real-time PCR (qPCR) was performed using TransStart Tip Green qPCR SuperMix (TransGen Biotech, Beijing, China) and the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The PCR mix was composed of 5 µL 2 × TransStart®Tip Green qPCR SuperMix, 1.0 µL of 1:10 diluted cDNA, 0.2 µL of each primer (10 mM), and 3.6 µL PCR grade water in a final volume of 10 µL. The reactions were incubated under the following conditions: 1 cycle at 94 °C for 30 s; 40 cycles at 94 °C for 5 s, 55 °C for 15 s, and 72 °C for 10 s; and then a single melt cycle from 65 to 95 °C. The primers used in qPCR are listed in Table S1 and the expression levels were calculated using the 2−ΔΔCt comparative CT method (Livak and Schmittgen 2001). The results were expressed as the mean ± standard deviation (SD) of three replicate experiments.
Subcellular localization analysis
The coding sequence of PcWRKY33 without the termination codon and the sequence of GFP were fusion expressed by overlapping PCR. Then, the fragment was cloned into the binary vector pBI121 and driven by the CaMV35S promoter, using seamless cloning technology. The recombined plasmid and the positive control 35S::GFP plasmid were, respectively, transferred into living onion epidermal cells via the biolistic bombardment transformation method (Shi et al. 2010), using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, Hercules, CA, USA) with gold particles (1.0 µL) and a helium pressure of 1,350 psi. Then, the treated onion epidermal cells were cultured on MS medium under dark conditions at 28 °C for 24 h. Before observation, the nuclei were stained with 0.1 µg/mL 4′6-diamidino-2-phenylindole (DAPI) in phosphate-buffered saline (PBS) for 15 min. The fluorescence was observed using an inverted fluorescence microscope (TCS SP5: Leica, Wetzlar, Germany).
Yeast one-hybrid and co-transfection experiments
For yeast one-hybrid experiments, triple tandem copies of the W-box (TTGACC) and mutant W-box (TAGACC) were, respectively, inserted into the pLacZi vector at the EcoRI and KpnI sites, using T4 DNA Ligase (NEB, Ipswich, MA, USA), and the coding sequence of PcWRKY33 was inserted into the pB42AD vector using the One-Step Cloning Kit (Vazyme, Nanjing, China). Both vectors were then co-transformed into the yeast strain EYG48, and the transformed yeast cells were grown on tryptophan (Trp) and uracil (Ura)-deficient synthetic dextrose (SD) medium (SD/-Trp/-Ura), to confirm the success of the transformation. Then, the confirmed yeast strains were smeared on SD/-Trp/-Ura medium with 20 mg/L X-gal and cultivated at 30 °C for less than 12 h.
For co-transfection experiments, the effector plasmid was obtained by replacing GUS in the binary vector pBI121 with the ORF of PcWRKY33 using the One-Step Cloning Kit (Vazyme, Nanjing, China), while the reporter vector was obtained by replacing CaMV35S in pCAMBIA1301 with 3 × W-box-35Smini and 3 × mW-box-35Smini, respectively, using the same method. Both recombinant plasmids were introduced into Agrobacterium tumefaciens strain GV3101 at a concentration equivalent to an OD600 of 0.2. The assay of Agrobacterium-mediated transient transformation was performed as previously described (Yang et al. 2010). GUS histochemical staining was detected using 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-gluc) as the substrate at 2 days after transformation. The leaf samples were soaked in GUS staining solution (2 mL 0.5 M Na2EDTA, 3.8 mL 1 M NaH2PO4·H2O, 12.4 mL 0.5 M Na2HPO4·12H2O, 0.5 mL 0.1 M K4Fe(CN)6, 0.5 mL 0.1 M K3Fe(CN)6, 25 mL methanol, 60 µL Triton X-100, and 100 mg X-gluc (dissolve in 500 µL DMFO), and distilled H2O to make the volume up to 250 mL, pH 7.0) and vacuumized at − 800 Pa for 5 min. After incubation overnight at 37 °C, the leaf samples were immersed in 70% (v/v) ethanol until the chlorophyll was removed.
Genetic transformation
The 35S::PcWRKY33-pBI121 recombinant plasmid was transferred into Agrobacterium tumefaciens strain GV3101, and A. thaliana was transformed with Agrobacterium-mediated DNA transfer by the floral dip method (Clough and Bent 1998). Transformed Arabidopsis seeds were selected on half-strength Murashige and Skoog (1/2 × MS) medium containing 40 mg/L kanamycin.
Semi-quantitative RT-PCR
For the semi-quantitative RT-PCR (RT-PCR) detection of transgenic A. thaliana, total RNA from the leaves of seven kanamycin resistance transgenic plants and control plants were isolated using the Plant Total RNA Purification Kit (GeneMark). First-strand cDNA synthesis was performed on total RNA using TransScript®One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). The resultant first-strand cDNA was used as a template, AtActin2 was used as an internal control, and its program was: 1 cycle at 94 °C for 10 min; 24 cycles at 94 °C for 30 s, 58 °C for 30 s, and 72 °C for 30 s; and then followed by 1 cycle at 72 °C for 5 min. After PCR, the products were detected using 2% (w/v) agarose gels. RT-PCR experiments were repeated at least three times using independent cDNA samples. All primers used in RT-PCR are listed in Table S1.
Salt sensitivity analyses
In the germination assay, seeds of three independent overexpressed PcWRKY33 lines (OE1, OE2, and OE3) from the T1 generation, and the wild-type (WT) seeds were surface-sterilized and spread on 1/2 × MS medium supplemented with different concentrations of NaCl (0, 100, or 125 mM), and the germination rates were counted daily. For the root elongation assay, the seeds were germinated on 1/2 × MS medium for 3 days before being transferred to 1/2 × MS medium containing different concentrations of NaCl (0, 100, or 125 mM), and the plates were placed vertically in the chamber. There were six independent samples of each of the four lines, and the root lengths were measured after 10 days treatment.
For salt stress treatment, 4-week-old transgenic and wild-type plants were irrigated every 3 days with 100 mM NaCl solution for 10 days and maintained under the same growth conditions as described above to record survival rates. The chlorophyll concentration was determined as previously described (Lichtenthaler and Wellburn 1983). In addition, the concentrations of Na+ and K+ were measured by inductively coupled plasma optical emission spectrometer (ICP-OES) (Perkin Elmer, Waltham, MA, USA); before measurement, the samples were nitrated with 8 mL concentrated nitric acid and 3 mL concentrated hydrogen peroxide. The salt stress treatment experiments were repeated at least three times and the results were expressed as the mean ± standard deviation (SD) (n = 6).
Oxidative stress analyses
To detect the accumulation of H2O2 and O2−, a histochemical staining procedure was performed using 3, 3′-diaminobenzidine (DAB) or nitrotetrazolium blue chloride (NBT), respectively, according to the method previously described (Hu et al. 2013). The content of MDA was detected using a maleic dialdehyde assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Plant tissue was extracted in a phosphate buffer (pH 7.8) as previously described (Li et al. 2013), and the ROS-scavenging enzymes activities of SOD, POD, and CAT were measured with kits produced by the Nanjing Jiancheng Bioengineering Institute (Nanjing, China). Protein concentration was quantified with the BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China), and specific enzyme activity was expressed on a protein basis.
Statistical analysis
The data were analyzed by analysis of variance (ANOVA) using the GraphPad software (La Jolla, CA, USA) and multiple comparisons were carried out using Tukey’s test. The results were expressed as the mean ± standard deviation (SD) of at least three independent replicate experiments, and the significance threshold was set at P < 0.05.
Results
Identification and sequence analysis of PcWRKY33
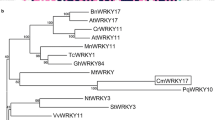
A cDNA fragment encoding PcWRKY33 was isolated using gene-specific primers designed from the sequences of the P. cuspidatum transcriptome database (SRA038892), with the rapid amplification of cDNA ends PCR (RACE-PCR) being used to amplify the 5′-untranslated region (UTR) and the 3′-UTR. The full-length cDNA sequence was 1572 bp and contained a complete open reading frame (ORF) of 1449 bp. The gene encoded a 482-amino-acid protein, with a predicted relative molecular mass of 53.354 kDa and a theoretical isoelectric point of 6.33. Due to the highest degree of homology with AtWRKY33, this new WRKY protein was designated as PcWRKY33 (MG779503). Multi-sequence alignment showed that PcWRKY33 showed 53.93, 48.79, and 47.11% homology with VvWRKY33 (AHG99400), MsWRKY33 (AKM20827), and AtWRKY33 (AAM34736), respectively (Fig. 1a). Structural analysis showed that the PcWRKY33 protein contained two WRKY domains and two zinc-finger motifs (C–X4–C–X23–H–X1–H), indicating that PcWRKY33 belongs to group I of the WRKY family. In Fig. 1a, the conserved WRKY sequence is outlined by a box, and the C and H residues in the zinc-finger motif are marked by triangles. A phylogenetic tree was constructed to evaluate the evolutionary position of PcWRKY33 within the WRKY protein family, using the MEGA 7.1 software, based on the amino acid sequences of different WRKY members from different species (Fig. 1b).
Sequence and phylogenetic analysis of PcWRKY33. a Alignment of the putative PcWRKY33 protein with its homologous proteins. Identical amino acids are shaded in black. The WRKYGQK conservative sequence is outlined by a box and the C and H residues in the zinc-finger motif are marked by triangles. b Phylogenetic analysis of PcWRKY33 and other WRKY proteins from different species. The amino acid sequences were subjected to Clustal W using the neighbor-joining method in MEGA 7.1. PcWRKY33 is boxed. At, Arabidopsis thaliana; Nt, Nicotiana tabacum; Pc, Polygonum cuspidatum; Ms, Medicago sativa; Vv, Vitis vinifera
Characterization of PcWRKY33 as a transcription factor
The presence of putative nuclear localization signals in the PcWRKY33 protein sequence predicted its potential location in the nucleus like most other transcription factors. To test this prediction, we constructed 35S::PcWRKY33-GFP and 35S::GFP vectors, with the green fluorescent protein (GFP) gene acting as the reporter gene in the subcellular localization experiment. The plasmids carrying 35S::PcWRKY33-GFP and 35S::GFP were introduced, respectively, into onion epidermal cells using a gene gun. Onion epidermal cells carrying the 35S::PcWRKY33-GFP plasmid emitted fluorescence only in the nucleus, which was further confirmed by DAPI staining, whereas the 35S::GFP control exhibited GFP signals in both the cytoplasm and the nucleus (Fig. 2a). These findings confirmed that the PcWRKY33 protein was localized in the nucleus.
Characterizations of PcWRKY33 as a transcription factor. a Subcellular localization of PcWRKY33. The images of fluorescence were visualized using an inverted fluorescence microscope. b Sequences of the triple tandem repeats of the W-box and mW-box. Restriction enzyme cutting sites are in bold font and the W-box and mW-box are underlined. c Transactivation activity analysis of PcWRKY33 by the yeast one-hybrid assay using the 3 × W-box or 3 × mW-box as bait. 1, pLacZi-W-box/pB42AD-PcWRKY33; 2, pLacZi-mW-box/pB42AD-PcWRKY33; 3, pLacZi-W-box/pB42AD; 4, pLacZi-mW-box/pB42AD. d GUS histochemical staining of co-transfected N. benthamiana leaves
On the basis of reports that WRKY transcription factors modulate protein expression by interacting with the cis-element W-box to regulate transcription, we performed the yeast one-hybrid assay to verify this characterization of the PcWRKY33 protein. Three tandem repeats of the W-box (TTGACC) and the mW-box (TAGACC) were, respectively, inserted into the pLacZi vector, and PcWRKY33 was cloned into the pB42AD vector (Fig. 2b). These two vectors were then co-transformed into the yeast strain EYG48, and the transformed yeast cells were grown on tryptophan- (Trp) and uracil- (Ura) deficient synthetic dextrose (SD) medium (SD/-Trp/-Ura), to test the success of transformation. After growth on SD/-Trp/-Ura medium containing 20 mg/L X-gal for less than 12 h, only the yeast strain carrying both the pLacZi-W-box and pB42AD-PcWRKY33 plasmids turned blue (Fig. 2c). These results demonstrated that PcWRKY33 specifically bound to the W-box element and functioned as a transcription factor in this yeast system.
For a further study on the transcriptional activities of PcWRKY33 in the plant cell, a transient co-expression experiment was performed in tobacco leaves. The W-box-35Smini and mW-box-35Smini sequence, respectively, replaced the promoter of GUS in the pCAMBIA1301 vector, and PcWRKY33 was inserted into the pBI121 vector. We transformed W-box-35Smini-GUS or mW-box-35Smini-GUS alone or with 35S::PcWRKY33 into Nicotiana benthamiana leaves using the Agrobacterium-mediated transient expression system, and used the pCAMBIA1301 vector as a positive control. After the GUS histochemical staining assay, we observed that tobacco leaves transformed with pCAMBIA1301 were stained dark blue and those co-transformed with W-box-35Smini-GUS and 35S::PcWRKY33 were stained light blue. In contrast, leaves co-transformed with mW-box-35Smini-GUS and 35S::PcWRKY33 were stained only slight blue, similar to that achieved in leaves transformed with only W-box-35Smini-GUS. As expected, the leaves transformed with only 35S::PcWRKY33 did not stain blue (Fig. 2d). These findings proved once again that PcWRKY33 could combine with the W-box to regulate gene expression, in a specific manner.
Promoter analysis of PcWRKY33
To clarify which biotic and abiotic stresses regulated the expression of PcWRKY33, we isolated the 1061 bp promoter region of this gene, using the genome walking method. Using the PlantCARE database for bioinformatics analysis, we found many response elements associated with biotic and abiotic stresses, and with pathogen- and light-induced, and with tissue-specific expression (Table 1). The presence of abiotic stress response elements, such as ARE, CGTCA-motif, ERE, HSE, P-box, TC-rich repeats, TCA-element, and WUN-motif, suggested that the expression level of PcWRKY33 might be regulated by methyl jasmonate (MeJA), ethylene (ET), heat, gibberellic acid (GA), salicylic acid (SA), and wounding. The presence of the W-box in the promoter of PcWRKY33 indicated that PcWRKY33 might have interactions with other WRKY transcription factors.
PcWRKY33 expression in P. cuspidatum influenced by various stresses
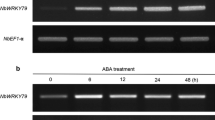
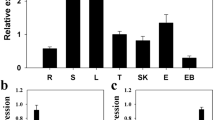
Quantification of the expression level of PcWRKY33 using qPCR revealed that expression was higher in the root than in either the stem or the leaf (Fig. 3a). Having identified cis-acting elements in the promoter of PcWRKY33, we used qPCR to analyze transcriptional changes in P. cuspidatum seedlings exposed to various abiotic stresses and hormone treatments. Results showed that ABA, GA, and salt markedly induced the expression of PcWRKY33, but its expression was significantly reduced under drought condition, while there was no significant change after MeJA treatment (Fig. 3b–f). Under GA and salt treatments, the expression profiles of PcWRKY33 showed two peaks, at 4 and 12 h, whereas, following ABA treatment, the two peaks appeared at 8 and 24 h. The greatest increases in expression level of PcWRKY33 under GA, ABA, and salt treatment were fourfold, 40-fold, and fivefold, respectively. Under drought treatment, the expression of PcWRKY33 initially increased at 2 days, but decreased rapidly at 3 days and then maintained a steady rise to the control level. These findings demonstrated that the expression of PcWRKY33 responded to various stress conditions.
Expression levels of PcWRKY33 in P. cuspidatum under various stresses. a Tissues expression, b MeJA, c GA, d ABA, e NaCl, and f Drought. The plants without any treatment were used as control, and PcActin2 was the reference gene. Each column stands for a point of time after treatment, and each experiment was repeated at least twice. Any two bars within a graph with a common letter mean they are not significantly different (P > 0.05). L leaf, S shoot, R root, CK under normal conditions, RW rewater
Increased salt sensitivity of transgenic Arabidopsis following PcWRKY33 overexpression
We constructed transgenic A. thaliana plants that overexpressed PcWRKY33 under the control of the CaMV35S promoter via Agrobacterium-mediated transformation, to further investigate the function of PcWRKY33 in plants. Seven transgenic lines were obtained using kanamycin selection (40 mg/L) and then confirmed by PCR. Then, we performed the semi-quantitative RT-PCR detection of kanamycin resistance Arabidopsis and the control plants (WT). The result showed that PcWRKY33 was expressed at the transcription level, and the expression was different in different transgenic lines (Fig. S1). Based on the result of RT-PCR detection, we selected line1, line3, and line4, which showed the highest expression of PcWRKY33, named as OE1, OE2, and OE3, respectively, for the subsequent experiments.
The results of promoter analyses and quantification of expression levels under the different treatments showed that PcWRKY33 might be involved in various stress defense responses, especially the salt stress response. To identify the function of PcWRKY33 during early plant development, the transgenic Arabidopsis seeds were surface-sterilized and germinated on 1/2 × MS agar medium containing 0, 100, or 125 mM NaCl to test the germination capacity. Under control conditions, there was no significant difference in germination rate between PcWRKY33 transgenic lines and the wild type (Fig. 4a, b). However, the PcWRKY33 transgenic lines showed a lower germination rate than the wild type under 100 and 125 mM NaCl treatments. In particular, on 1/2 × MS medium containing 125 mM NaCl, the seed germination rate of the wild type was 83.3%, while those of the three PcWRKY33 transgenic lines were only 54.8% (OE1), 66.7% (OE2), and 59.5% (OE3), respectively. Seedlings of the transgenic lines and wild type were cultured vertically on 1/2 × MS medium containing different NaCl concentrations (0, 100, or 125 mM) for 10 days, before the root lengths of the seedlings were measured. The root lengths of the wild-type and transgenic lines were similar on 1/2 × MS medium lacking NaCl, but the root lengths of the three transgenic lines were shorter than those of the wild type growing on medium containing NaCl (Fig. 4c, d). Statistical analysis confirmed that there was a significant difference in root length between the transgenic lines and wild type under conditions of salt stress.
Expression of PcWRKY33 enhanced the salt sensitivity of transgenic plants. a Germination of transgenic and wild-type plants grown on 1/2 × MS medium containing 0, 100, 125 mM NaCl. b Statistical analysis of the seed germination rate in a. c Root elongation of transgenic and wild-type plants grown on 1/2 × MS medium containing 0, 100, and 125 mM NaCl. d Graphical representation of the root length in c. e Phenotype of 4-week-old transgenic and wild-type plants after salt treatment (100 mM NaCl) for 10 days. f Statistical analysis of the survival rate in e. g Chlorophyll content of 4-week-old transgenic and wild-type plants under normal conditions and salt treatment. The mean values and standard errors were calculated based on the results of three independent experiments. One, two, and three asterisks represent significance level P < 0.05, P < 0.01, and P < 0.001, respectively. Any two bars within a graph with a common letter mean that they are not significantly different (P > 0.05). WT wild-type plants, OE overexpressed plants, CK under normal conditions, NaCl salt treatment
To confirm the function of PcWRKY33 during vegetative growth, 1-month-old plants grown in soil were watered with 100 mM NaCl solution for 10 days. The transgenic plants showed serious leaf curling and chlorosis, compared with wild-type plants under NaCl treatment (Fig. 4e). The average survival rate of transgenic plants under salt stress was only 37.4%, which was considerably lower than that of the wild-type plants (58.3%) (Fig. 4f). In addition, the leaves of transgenic and wild-type plants grown under control or salt stress were detached to measure chlorophyll concentrations. Compared with the values under control conditions, the chlorophyll concentrations of both transgenic and wild-type plants growing under salt stress declined, but the decrease in transgenic plants was greater than that in the wild-type plants (Fig. 4g).
K+/Na+ homeostasis disruption and down-regulation of stress-responsive gene expression by PcWRKY33 overexpression in Arabidopsis under salt stress treatment
Long-term exposure to salt stress can cause accumulation of Na+ in plant cells, inhibiting the absorption of K+. Since the K+/Na+ ratio is an important indicator of salt tolerance (Zhu 2003), to clarify the possible mechanism of the reduced salt tolerance exhibited by plants overexpressing PcWRKY33, we measured the concentrations of Na+ and K+ ions by ICP-OES in transgenic and wild-type plants under control conditions and under 100 mM salt treatment. The results showed that there was no obvious difference in the concentrations of Na+ and K+ between transgenic and wild-type plants under control conditions. Under the 100 mM salt treatment, however, transgenic plants accumulated more Na+, while the K+ concentration remained largely unchanged. As a consequence, the Na+/K+ ratio in transgenic plants was significantly higher than that in wild-type plants under salt stress (Fig. 5a–c).
Salt tolerance of transgenic and wild-type plants in the vegetative stage. a–c Na+ content, K+ content, and Na+/K+ ratio of transgenic and wild-type plants under normal conditions and salt treatment. The Na+/K+ ratio calculated from the rate of Na+ content and K+ content. Four-week-old transgenic and wild-type plants were irrigated every 3 days with 100 mM NaCl solution for 10 days. d–i Expression levels of stress-related genes in transgenic and wild-type plants under normal conditions and salt treatment. The mean values and standard errors were calculated based on the results of three independent experiments. One, two, and three asterisks represent significance level P < 0.05, P < 0.01, and P < 0.001, respectively. WT wild-type plants, OE overexpressed plants, CK under normal conditions, Salt after salt treatment
To further elucidate the molecular mechanism involved in the increased salt sensitivity in PcWRKY33 overexpressed (OE) plants, the expression levels of stress-related genes (AtRD29A, AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1) were determined by qPCR in the transgenic and wild-type plants exposed to 100 mM salt stress. There was no significant difference in the expressions of these genes between transgenic and wild-type plants under control conditions (Fig. 5d–i). However, under salt stress, the expressions of these genes were induced in both the transgenic and wild-type plants, but the magnitude of the induction in the transgenic plants was notably lower than that in the wild-type plants. The maximum inhibition (in the transgenic relative to the wild type) of expression of AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 occurred at 6 h after salt treatment commenced, while that of AtRD29A occurred at 4 h after treatment. The expression in the overexpressed plants of AtSOS3, the most inhibited of the stress-related genes, decreased by about 16.6 times, compared with that in the wild type (Fig. 5d–i). These findings indicated that overexpression of PcWRKY33 negatively regulated salt tolerance, possibly through decreasing the expression of stress-related genes.
The increased accumulation of ROS and oxidative damage associated with PcWRKY33 overexpression
Salt stress leads to the accumulation of ROS in plants, mainly H2O2 and O2− (Mittler 2002). We treated the detached leaves of transgenic PcWRKY33-overexpressed and wild-type plants with 3, 3′-diaminobenzidine (DAB: to detect hydrogen peroxide) or nitroblue tetrazolium (NBT: to detect the superoxide anion) to visualize the accumulation of ROS in PcWRKY33-overexpressed plants. There was no obvious staining in wild-type or transgenic plants under control conditions. However, under salt stress treatment, staining in the transgenic lines was darker than that in the wild-type plants (Fig. 6a, b). The previous studies have shown that excessive accumulation of ROS results in oxidative damage to plant cells, with the concentration of MDA being an important physiological index of oxidative damage to cell membranes. After the salt stress treatment in the present work, the MDA concentration increased in both transgenic and wild-type plants, but the increase was higher in transgenic plants than in wild-type plants (Fig. 6c).
Overexpression of PcWRKY33 increased the accumulation of ROS and reduced the activities of antioxidant enzymes. a Leaves of transgenic and wild-type plants after DAB staining for H2O2 detection under normal conditions and salt treatment. b Leaves of transgenic and wild-type plants after NBT staining for O2− detection under normal conditions and salt treatment. c MDA content of transgenic and wild-type plants under normal conditions and salt treatment. d–f Enzyme activities of SOD, POD, and CAT. The results came from three independent experiments (n = 6). The mean values and standard errors were calculated based on the results of three independent experiments. One and two asterisks represent significance level P < 0.05 and P < 0.01, respectively. Control and CK under normal conditions, salt under salt treatment, WT wild-type plants, OE overexpressed plants
Increasing the activities of ROS-scavenging enzymes is one of the main ways to alleviate oxidative damage (Bartels and Sunkar 2005). Therefore, we analyzed the activities of SOD, POD, and CAT under control and salt stress conditions. There was no significant difference between the enzyme activities of the transgenic and wild-type plants under control conditions (Fig. 6d–f). However, after exposure to salt stress, the activities of SOD, POD, and CAT in both transgenic and wild-type plants increased considerably, with the increases in activities of the ROS-scavenging enzymes being higher in the wild-type plants than in the transgenic plants (Fig. 6d–f).
Discussion
As a unique transcription factor family in plants, the WRKY transcription factor family has attracted an increasing attention in recent years. Although WRKY transcription factors have been reported to be involved in various activities in many plants, such as growth and senescence (Miao et al. 2004), responses to biotic and abiotic stresses (Li et al. 2004; Zhou et al. 2008), and development and metabolism (Devaiah et al. 2007; Kato et al. 2007), there have been no reports of WRKY transcription factors in P. cuspidatum. In the present study, the results of gene structure and phylogenetic analysis showed that PcWRKY33 belongs to the group I WRKY family (Fig. 1b). Further studies of multi-sequence alignment showed that PcWRKY33 showed the greatest similarity to VvWRKY33, MsWRKY33, and AtWRKY33 (Fig. 1a). These results confirmed that PcWRKY33 is a group I WRKY transcription factor from P. cuspidatum.
Members of the WRKY family have been reported to bind specifically to the W-box (TTGACC/T) cis-acting elements (Eulgem et al. 2000). In our studies, the results of the yeast one-hybrid assay and the transient co-expression experiment in tobacco leaves showed that PcWRKY33 has the ability to specifically bind to the W-box and to regulate the expression of downstream reporter genes (Fig. 2c, d). In addition, the PcWRKY33 protein was localized to the nucleus, also confirming its role as a transcription factor (Fig. 2a). These results suggested that PcWRKY33 might activate the expression of target genes in the nucleus by binding to the W-box in the promoters of these genes.
It has been reported that the expression of WRKY genes can be induced by various abiotic stresses (Matsui et al. 2008). Using northern hybridization, the expression profiles of AtWRKY26, AtWRKY25, and AtWRKY33 were monitored (Fu and Yu 2010), and their results showed that expression of these three genes was induced by a range of abiotic stresses, such as temperature, high salinity, and osmotic stress, as well as by ABA. In the present study, promoter analysis and expression pattern detection under various treatments indicated that the expression profiles of PcWRKY33 and AtWRKY33 were very similar, being induced by a number of abiotic stresses (Fig. 3b–f). In addition, the presence of the W-box in the promoter of PcWRKY33 suggested that transcription of PcWRKY33 might be mediated by other WRKY transcription factors to regulate gene expression (Table 1).
Salinity has become one of the most important environmental threats to plants, and its most obvious effect is on plant growth and development. In the present study, under salt stress treatment, several growth parameters (such as root length, seed germination rate, seedling survival rate, and chlorophyll concentration) in the overexpressing transgenic lines were significantly lower than those in the wild type (Fig. 4). These results indicated that PcWRKY33 might play a negative role in a plant’s response to salt stress. A high salt concentration in the soil usually disrupts K+/Na+ homeostasis in the plant. Since an environment with a high concentration of Na+ forces plants to absorb large amounts of Na+, instead of K+, which is toxic to plant growth, and which is an essential nutrient for plant cells (Hasegawa et al. 2000), the K+/Na+ ratio is an important indicator of plant tolerance to salt. Under salt stress, the extremely high Na+/K+ ratio in transgenic plants (compared with wild-type plants), caused by excessive accumulation of Na+ instead of K+, indicated that the increase in salt sensitivity associated with overexpression of PcWRKY33 was probably caused by its relatively weak ability to maintain K+/Na+ homeostasis (Fig. 5a–c).
One of the important aspects of plant stress response is the modification of the temporal and spatial expression patterns of stress-related genes (Rushton and Somssich 1998). The salt overly sensitive (SOS) signaling pathway is an important regulatory system for plant survival under salt stress. In the SOS signaling pathway, SOS3 and SOS2 interact to form a complex, and then activate SOS1. SOS1 encodes a Na+/H+ antiporter located on the plasma membrane, which can eliminate the Na+ from the cell, and transport it to various tissues of the plant simultaneously (Shi et al. 2000). Similarly, overexpression of AtNHX1 can sharply improve salt tolerance (Apse et al. 1999). There are two ways that salt stress induces the expression of stress-related genes; one is via an ABA-dependent pathway and the other is via an ABA-independent pathway (Zhu 2002). In the ABA-dependent pathway, RD29A is usually considered to be a marker gene for plant adaptation to stress, and it was reported to function in the detoxification or alleviation of stress-related damage (Zhu 2001). In the ABA-independent pathway, the expression of the DREB transcription factor could simultaneously regulate the expression of multiple stress-related genes, and participate extensively in the physiological and biochemical processes of plants (Zhu 2001).
In our study, the qPCR results showed that the increased expressions of AtRD29A, AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1, when the plant was exposed to salt stress, were inhibited in transgenic plants (Fig. 5d–i). Under salt stress treatment, AtRD29A exhibited the most marked and rapid inhibition effect, with its expression peaking at 2 h after exposure to salt stress, while the greatest inhibition of AtDREB2B, AtSOS1, AtSOS2, AtSOS3, and AtNHX1 occurred at 6 h. The time course of the induced expressions of these genes indicated that the regulation of expressions of stress-related genes may occur through both the ABA-dependent and ABA-independent pathways, with the ABA-dependent pathway responding first. In addition, after promoter analysis of these genes, we found four W-box cis-elements in the AtSOS2 promoter region, suggesting that PcWRKY33 might bind directly to the W-box in the promoter of AtSOS2 in response to salt stress. All these findings suggest that overexpression of PcWRKY33 can increase salt sensitivity through both the ABA-dependent and ABA-independent pathways, to regulate the expressions of stress-related genes. In A. thaliana, AtWRKY33 increased salt tolerance, via a route which depends not on the SOS signaling pathway, but partially on the ABA signaling pathway (Jiang and Deyholos 2009). These results indicate that the same member of the WRKY family from different plants may have different functions in response to salt stress.
Note that our reported generally decreased expression effects of stress-related genes in PcWRKY33 transgenic plants do not necessarily mean that PcWRKY33 directly inhibits the expression of genes; it is a result of complex multi-stage interaction. At this stage, whether PcWRKY33 plays a direct activation or inhibition role in transgenic plants is still an open issue, and warrants further investigation in the future.
Another mechanism that increased salt sensitivity was the accumulation of ROS and the associated decrease in the activities of ROS-scavenging enzymes. High salt concentrations in the soil affect plants through not only ionic pressure but also osmotic stress, which results in the production of ROS, mainly H2O2 and O2− (Zhu 2002). In response to drought and salt stress, ROS act as crucial signal molecules (Bhattacharjee 2005). The accumulation of ROS causes severe oxidative damage to plants, especially peroxidation of the lipids in cell membranes, resulting in a large number of degradation products, most importantly the accumulation of MDA (Moore and Roberts 1998). Our results of DBA and NBT staining in detached leaves from transgenic plants and wild-type plants showed that the transgenic plants accumulated higher levels of O2− and H2O2 than did the wild-type line (Fig. 6a, b). In addition, MDA accumulation in transgenic plants was also significantly higher than that in the wild type (Fig. 6c). All these results demonstrate that salt stress inflicts greater osmotic stress on the transgenic plants, as evidenced by the observation that the transgenic leaves accumulated more ROS. To remove excessive ROS, plants have an antioxidant enzyme system composed of multiple ROS-scavenging enzymes, such as SOD, CAT, and POD. The superoxide anion radicals in the cell are catalyzed by SOD to form O2 and H2O2, with CAT degrading H2O2 into water and molecular oxygen, while POD oxidizes co-substrates such as phenolic compounds and/or antioxidants to break down H2O2 (Gill and Tuteja 2010). In the present study, there was no obvious difference in the activity of SOD, POD, and CAT under control conditions, but, under salt stress, the activities of these enzymes increased considerably in both transgenic and wild-type plants, with the increase in the wild-type plants being higher than in the transgenic plants (Fig. 5d–f). Higher concentrations of ROS and lower activities of ROS-scavenging enzymes in transgenic plants under salt stress indicated that PcWRKY33 might alter salt sensitivity via regulation of ROS levels.
In brief, our findings suggest that PcWRKY33 may increase salt sensitivity through decreasing the expression of stress-related genes and increasing the cellular levels of ROS. Our present work seems to be the first report of WRKY transcription factors from P. cuspidatum, and provides some new insights into the functionality of WRKY transcription factors.
Author contribution statement
WQB and HW conceived and designed the research. WQB and XWW conducted the experiments. MC contributed new reagents and analytical tools. WQB analyzed the data. WQB, TYC, and HW wrote the manuscript. All authors revised and approved the manuscript.
References
Agarwal SK, Singh SS, Verma S, Kumar S (2000) Antifungal activity of anthraquinone derivatives from Rheum emodi. J Ethnopharmacol 72:43–46
Apse MP, Aharon GS, Snedden WA, Blumwald E (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285:1256
Barnard DL, Fairbairn DW, O’Neill KL, Gage TL, Sidwell RW (1995) Anti-human cytomegalovirus activity and toxicity of sulfonated anthraquinones and anthraquinone derivatives. Antivir Res 28:317–329
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Crit Rev Plant Sci 24:23–58
Bhattacharjee S (2005) Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr Sci India 89:1113–1121
Chao D, Zhao P, Zhang H, Li N, Zheng L, Wang Y (2017) The Reaumuria trigyna transcription factor RtWRKY1 confers tolerance to salt stress in transgenic Arabidopsis. J Plant Physiol 215:48–58
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cui MH et al (2013) An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett 587:1773–1778
Devaiah BN, Karthikeyan AS, Raghothama KG (2007) WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol 143:1789–1801
Duan Y et al (2015) PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep 34:831–841
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206. https://doi.org/10.1016/S1360-1385(00)01600-9
FAO (2005) Global network on integrated soil management for sustainable use of salt-affected soils. Rome, Italy: FAO Land and Plant Nutrition Management Service. http://www.fao.org/ag/agl/agll/spush
Fu QT, Yu DQ (2010) Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Hereditas 32:848–856
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Bioch 48:909–930
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Phys 51:463–499
Hu W et al (2013) TaASR1, a transcription factor gene in wheat, confers drought stress tolerance in transgenic tobacco. Plant Cell Environ 36:1449–1464
Jiang Y, Deyholos MK (2009) Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 69:91–105
Kato N et al (2007) Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol 48:8–18
Li J, Brader G, Palva ET (2004) The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16:319–331
Li HX et al (2013) Cerebroside C increases tolerance to chilling injury and alters lipid composition in wheat roots. Plos One 8:e73380
Li P et al (2015) Chrysanthemum WRKY gene CmWRKY17 negatively regulates salt stress tolerance in transgenic chrysanthemum and Arabidopsis plants. Plant Cell Rep 34:1365–1378
Liang QY et al (2017) Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci Rep 7:4799
Lichtenthaler HK, Wellburn AR (1983) Determination of total carotenoids and chlorophylls a and b of leaf in different solvents. Biochem Soc Trans 11:591–592
Liu QL, Zhong M, Li S, Pan YZ, Jiang BB, Jia Y, Zhang HQ (2013) Overexpression of a chrysanthemum transcription factor gene, DgWRKY3, in tobacco enhances tolerance to salt stress. Plant Physiol Bioch 69:27–33
Liu QL, Xu KD, Pan YZ, Jiang BB, Liu GL, Jia Y, Zhang HQ (2014) Functional analysis of a novel chrysanthemum WRKY transcription factor gene involved in salt tolerance. Plant Mol Biol Rep 32:282–289
Liu X, Song Y, Xing F, Wang N, Wen F, Zhu C (2016) GhWRKY25, a group I WRKY gene from cotton, confers differential tolerance to abiotic and biotic stresses in transgenic Nicotiana benthamiana. Protoplasma 253:1265–1281
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
Matsui A et al (2008) Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49:1135–1149
Merz PR, Moser T, Höll J, Kortekamp A, Buchholz G, Zyprian E, Bogs J (2015) The transcription factor VvWRKY33 is involved in the regulation of grapevine (Vitis vinifera) defense against the oomycete pathogen Plasmopara viticola. Physiol Plant 153:365–380
Miao Y, Laun T, Zimmermann P, Zentgraf U (2004) Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol Biol 55:853–867
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Moore K, Roberts LJ (1998) Measurement of lipid peroxidation. Free Radic Res 28:659–671
Rushton PJ, Somssich IE (1998) Transcriptional control of plant genes responsive to pathogens. Curr Opin Plant Biol 1:311–315
Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15:5690–5700
Rushton PJ et al (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the solanaceae. Plant Physiol 147:280–295
Shi H, Ishitani M, Kim C, Zhu JK (2000) The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. PNAS 97:6896–6901
Shi J, An HL, Zhang L, Gao Z, Guo XQ (2010) GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol Biol 74:1–17
Siebert PD, Chenchik A, Kellogg DE, Lukyanov KA, Lukyanov SA (1995) An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res 23:1087–1088
Su YT, Chang HL, Shyue SK, Hsu SL (2005) Emodin induces apoptosis in human lung adenocarcinoma cells through a reactive oxygen species-dependent mitochondrial signaling pathway. Biochem Pharmacol 70:229–241
Ulker B, Somssich IE (2004) WRKY transcription factors: from DNA binding towards biological function. Curr Opin Plant Biol 7:491–498
Ullah A, Sun H, Yang X, Zhang X (2017) A novel cotton WRKY-gene, GhWRKY6-like, improves salt tolerance by activating the ABA signalling pathway and scavenging of reactive oxygen species. Physiol Plant. https://doi.org/10.1111/ppl.12651
Wang Y, Shu Z, Wang W, Jiang X, Li D, Pan J, Li X (2016) CsWRKY2, a novel WRKY gene from Camellia sinensis, is involved in cold and drought stress responses. Biol Plantarum 60:1–9
Wang K et al (2017) Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front Plant Sci 8:1592. https://doi.org/10.3389/fpls.2017.01592
Xu Z, Wang C, Xue F, Zhang H, Ji W (2015) Wheat NAC transcription factor TaNAC29 is involved in response to salt stress. Plant Physiol Bioch 96:356–363
Yan H, Jia H, Chen X, Hao L, An H, Guo X (2014) The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant Cell Physiol 55:2060–2076
Yang YO, Li RG, Min Q (2010) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22:543–551
Yen GC, Duh PD, Chuang DY (2000) Antioxidant activity of anthraquinones and anthrone. Food Chem 70:437–441
Zhang Y, Wang L (2005) The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol Biol 5:1
Zhou QY et al (2008) Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J 6:486–503. https://doi.org/10.1111/j.1467-7652.2008.00336.x
Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY (2009) Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol 166:1296–1306
Zhou L, Wang NN, Gong SY, Lu R, Li Y, Li XB (2015) Overexpression of a cotton (Gossypium hirsutum) WRKY gene, GhWRKY34, in Arabidopsis enhances salt-tolerance of the transgenic plants. Plant Physiol Bioch 96:311–320
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2003) Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol 6:441–445
Zou C, Jiang W, Yu D (2010) Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J Exp Bot 61:3901–3914
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No.61672489; 61379081).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest regarding the publication of this paper.
Additional information
Communicated by Kang Chong.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bao, W., Wang, X., Chen, M. et al. A WRKY transcription factor, PcWRKY33, from Polygonum cuspidatum reduces salt tolerance in transgenic Arabidopsis thaliana. Plant Cell Rep 37, 1033–1048 (2018). https://doi.org/10.1007/s00299-018-2289-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-018-2289-2