Abstract
The increasing interest in renewable energy has attracted more research attention on biofuels. In order to generate sustainable amount of biomass feedstock from dedicated biofuel crops such as switchgrass they need to be genetically improved. Genetic transformation is one of the techniques to achieve this goal. The aim of our study was to devise a simplified protocol for switchgrass genetic transformation. We have used NB0 as the basal medium and mature seeds of the cultivar Alamo as the starting material. The nutrient medium used and scutellum-derived callus are fashioned after rice genetic transformation protocols. We obtained friable calluses, which were similar to the type II calluses in other monocotyledonous species. Calluses were amenable for Agrobacterium-mediated genetic transformation with at least 6 % transformation efficiency. The concentration of hygromycin was optimized for successful selection of transgenic calluses. The Green Fluorescent Protein gene was used to monitor and demonstrate successful genetic transformation. Compared to the previously published methods for genetic transformation of switchgrass, our protocol is simpler and equally efficient.
Key message
An efficient, simplified switchgrass genetic transformation method with NB0 basal medium and mature seeds as inoculum was developed. The appropriate concentrations of hormones and selection agent are described.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biofuels are becoming an essential alternative for fossil fuels due to their renewable nature and reduced emission of greenhouse gases. The first generation biofuels were produced mainly from corn starch, sugarcane and sugar beet. Since these food crops are cultivated in arable land, it led to socioeconomic conflicts of “food versus fuel” use of crops. Currently, the second generation biofuels from cellulosic biomass have become a vital source of alternative fuel, which are produced from agricultural waste and non-food cellulosic biofuel crops such as Miscanthus and switchgrass (Panicum virgatum L.). Among them switchgrass has been identified as a dedicated biofuel crop by US Department of Energy (Vogel and Jung 2001), which is a North American warm-season C4 perennial grass. Switchgrass grows in marginal land and gives high perennial productivity with low maintenance cost, which makes it a desirable biomass source for biofuel production (Young et al. 2010). It was reported that farm-scale production of switchgrass biomass would cost $50 per ton, which translates to $0.13 l−1 of ethanol (Perrin et al. 2008). It is important to increase the yield per unit land area in order to generate sustainable amounts of biomass and keep the biofuel production cost low. Breeding is the traditional way of improving crop plants for agronomic traits such as high yield, biotic and abiotic stress resistance. In the case of switchgrass, which is a wind-pollinated polyploid grass species with high level of self-incompatibility, it may be difficult to introduce desirable traits through the conventional breeding approach. Genetic improvement of switchgrass through biotechnological approaches may be the choice technique. Plant tissue culture and genetic transformation methods are important to genetically improve any plant species (Kumar and Loh 2012). There are numerous reports of successful genetic transformation of dicotyledonous and monocotyledonous species. More than 120 plant species from at least 35 different families were genetically modified, which includes major crops, vegetables, fruits, medicinal, ornamental and pasture plants using different transformation methods such as Agrobacterium-mediated genetic transformation, direct gene transfer to protoplast and particle bombardment (Birch 1997; Abalaka and Mohammed 2011). A few genetic transformation methods have been described for switchgrass (Burris et al. 2009; Li and Qu 2011; Somleva et al. 2002; Xi et al. 2009). Each protocol has its merits and demerits, with consistency of yielding transgenic plants in a reproducible manner in different laboratories as the main issue. Also, different protocols recommend the same media component in different amounts, for example Burris et al. (2009) recommended to use 100 mg l−1 l-proline in the medium, while Li and Qu (2011) recommended to use 2 g l−1 l-proline in the medium. Likewise different concentrations of hygromycin (50–200 mg l−1) were recommended in different protocols (Burris et al. 2009; Li and Qu 2011; Somleva et al. 2002; Xi et al. 2009). Furthermore, some protocols require multiple handling steps such as vacuum infiltration and desiccation treatments, which are not only time consuming but also may introduce contaminants to the culture. Hence, there is a need to simplify the transformation protocol for switchgrass that may be used in laboratory-scale transformation efforts.
Here, we describe a simplified method for switchgrass genetic transformation adapted from rice transformation protocol. In our protocol we use NB0 mineral formulation (Li et al. 1993; Yin and Wang 2000) as the basal medium, which includes N6 major salts (Chu et al. 1978), B5 minor salts and vitamins (Gamborg et al. 1968), 300 mg l−1 casein enzymatic hydrolysate, 500 mg l−1 l-proline, 100 mg l−1 myo-inositol, 30 g l−1 sucrose, 3 g l−1 Phytagel™, pH 5.8 for all stages of culture including callus initiation and maintenance, co-cultivation, selection of transgenic calluses and regeneration of shoots. Besides the use of a single basal medium and readily available mature seeds as inoculum, this simplified protocol does not involve any vacuum or desiccation treatments. The appropriate hormones, antibiotics and selection agents to be used are described.
Materials and methods
Plant materials
Mature dry seeds of switchgrass, Panicum virgatum L. cv. Alamo (accession 422006) obtained from US Department of Agriculture, National Plant Material Center Beltsville, MD, USA, were used in our experiments.
Experimental procedure
Induction and maintenance of callus from mature dry seeds
The mature dry seeds were scarified and surface sterilized as described by Somleva et al. (2002). The seeds were scarified with 60 % sulfuric acid (H2SO4) for 30 min in a glass flask on a shaker and the H2SO4 carefully discarded. The scarified seeds were rinsed with water and surface sterilized with 50 % (v/v) Clorox® containing 0.1 % Tirton X-100 for 30 min. The seeds were washed about ten times with sterile distilled water for complete removal of the chemicals. The aseptic seeds were blotted on sterile tissue paper and placed on NB0 basal medium (Li et al. 1993; Yin and Wang 2000) which consists of N6 major salts (Chu et al. 1978), B5 minor salts and vitamins (Gamborg et al. 1968), 300 mg l−1 casein enzymatic hydrolysate (Sigma Cat. No. C7290), 500 mg l−1 l-proline (Sigma Cat. No. P0380), 100 mg l−1 myo-inositol (Sigma Cat. No. I3011), 30 g l−1 sucrose, 3 g l−1 Phytagel™. The pH was adjusted to 5.8 with 0.1 M KOH prior to autoclaving. This NB0 basal medium was used for callus induction, selection and regeneration with appropriate plant growth hormones. The hormones were filter sterilized and added after autoclaving. Different concentrations of 2,4-dichlorophenoxyacetic acid (2,4-D) at 2, 3, 4, 5 and 6 mg l−1 plus 0.2 mg l−1 N 6-benzyladenine (BA) were tested. Ten to 12 seeds were placed per plate and at least 10 plates were used per experiment, which was repeated twice. The culture plates were sealed with 3 M Micropore™ tape (3 M Health Care, Ref. No. 1530-0) and incubated in the dark at 26 °C. Calluses arising from the zygotic embryo region were subcultured after 8 weeks on NB0 medium with 3 mg l−1 2,4-D plus 0.2 mg l−1 BA. These were maintained by subculturing onto fresh plates containing callus induction medium (25 callus pieces per plate, each callus piece was 3–5 mm across) at 2- to 3-week intervals.
Regeneration of switchgrass plantlets from calluses
The subcultured calluses were transferred to regeneration medium consisting of NB0 basal medium with 2 mg l−1 BA, 1 mg l−1 indole-3-acetic acid (IAA), 1 mg l−1 kinetin and 1 mg l−1 α-naphthaleneacetic acid (NAA) and incubated at 26 °C under 16-h light (about 2,000 lx) and 8-h dark period for 3–4 weeks. Subsequently, the regenerated shoots were transferred to ½ Murashige and Skoog (MS) medium for rooting (Murashige and Skoog 1962). Once roots developed in 1–2 weeks, the plantlets were acclimatized (by gradual opening of the lids) and transferred to the greenhouse with natural light. Three petri dishes, each with five callus pieces as the inoculum, were used per experiment and the experiment was repeated once.
Hygromycin sensitivity of switchgrass callus
We determined the hygromycin sensitivity of switchgrass cv. Alamo callus by growing callus pieces on medium containing hygromycin (the selection agent) at various concentrations 0, 25, 50, 75, 100 and 200 mg l−1. The fresh weights of the callus pieces at the start of culture and at the end of 2 weeks were recorded. Subsequently, the increase in fresh weight was calculated to serve as an indication of the concentration of hygromycin that can effectively block cell proliferation. Three petri dishes, each with 16 callus pieces as the inoculum, were used per experiment and the experiment was repeated twice. Mean ± SD values from three independent experiments are presented. The data were subjected to Student’s t test to determine the mean values that were significantly different from each other at P ≥ 0.05.
Agrobacterium-mediated genetic transformation
Agrobacterium-mediated transformation protocol was modified from that described for rice by Hiei et al. (1994). Actively growing calluses were used as explants for transformation with Agrobacterium tumefaciens (strain AGL1) harboring the synthetic Green Fluorescent Protein gene (sGFP as the reporter gene) driven by maize UBIQUITIN promoter in binary vector pCAMBIA1300 (Chiu et al. 1996; Kolesnik et al. 2004). Agrobacterium harboring sGFP construct was grown in 3 ml of LB medium with appropriate antibiotics for 48 h at 28 °C with rotary shaking (250 rpm) then transferred to 50 ml of LB medium and grown under the same conditions as the 3 ml culture for 5–6 h. This Agrobacterium culture was centrifuged at 4,000 rpm for 15 min at 28 °C. The pellet was washed and resuspended in 25 ml of liquid NB0 medium and induced with 100 μM acetosyringone (3′,5′-dimethoxy-4′-hydroxyacetophenone, Sigma Cat. No. D134406) for 15 min. The calluses were immersed in the induced Agrobacterium suspension for 15 min and excess bacteria were removed by blotting with sterile tissue paper then transferred onto the co-cultivation medium (NB0 with 3 mg l−1 2,4-D, 0.2 mg l−1 BA and 100 μM acetosyringone) in the dark at 26 °C for 3 days. Co-cultivation was carried out with two independent experiments with 150 and 300 callus pieces in the first and second round, respectively. After co-cultivation, the calluses were washed with sterile water to remove the surface-dwelling Agrobacterium (during this process there were losses of a few callus pieces) and blotted dry on sterile tissue paper. Subsequently, callus pieces were transferred (25 pieces per petri dish) onto selection medium [NB0 with 3 mg l−1 2,4-D, 0.2 mg l−1 BA, 400 mg l−1 cefotaxime and 75 mg l−1 hygromycin B (A. G. Scientific, San Diego, CA, USA, CAS 31282-04-9)]. The plates were incubated in the dark at 26 °C. Calluses were transferred onto fresh selection plates every 2 weeks or earlier if Agrobacterium growth was seen. Once hygromycin resistant calluses (putative transgenic) were obtained, they were transferred to fresh selection plates. Calluses from 10 independent transformation events were inoculated per petri dish for another round of selection. After the final round of selection, they were transferred (5 calluses per plate) to regeneration medium (NB0 basal medium with 2 mg l−1 BA, 1 mg l−1 IAA, 1 mg l−1 kinetin, 1 mg l−1 NAA and 75 mg l−1 hygromycin) and placed at 26 °C under 16-h light (about 2,000 lx) and 8-h dark period for 3–4 weeks. Developed shoots were then rooted in ½ MS medium with 50 mg l−1 hygromycin. The rooted plantlets were acclimatized and transferred to pots filled with Ready Mixed Potting Medium (GREEN & SCENE, Jakarta, Indonesia) and grown in the greenhouse.
Visualization of GFP expression
The GFP fluorescence was visualized using OLYMPUS SZX12 microscope and photographed using either Nikon Coolpix 4500 camera attached to the microscope or a Nikon Digital camera connected to a computer with Nikon ACT-1 software.
We observed masking of the green fluorescence in the green plantlets by the chlorophyll in the early trials. In order to avoid the interference, we subjected the plantlets to etiolation by incubating them in the dark. Putative transgenic plantlets were transferred to ½ MS medium for rooting and kept in a cardboard box and covered with black polyethylene bag for 2 weeks. Subsequently, the green fluorescence was readily viewable under the fluorescence microscope.
Genomic Southern blot analysis of the transgenic plants
Southern blot analysis was performed to confirm stable integration of the transgene and to check the copy number of T-DNA insertion in the transgenic plants. Genomic DNA (5 μg per lane) was digested with HindIII restriction enzyme, fractionated on 1 % (w/v) agarose gel and transferred onto a Hybond-N+ membrane (Amersham). The DNA blot was hybridized with digoxigenin-labeled (DIG) HPT fragment (745 bp) as the probe, which was synthesized using PCR DIG Probe Synthesis Kit (Roche Cat. No. 11636090910) with primers HPT-F: CAACCAAGCTCTGATAGAGT, HPT-R: GAAGAATCTCGTGCTTTCA in DIG Easy Hyb solution (Roche Cat. No. 116035580 01) at 42 °C. After hybridization, the membrane was washed twice with 2× SSC and 0.1 % SDS for 15 min, then twice with 0.5× SSC and 0.1 % SDS for 15 min at 68 °C. Detection was carried out according to the manufacturer’s protocol using DIG Wash and Block Buffer Set and chemiluminescent substrate CDP-Star™ (Roche Cat. No. 11685627001).
Gene expression analysis by real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using Qiagen RNeasy Mini Kit (Cat. No. 74904) from leaves according to the manufacturer’s instructions. First strand cDNA synthesis was carried out from 2 μg of total RNA using Maxima® First Strand cDNA Synthesis Kit (Fermentas, Cat. No. K1641) as per the manufacturer’s protocol.
RT-qPCR analyses were performed to check the expression of sGFP using Applied Biosystems (ABI) StepOne™ Real-Time PCR System with denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 1 min. Triplicate quantitative assays were performed on 1 μl (~20 ng) of each cDNA dilution using the Fast SYBR® Green Master Mix (ABI, P/N 4385612) with primers sGFP-F: GCCACAAGTTCAGCGTGTCC, sGFP-R: CCAGCAGGACCATGTGATCG according to the manufacturer’s protocol. The threshold cycle (C T) value was automatically calculated based on the changes in fluorescence of SYBR® Green in every cycle monitored by the StepOne™ Real-Time PCR Software v2.0. The amplification of switchgrass 18S ribosomal RNA gene (Pv18S rRNA) with primers Pv18SrRNA-F: CATGATAACTCGATGGATCGCA and Pv18sRNA-R: TCCTTGGATGTAGTAGCCGTTT was used as an endogenous control. The expression values were calculated as relative quantity (RQ) using the StepOne™ Real-Time PCR System.
Results and discussion
Optimization of callus induction and regeneration
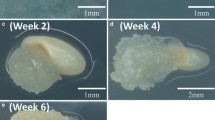
We routinely use NB0 basal medium with 2 mg l−1 of 2,4-D to initiate rice callus in our laboratory. Also, previous reports of switchgrass transformation mostly used MS basal medium with 5 mg l−1 of 2,4-D with or without BA. Based on these information, scarified and surface sterilized switchgrass seeds were cultured on NB0 basal medium containing 2, 3, 4, 5 or 6 mg l−1 of 2,4-D with 0.2 mg l−1 of BA for callus induction screen. Friable calluses were seen in all concentrations above 3 mg l−1 of 2,4-D with 0.2 mg l−1 of BA (Fig. 1a). These were morphologically similar to type II calluses described earlier (Burris et al. 2009). Based on this observation, NB0 basal medium containing 3 mg l−1 of 2,4-D with 0.2 mg l−1 of BA was selected for further use. Only 57–66 % of the inoculated seeds responded by germination, which reflects the percentage viability of the batch of seeds used. Up to 90 % of the germinated seeds produced calluses, out of which about 60 % were type II friable calluses. Calluses were competent to regenerate shoots on regeneration medium (Fig. 1b, c). All calluses tested for regeneration produced shoots, but up to 10 % of them were albino shoots. Green shoots were successfully rooted on ½ MS medium and plantlets were acclimatized and planted in pots as mentioned in the methods and grown in the greenhouse. All the acclimatized plantlets survived (Fig. 1d, e).
Different stages of switchgrass callus induction and regeneration of wild-type plantlets. a Induction of friable calluses from mature seeds at the end of 8 weeks of culture. b Early stages of shoot initiation after 1-week incubation of the callus on regeneration medium. Shoot buds were visible initially as purple spots and later as green buds. c Plantlets produced at the end of 4 weeks on the regeneration medium. d Rooted plantlets 4 weeks after transfer of the shoots to ½ MS medium. e Rooted plantlets were acclimatized and transferred to the greenhouse. Two-month-old plants in pots of 23 cm internal diameter
Optimization of hygromycin concentration for selection of transgenic calluses
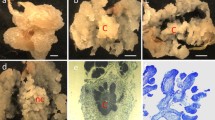
The desirable hygromycin concentration for efficient selection of transgenic plants was optimized by growing callus pieces on medium supplemented with hygromycin ranging from 0 to 200 mg l−1 (Fig. 2). Cell proliferation was observed in the media with low hygromycin concentrations such as 25 and 50 mg l−1 compared to the control with no hygromycin (Fig. 2a–c) and inhibition of callus growth was observed from 75 mg l−1 onwards (Fig. 2d). Hence, 75–100 mg l−1 will be ideal concentration for selection of switchgrass (Alamo) transgenic calluses. Based on this, we used 75 mg l−1 of hygromycin for our selections.
Growth inhibition analysis of wild-type callus on medium containing different concentrations of hygromycin. a 0, b 25, c 50 and d 75 mg l−1 of hygromycin. Wild-type calluses were able to proliferate when the hygromycin concentration was lower than 75 mg l−1. e Increase in fresh weight of wild-type callus cells after 2 weeks of culture on medium containing various concentrations of hygromycin. Data are mean ± SD of one experiment (n = 48), and Mean values are significantly different (*P ≤ 0.05) according to Student’s t test
Agrobacterium-mediated genetic transformation
We carried out two independent rounds of transformation experiments. In the first round of transformation 10 independent (putative) transgenic events were obtained from 100 callus pieces infected with Agrobacterium harboring the sGFP gene. Six of the ten independent events regenerated transgenic plants. The calculated transformation efficiency for callus formation is 10 %, but if we consider regenerated plants the efficiency is 6 %, which is better than that of the earlier reports of 3.4 % (Somleva et al. 2002) and 4.4 % (Burris et al. 2009) transformation efficiencies. In the second round of transformation experiment, 30 independent (putative) transgenic calluses were obtained from 250 calluses infected. From these, 18 events yielded transgenic plants, and among them 3 lines exhibited albino phenotype. Such regeneration of albino shoots was also reported earlier in switchgrass cv. Alamo (Li and Qu 2011).
Selection of transgenic calluses using GFP fluorescence and regeneration of shoots
The stable transgene expression was monitored using the reporter gene sGFP. After 1 week of selection, the co-cultivated calluses were observed under fluorescence microscope. A 2-week-old transgenic callus on selection medium showing GFP fluorescence is shown in Fig. 3 under white (Fig. 3a) and blue lights (Fig. 3b). These GFP fluorescing calluses were subcultured onto fresh selection medium for further growth. After 2 weeks, actively growing calluses with GFP fluorescence throughout the callus mass were obtained (Fig. 3c). These calluses were then transferred to regeneration medium for shoot induction. The regenerated shoots were examined under white (Fig. 3d, f, h) and blue lights (Fig. 3e, g, i) for GFP fluorescence. The green fluorescence observed in the developing shoots was not as bright as that seen in the calluses (Fig. 3c). Masking of GFP fluorescence was observed in the developed shoots due to the accumulation of chlorophyll as shown in Fig. 3, under white (Fig. 3j) and blue lights (Fig. 3k). To avoid the masking of GFP by chlorophyll, we etiolated the plantlets during rooting. After 2 weeks of etiolation, higher levels of GFP fluorescence were observed in the plantlets (Fig. 4a–d). Plantlets were then grown under 16 h/8 h light/dark cycle for 2 more weeks before acclimatization and potting. Green fluorescence was also observed in the roots of plantlets (Fig. 4e–g), indicating that the transgene was stably integrated.
Transgenic calluses and plantlets expressing green fluorescent protein (GFP). Two-week-old transgenic calluses on selection medium a under white light and b under blue light. c Four-week-old transgenic callus under blue light. Plantlets expressing GFP protein observed d, f, h under white light and e, g, i under blue light. Masking of the GFP fluorescence at the base of a plantlet due to the presence of chlorophyll j under white light and k blue light
We used genomic Southern blot analysis to confirm the stable integration of the transgene into 11 independent lines. Hybridization data showed that different lines had one to four copies of transgene integrated (Fig. 5a). Furthermore, the expression of sGFP was analyzed at the transcript level by RT-qPCR. The sGFP transcripts were detected and quantified over the endogenous control Pv18S rRNA transcripts (Fig. 5b). Our transgene integration and expression analyses confirmed stable integration of sGFP demonstrating the effectiveness of the transformation protocol described.
Switchgrass is a desirable bioethanol species mainly because of the relatively high growth rate and also due to the fact that the net energy derived from its biomass is about 13 times higher than input energy resulting in 94 % lower net emission of greenhouse gases (Schmer et al. 2008). Consideration of switchgrass as a dedicated biomass crop for bioethanol production by US Department of Energy (Vogel and Jung 2001) and due to its high perennial productivity with low maintenance cost (Young et al. 2010) have drawn more attention towards improving the crop. These include introduction of desirable traits such as biotic and abiotic stress tolerance, modification of cell wall content with less lignin, which leads to easy release of cellulose for ethanol production and biomass increase (Fu et al. 2011, 2012; Mann et al. 2011; Shen et al. 2012). This species had not received adequate attention from plant breeders in the past. The traditional plant breeding approaches are lengthy processes that are well established for the improvement of food crops such as rice and wheat (Peng et al. 1999). Thus far, there have been only limited efforts to improve switchgrass through breeding, such as for its forage nutritional quality (McLaughlin et al. 1999; Vogel et al. 1989). Also, there are other issues like self-incompatibility in switchgrass (Martinez-Reyna and Vogel 2002) which makes cross hybridization difficult to achieve. Hence, biotechnological approaches such as those described here will significantly speed up the process of improvement of biofuel crops such as switchgrass.
A few genetic transformation protocols have been reported for switchgrass with differing results (Burris et al. 2009; Li and Qu 2011; Somleva et al. 2002; Xi et al. 2009). Also, the protocols require additional handling steps, making them more involved procedures. Therefore, we have established a simplified genetic transformation protocol based on rice transformation experience, which utilizes NB0 (Li et al. 1993; Yin and Wang 2000) as the basal medium. The overview of the genetic transformation protocol is shown as a flow chart (Fig. 6). The efficiency for transgenic callus production was 10–12 % when calculated as putative transgenic calluses obtained versus the number of co-cultivated calluses in the first and second rounds, respectively. The transformation efficiency is 7.2 % when calculated for recovery of all transgenic plants including the albino plantlets. However, when the albino plantlets are excluded the efficiency is 6 %, which is significantly higher than the earlier reported transformation efficiencies. Meanwhile, one of the recent reports of a high throughput Agrobacterium-mediated transformation of switchgrass cv. Alamo (Li and Qu 2011) claimed up to 50 % transformation efficiency. However, their method used several additional steps such as vacuum application and desiccation during the Agrobacterium co-cultivation. The simplified protocol presented here is ideal to generate transgenic plants in routine laboratory-scale experiments.
Abbreviations
- 2,4-D :
-
2,4-Dichlorophenoxyacetic acid
- BA:
-
N 6-Benzyladenine
- sGFP:
-
Synthetic green fluorescent protein
- HPT:
-
Hygromycin phosphotransferase
- KOH:
-
Potassium hydroxide
- MS:
-
Murashige and Skoog
References
Abalaka ME, Mohammed A (2011) Agrobacterium transformation: a boost to agricultural biotechnology. J Med Genet Genomics 3:126–130
Birch RG (1997) Plant transformation: problems and strategies for practical application. Annu Rev Plant Physiol Plant Mol Biol 48:297–326
Burris JN, Mann DGJ, Joyce BL, Stewart CN (2009) An Improved tissue culture system for embryogenic callus production and plant regeneration in switchgrass (Panicum virgatum L.). Bioenerg Res 2:267–274
Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Chu CC, Wang CC, Sun CS (1978) The N6 medium and its application to anther culture of cereal crops. In: Proceedings of symposium on plant tissue culture. Science Press, Peking, pp 45–50
Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M, Chen F, Foston M, Ragauskas A, Bouton J, Dixon RA, Wang ZY (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108:3803–3808
Fu C, Sunkar R, Zhou C, Shen H, Zhang JY, Matts J, Wolf J, Mann DGJ, Stewart CN Jr, Tang Y, Wang ZY (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6:271–282
Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S (2004) Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J 37:301–314
Kumar PP, Loh CS (2012) Plant tissue culture for biotechnology. In: Altman A, Hasegawa PM (eds) Plant biotechnology and agriculture. Academic Press, Amsterdam, pp 131–138
Li R, Qu R (2011) High throughput Agrobacterium-mediated switchgrass transformation. Biomass Bioenerg 35:1046–1054
Li L, Qu R, Kochko A, Fauquet C, Beachy RN (1993) An improved rice transformation system using the biolistic method. Plant Cell Rep 12:250–255
Mann DGJ, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, Bennetzen JL, Parrott WA, Stewart CN (2011) Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol 11:74
Martinez-Reyna JM, Vogel KP (2002) Incompatibility systems in switchgrass. Crop Sci 42:1800–1805
McLaughlin S, Bouton J, Bransby D, Conger B, Ocumpaugh W, Parrish D, Taliaferro C, Vogel K, Wullschleger S (1999) Developing switchgrass as a bioenergy crop. In: Janick J (ed) Perspectives on new crops and new uses. ASHS Press, Alexandria, pp 282–299
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio-assay with tobacco tissue cultures. Physiol Plant 15:473–497
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400:256–261
Perrin R, Vogel K, Schmer M, Mitchell R (2008) Farm-scale production cost of switchgrass for biomass. BioEnerg Res 1:91–97
Schmer MR, Vogel KP, Mitchell RB, Perrin RK (2008) Net energy of cellulosic ethanol from switchgrass. Proc Natl Acad Sci USA 105:464–469
Shen H, He X, Poovaiah CR, Wuddineh WA, Ma J, Mann DGJ, Wang H, Jackson L, Tang Y, Neal Stewart C, Chen F, Dixon RA (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193:121–136
Somleva MN, Tomaszewski Z, Conger BV (2002) Agrobacterium-mediated genetic transformation of switchgrass. Crop Sci 42:2080–2087
Vogel KP, Jung HG (2001) Genetic modification of herbaceous plants for feed and fuel. Crit Rev Plant Sci 20:15–49
Vogel KP, Gon HJ, HF A (1989) Breeding grasses for the future. In: Sleper DA, Asay KH, Pedersen JF (eds) Contributions from breeding forage and turf grasses. Crop Society of America, Madison, pp 105–122
Xi Y, Fu C, Ge Y, Nandakumar R, Hisano H, Bouton J, Wang Z-Y (2009) Agrobacterium-mediated transformation of switchgrass and inheritance of the transgenes. BioEnerg Res 2:275–283
Yin Z, Wang G-L (2000) Evidence of multiple complex patterns of T-DNA integration into the rice genome. Theor Appl Genet 100:461–470
Young HA, Hernlem BJ, Anderton AL, Lanzatella CL, Tobias CM (2010) Dihaploid stocks of switchgrass isolated by a screening approach. BioEnerg Res 3:305–313
Acknowledgments
This work was funded by the Science and Engineering Research Council (SERC Grant No.: 0921390036) of the Agency for Science Technology and Research, Singapore; and the National University of Singapore.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Lakshmanan.
Rights and permissions
About this article
Cite this article
Ramamoorthy, R., Kumar, P.P. A simplified protocol for genetic transformation of switchgrass (Panicum virgatum L.). Plant Cell Rep 31, 1923–1931 (2012). https://doi.org/10.1007/s00299-012-1305-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-012-1305-1