Abstract
Chromatin remodeling-related proteins, such as linker histone H1, involving in fruit ripening and stress responses are poorly understood. In the present study, a novel cDNA encoding linker histone H1 gene, designated as MaHIS1 was isolated and characterized from banana fruit. The full-length cDNA sequence was 1,253 bp with an open-reading frame (ORF) of 948 bp, encoding 315 amino acids with a molecular weight of 31.98 kDa and a theoretical isoelectric point of 10.67. Subcellular localization analysis showed that MaHIS1 was a nucleus-localized protein. Real-time PCR analysis indicated that expression of MaHIS1 gene is induced by external and internal ethylene during fruit postharvest ripening. Accumulation of MaHIS1 transcript was also obviously enhanced by exogenous hormones, including methyl jasmonate, abscisic acid, and hydrogen peroxide (H2O2), as well as stresses, such as chilling and pathogen Colletotrichum musae infection. Moreover, yeast two-hybrid and bimolecular fluorescence complementation assays showed that MaHIS1 could interact with a transcription factor (TF) MaWRKY1. Taken together, our results suggest that MaHIS1 may be related to ripening and stress responses of banana fruit, and be likely functionally coordinating with MaWRKY1 in these physiological processes.
Key message MaHIS1 may be related to ripening and stress responses of banana fruit, and it also could interact with WRKY TF, which expands the very limited information regarding the functions of linker histone H1 in fruits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plants, as sessile organisms, have evolved complicated adaptation mechanisms to cope with regular environmental variations and to tolerate environmental stresses. Molecular and genomic studies have revealed that thousands of genes respond to stresses at the transcriptional level. Among them, transcription factors (TFs), many stress-inducible genes with various functions, protein phosphatases, and protein kinases are involved in the plant stress responses (Lee et al. 2005; Mitsuda and Ohme-Takagi 2009; Bartels et al. 2010; Kim et al. 2010). Increasing evidences have suggested that regulation of stress-responsive genes often depends on chromatin remodeling, i.e., the process inducing changes in chromatin structure (Chinnusamy et al. 2008; Chinnusamy and Zhu 2009; Kim et al. 2010; Luo et al. 2011; Zhu et al. 2011). These changes in chromatin status could have either positive or negative impact on gene expression. Changes in chromatin structure are related with modification of the N-terminal tails of histones (Strahl and Allis 2000; Fischle et al. 2003). Several chromatin-related proteins, such as histone modification enzymes, components of chromatin remodeling complex, and linker histone H1, are involved in this process (Kadonaga 1998; Verbsky and Richards 2001; Kim et al. 2010; Zhu et al. 2011). Histone modification enzymes, including histone acetyltransferase (HAT), histone deacetylase (HDAC), histone methyltransferase (HMT), and histone demethylase (HDM), which are essential factors for chromatin remodeling have been well documented in Arabidopsis (Kim et al. 2010; Luo et al. 2011). However, less attention has been paid to linker histone H1.
Linker histone H1 or Histone H1 is the fifth class of histones, with highly basic proteins, belonging to histone H1 family. The linker histone H1 is more variable than the core histones (Histones H2A, H2B, H3, and H4), and exhibits significant variations in sequence (Zhu et al. 2011). Linker histone H1 has been thought to play a structural role in chromatin organization and in the general repression of gene expression (Scippa et al. 2000; Izzo et al. 2008). It binds to the linker DNA between nucleosome cores, thus promoting the compaction of chromatin (Ivanchenko et al. 1997; Bassett et al. 2009; Kim et al. 2010). It may control specific processes during development and in response to the environment through alterations in expression of cell-type or stage-specific H1 variants (Jerzmanowski et al. 2000; Scippa et al. 2004). Moreover, some reports have suggested that linker histone H1 belongs to stress-induced genes and plays an important role in adaptive stress responses to environmental stimuli in plants (Wei and O’Connell 1996; Ascenzi and Gantt 1997; Scippa et al. 2000; Kreps et al. 2002; Neilson et al. 2011). However, whether linker histone H1 is involved in fruit ripening or stress responses, especially those of economic fruits, is largely unknown.
Banana (Musa acuminate) is the second ranking fruit crop in the world, and is the staple food of over 400 million people in the developing countries, as not only a popular dessert fruit, but also a source of vital carbohydrate. However, postharvest problems, including rapid ripening, particularly susceptible to biotic and abiotic stresses, are generally observed and account for severe losses (Chen et al. 2008, 2011). As a result, the resolve of postharvest problems is important for maintaining banana quality and extending shelf life. Thus, understanding the molecular mechanisms of banana fruit ripening and stress responses is highly desirable; nevertheless, no evidence of linker histone H1 involving in fruit ripening and stress responses has been reported. In the present study, a linker histone H1 gene, designated as MaHIS1 was isolated and characterized from banana fruit for the first time. The expression patterns of MaHIS1 during natural, ethylene-induced and 1-methylcyclopropene (1-MCP) inhibited ripening, as well as in response to hormones, including methyl jasmonate (MeJA), abscisic acid (ABA) and hydrogen peroxide (H2O2), and stresses, such as chilling and pathogen Colletotrichum musae infection, were also investigated. Moreover, the direct interaction between MaHIS1 and a WRKY TF, MaWRKY1 was detected by yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays. Our results suggest that MaHIS1 may be related to ripening and stress responses of banana fruit, and be likely functionally coordinating with WRKY TF in these physiological processes, which expand the very limited information regarding the possible functions of linker histone H1 in fruits.
Materials and methods
Plant materials
Pre-climacteric banana (Musa acuminata AAA Group, cv. Cavendish) fruit at the 75–80 % plump stage were obtained from a local commercial plantation near Guangzhou, south-eastern China. Hands were separated into individual fingers. Fruit were selected for freedom from visual defects and for uniformity of weight, shape, and maturity. The selected fruit were first surface-sterilized by dipping in a 1 % hypochlorite solution for 1 min and then immersed in 0.05 % Sporgon (with 46 % Prochloraz–Mn; Aventis, Valencia, Spain) for 3 min to prevent fungal disease. They were then allowed to air-dry at 25 °C for 2 h and treated as follows. In addition, root, leaf, petal, and the whole fruit (about 3 weeks after anthesis) were also collected to compare the expression difference of MaHIS1 in different tissues.
Treatments
For postharvest ripening samples, the selected banana fruit were randomly divided into three groups of 100 fingers each for the following treatments, and 10 bags of 10 fingers each were used for each treatment. Group 1 (control, non-conditioned for natural ripening) fruit were directly stored at 22 °C and 90 % relative humidity (RH) for 25 days; samples were taken at 0, 1, 3, 5, 7, 15, 19, 21, and 25 days until the fruit ripened completely. Fruit of group 2 were treated with 100 μl/L ethylene for 18 h in a closed chamber and then stored at 22 °C for 7 days; samples were taken at 0, 1, 3, 5, and 7 days of storage. Fruit of group 3 were treated with 0.5 μl/l 1-MCP for 18 h and then stored at 22 °C for 35 days, samples were taken at 0, 1, 3, 5, 7, 21, 25, 30, and 35 days of storage. For each treatment, the samples were taken based on the rate of internal ethylene production and fruit firmness changes during ripening (data not shown), and the pulp of banana fruit were separated and collected.
For hormone treatments, the selected banana fruit were randomly divided into three groups of 50 fingers each. These three group fruit were treated for 30 min in 10 l of distilled water containing 0.1 mM MeJA, 0.1 mM ABA or 2 mM H2O2 respectively, under a reduced pressure of about 0.1 MPa as described by Chen et al. (2008). After treatment, fruit were subsequently placed into five individual unsealed polyethylene plastic bags (0.01 mm thickness) and stored at 22 °C and 90 % relative humidity (RH) for 24 h. Samples were taken at 0 (control), 6, 12, 24 h, and peel of banana fruit were collected.
For stress treatments, the selected banana fruit were placed into unsealed plastic bags and transferred to 7 °C for chilling stress. For biotic stress treatment, fruit were inoculated with 20 μl (1.0 × 105 spores/ml) of C. musae spores in suspension as described by Chen et al. (2011). The samples were taken at 0 (control), 1, 3, 5, and 7 days after each treatment and the peel of banana fruit were collected.
All of the samples were frozen in liquid nitrogen immediately after sampling, and stored at −80 °C for further use. All assessments were conducted in three biological replicates.
RNA extraction, gene isolation, and bioinformatics analysis
Frozen tissues were ground in liquid nitrogen using a mortar and a pestle. Total RNA was extracted using the hot borate method of Wan and Wilkins (1994). Potentially contaminating DNA was eliminated by treatment with DNAse I digestion using the RNAse-free kit (Promega, Madison, WI, USA). The DNA-free total RNA was then used as template for reverse transcription-PCR (RT-PCR). The first-strand cDNA of the product was subjected to PCR amplification.
MaHIS1 full-length cDNA sequence was selected from our transcriptome database obtained by high-throughput Solexa/Illumina sequencing platform (Beijing Genomics Institute, Shenzhen, China) and the sequence was firstly verified by re-cloning and re-sequencing. Alignments were carried out on Clustalx 1.83 and GeneDoc software. The theoretical isoelectric points (pIs) and mass values for mature peptides were calculated using the PeptideMass program (http://us.expasy.org/tools/peptidemass.html).
Subcellular localization analysis
The coding region sequence of the MaHIS1 gene without stop codon was amplified by PCR with the specific primers (forward, 5′- CGGGATCCATGGTGCAGGAGGAGACTGAC -3′, BamHI site underlined, and reverse, 5′- CCCAAGCTTCTTCTTGGCTTTCTTCGTCGTCG -3′, HindIII site underlined) and subcloned into the BamHI/HindIII sites of pBI221-GFP vector, in-frame with the green fluorescent protein (GFP) sequence, resulting in the 35S:: gene-GFP vector under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The fusion construct and the control GFP vector were used for transient assays by a modified PEG transfection of tobacco BY-2 suspension culture cell protoplasts as described previously (Abel and Theologis 1994). GFP fluorescence was observed with a fluorescence microscope (Zeiss Axioskop 2 Plus). All transient expression assays were repeated at least three times.
Quantitative real-time PCR analysis
Total RNA from the samples was isolated and first-strand cDNA was synthesized as described above. The synthesized cDNA was diluted 1:40 with water, and 2 μl of the diluted cDNA was used as a template for quantitative real-time PCR analysis. PCR was performed in a total volume of 20 μl, 1 μl for each primer (10 μM, final concentration 200 nM) and 10 μl for SYBR®Green PCR Supermix (Bio-Rad Laboratories) on an Bio-Rad CFX96 Real-Time PCR System according to the manufacturer’s instructions. The RT-qPCR programme included an initial denaturation step at 94 °C for 5 min, followed by 40 cycles of 10 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C. No-template controls for each primer pair were included in each run. The oligonucleotide primers for RT-qPCR analysis were designed on the basis of the 3′-untranslated region using Primer5.0 software. The primer sequences used for MaHIS1 were as follows: forward, 5′-GCGAAGAAGAATACCAGGAAGG-3′ and reverse, 5′-ACAACAGACTACCCCCGTGA-3′. RPS2 and CAC were selected as reference genes under different series samples according to our previous study on the selection of reliable reference genes for expression study by RT-qPCR in banana fruit (Chen et al. 2011). Each assay using the gene-specific primers amplified a single product of correct size with high PCR efficiency (90–110 %). All RT-qPCR reactions were normalized using Ct value corresponding to the reference gene. The relative expression levels of target gene were calculated with formula 2−ΔΔCT (Livak and Schmittgen 2001). Values represented the average of three biological replicate.
Yeast two-hybrid assay
Yeast two-hybrid assays were performed using the Matchmaker GAL4-based two-hybrid system 3 (Clontech, USA). The ORFs of MaHIS1 and MaWRKY1 were amplified by PCR and subcloned into pGBKT7 and pGADT7 vectors as bait and prey, respectively. Construct pairs including pGBKT7-53 + pGADT7-T (positive control), pGBKT7-Lam + pGADT7-T (negative control), pGBKT7-MaHIS1 + pGADT7-MaWRKY1, pGBKT7-MaHIS1 + pGADT7 and pGBKT7 + pGADT7-MaWRKY1 were transformed into the yeast strain AH109 by the lithium acetate method. Transformed yeast cells are first grown up on a minimal medium/-Leu-Trp (SD/-Leu-Trp) according to the manufacturer’s instructions (Clontech), and then plated onto a minimal medium/-Leu-Trp-His-Ade (SD/-Leu-Trp-His-Ade) and SD/-Leu-Trp-His-Ade containing 4 mg/ml X-α-Gal at 30 °C to test the possible interactions between MaHIS1 and MaWRKY1.
BiFC assay
To generate the constructs for BiFC assay, full-length coding sequences of MaHIS1 and MaWRKY1 (without their stop codons) were subcloned into pUC-pSPYNE or pUC-pSPYCE vectors obtained from the Harter and Kudla’s laboratory (Walter et al. 2004), respectively. Expressions of target genes alone were used as negative controls. The resulting constructs were used for transient assays by a modified PEG transfection of tobacco BY-2 suspension-culture cell protoplasts as described previously (Abel and Theologis 1994). The transformed protoplasts were then grown in liquid MS medium containing 0.4 M sucrose for 24–48 h, and transfected cells were imaged using the a fluorescence microscope with a YFP filter (Zeiss Axioskop 2 Plus).
Statistical analysis
The experiment was arranged in a completely randomized design. Each sample time point for each treatment was comprised of three independent biological replicates. Data were plotted in figures as means ± standard errors (SE). Least significant difference (LSD) at the 5 % level was analyzed by DPS software (version 3.01; Zhejiang University, Hangzhou, China).
Results and discussions
Characterization of MaHIS1 gene
Linker histone H1 genes have been isolated from many plants, including Arabidopsis (Ascenzi and Gantt 1997), tomato (Jayawardene and Riggs 1994; Bray et al. 1999), Maize (Razafimahatratra et al. 1991), pea (Gantt and Key 1987) and tobacco (Szekeres et al. 1995). In this study, one full-length cDNA of linker histone H1 gene, was obtained from banana fruit, then termed as MaHIS1 and deposited in GenBank (accession number JQ772499). MaHIS1 was 1,253 bp in length with an open-reading frame (ORF) of 948 bp, encoding 315 amino acids with a predicted molecular weight of 31.98 kDa and a calculated pI of 10.67. The amino acid composition of deduced MaHIS1 protein is very similar to other plant histone H1 proteins (Smith et al. 1984; Gantt and Key 1987; Jayawardene and Riggs 1994), with high numbers of lysine (67), alanine (88) and proline (41) residues, and a very high lysine to arginine ratio (Fig. 1). Moreover, MaHIS1 polypeptide also contained three distinct regions of known plant histone H1 proteins. The amino-terminal “nose” composes of an acidic domain (residues 1–42, including 12 acidic amino acids), a basic domain (residues 43–69, with 12 basic amino acids) and a conserved central globular domain extends from amino acid 73 through 136, showing 60–79 % identity at amino acid level with those of other histone H1 (Fig. 1) (Smith et al. 1984; Gantt and Key 1987; Jayawardene and Riggs 1994). It has been suggested that the conserved central globular domain is responsible for sealing the points of entry and exit of DNA from the core particle, while the “nose” region may be important for the proper positioning of the central globular domain (Jayawardene and Riggs 1994; Bharath et al. 2002). In addition, several positively charged lysine residues were also found in the carboxy-terminal tail of MaHIS1 protein (Fig. 1), which is generally thought to neutralize the negative phosphates of linker DNA to permit compaction of the chromatin fiber (Jayawardene and Riggs 1994; Bharath et al. 2002). Overall, these observations indicate that the MaHIS1 belongs to the plant histone H1 protein family.
Amino acid sequence alignment of the MaHIS1 protein with other plant HIS proteins. MaHIS1 was aligned with Arabidopsis AtHIS1 (CAA44314), tomato SlHIS1 (AAA50578), tobacco NtHIS1 (AAC41651), and pea PsHIS1 (P08283). Identical and similar amino acids are represented by black and gray shading, respectively. Gaps were introduced to optimize alignment. The acidic domain and basic domain consist of an amino-terminal “nose”, and the conserved central globular domain are included in the rectangles. The positively charged lysine residues at carboxy-terminal tail are indicated by asterisks
MaHIS1 was a nucleus-localized protein
Some reports have shown that linker histone H1 were preferentially localized in the cell nucleus (Ascenzi and Gantt 1999; Tanaka et al. 1999; Folco et al. 2003). To determine the subcellular localization of MaHIS1 protein, the coding region of MaHIS1 was fused in-frame with the GFP gene, and the resulting constructs were then transfected into tobacco BY-2 protoplasts. As shown in Fig. 2, similar with previous studies, the fluorescence of MaHIS1–GFP was localized exclusively to the nucleus, while the fluorescence of GFP alone was observed in the entire cells. It has been suggested that linker histone H1 influences core histone–DNA interactions in the nucleosome, which may increase the stability of individual nucleosomes (Usachenko et al. 1996). Thus, linker histone H1 is localized exclusively in the nucleus, consistent with its function of chromatin regulation.
Subcellular localization of MaHIS1 in tobacco BY-2 protoplasts. The BY-2 protoplasts were transiently transformed with MaHIS1-GFP or GFP vector by a modified polyethylene glycol method. GFP fluorescence was observed with a fluorescence microscope. The mCherry-VirD2NLS was included in each transfection to serve as a control for successful transfection as well as for nuclear localization. The MaHIS1-GFP fusion protein was observed in the nucleus. Images were taken in the dark field for green fluorescence, while the outline of the cell and the combination were photographed in a bright field. The length of the bar indicated in the photographs is 25 μm
Expression of MaHIS1 during three different time course of postharvest fruit ripening
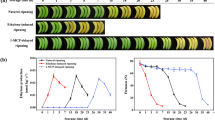
Plant development is an intricate process involving a series of highly organized mechanisms, including both genetic and epigenetic regulation. There is extensive evidence to show that plant HDACs, one of the chromatin-related proteins, act as global transcriptional regulators to play crucial roles in a range of plant developmental processes (Hollender and Liu 2008; Chung et al. 2009). Recently, histone deacetylase HD2 has also been shown to be involved in longan fruit senescence (Kuang et al. 2012). To understand the possible role of the MaHIS1 during banana fruit ripening, the expression patterns of MaHIS1 during three different time course of ripening, including natural, ethylene-induced, and 1-MCP-inhibited were investigated by RT-qPCR. As shown in Fig. 3, MaHIS1 transcript levels in natural-ripening fruit were low at the early storage (0–7 days), while began to obviously increase, with the increase in internal ethylene production at day 15, as the fruit started to ripen, and reached a maximum at day 21, following the ethylene production peak (Fig. 3a). MaHIS1 transcripts began to increase after 1 day of ethylene treatment, reached a maximum after 3 day, with 5.0-fold higher than those in the fruit at day 0, and finally decreased at day 5 (Fig. 3b). In addition, MaHIS1 transcripts in 1-MCP-inhibited ripening fruit were inhibited during 0–7 days, and also started to increase, with the increase in internal ethylene production at day 21, following, fruit began to ripen (Fig. 3c). These results indicate that expression of MaHIS1 gene is induced by external and internal ethylene and might be positively involved in regulating banana fruit postharvest ripening.
Expression of MaHIS1 during three different time course of banana fruit ripening, including natural (a), ethylene-induced (b) and 1-MCP-inhibited (c) ripening. MaRPS2 gene was used as reference to normalize expression of MaHIS1 under identical condition, and expression level at different time point was expressed as a ratio relative to the harvest time (0 day of natural ripening), which was set to 1. Each value represents the means of three replicates, and vertical bars indicate the SE. An asterisk indicates a statistical difference at the 5 % level between each time point and the harvest time. The broken arrow and full arrow represent the increasing time point of ethylene production and its peak, respectively
Expression of MaHIS1 in response to hormones and stresses
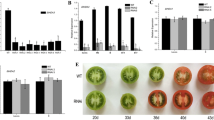
To examine the effect of exogenous hormones and stresses on the MaHIS1 expression, the transcriptional levels of MaHIS1 under hormones treatments (ABA, MeJA, and H2O2) and two main postharvest stresses of banana fruit (chilling and pathogen infection) were investigated. The results of RT-qPCR analysis showed that MaHIS1 was significantly induced by MeJA (Fig. 4a), ABA (Fig. 4b), and H2O2 (Fig. 4c) treatments at 6 and 12 h. In addition, MaHIS1 was also strongly induced by chilling (Fig. 4d) and pathogen infection stresses (Fig. 4e). Previous studies have shown that Arabidopsis HIS1-3 (Ascenzi and Gantt 1997) and tomato HIS1 (Wei and O’Connell 1996; Scippa et al. 2000) were induced by drought and ABA treatments. However, cold stress caused different influence on the expression of linker histone H1 gene. For e.g., cold stress down-regulated the expression of linker histones H1 in rice (Neilson et al. 2011), while up-regulated the transcript level of linker histone H1 in Arabidopsis (Kreps et al. 2002). Our results further confirm that the HIS1 can be induced by many hormones, and may be involved in abiotic and biotic stress responses.
Expression profiles of MaHIS1 in response to MeJA (a), ABA (b) or H2O2 (c) treatments, and to chilling (d) and pathogen infection (e) stresses. MaCAC gene was used as reference to normalize expression of MaHIS1 under identical condition, and expression level at different time point was expressed as a ratio relative to the harvest time 0 (control), which was set to 1. Each value represents the means of three replicates, and vertical bars indicate the SE. An asterisk indicates a statistical difference at the 5 % level between treated and control fruit
Interaction between MaHIS1 and MaWRKY1
WRKY proteins comprise one of the largest families of TFs found in plants (Rushton et al. 2012), and have been well documented as key regulators of many plant processes, including the responses to biotic and abiotic stresses, ABA and salicylic acid (SA) signaling, and senescence (Chen et al. 2010; Verk et al. 2011; Rushton et al. 2012). It has been demonstrated that chromatin structure including the structure imposed by the nucleosome implies that TFs work together with large multi-subunit complexes that remodel nucleosomes to facilitate DNA accessibility and to enable transcription (Depège-Fargeix et al. 2011). Recently, histone deacetylase 19 (HDA19) interacted with WRKY38 and WRKY62 TFs has been identified in Arabidopsis (Kim et al. 2008a, b). Banana fruit MaWRKY1 is also involved in hormone signaling and postharvest stress responses (unpublished data). In this work, to analyze the possible interaction between MaHIS1 and MaWRKY1, MaHIS1 and MaWRKY1 coding sequences were subcloned into pGBKT7 and pGADT7 vectors for yeast two-hybrid assay. Similar to pGBKT7-53 + pGADT7-T (positive control), yeast cells co-transformed with pGBKT7-MaHIS1 + pGADT7-MaWRKY1 could grow on selective medium lacking Trp, Leu, His, and Ade and turn blue in the presence of the chromogenic substrate X-α-Gal (Fig. 5a). In contrast, yeast cells harbouring pGBKT7-Lam + pGADT7-T (negative control), pGBKT7-MaHIS1 + pGADT7, and pGBKT7 + pGADT7-MaWRKY1 could not grow on the selective medium and turn blue under the same condition (Fig. 5a). These results indicated that MaHIS1 may physically interact with MaWRKY1.
Interaction between MaHIS1 and MaWRKY1 detected in yeast two-hybrid and BiFC assays. a Interaction between MaHIS1 and MaWRKY1 in the yeast two-hybrid assay. AH109 yeast strains were transformed with plasmids pGBKT7-53 + pGADT7-T (positive control), pGBKT7-Lam + pGADT7-T (negative control), pGBKT7-MaHIS1 + pGADT7-MaWRKY1, pGBKT7-MaHIS1 + pGADT7, or pGBKT7 + pGADT7-MaWRKY1. The ability of yeast cells to grow on synthetic medium lacking tryptophan, leucine, histidine, and adenine (SD/-Leu-Trp-His-Ade), and to turn blue in the presence of the chromagenic substrate X-α-Gal (SD/-Leu-Trp-His-Ade/X-α-Gal) was scored as a positive interaction. b BiFC visualization of MaHIS1 and MaWRKY1 interaction in transiently co-expressed tobacco BY-2 protoplasts. MaHIS1 or MaWRKY1 proteins were fused with the N- and C-terminus of YFP, respectively. Expressions of MaHIS1 or MaWRKY1 alone were used as negative controls. YFP indicates fluorescence of YFP, Merge is digital merge of bright field and fluorescent images. The length of the bar indicated in the photographs is 25 μm
To further confirm the interaction between MaHIS1 and MaWRKY1 observed in the yeast two-hybrid assays, we performed BiFC assays in tobacco BY-2 protoplasts. MaHIS1 and MaWRKY1 tagged with pSPYNE (split YFP N-terminal fragment expression) or pSPYCE (split YFP C-terminal fragment expression), respectively, then these constructs were transiently co-expressed into tobacco BY-2 protoplasts by PEG transfection. As shown in Fig. 5b, a robust YFP fluorescent signal was detected in the nucleus of BY-2 cells expressing either MaHIS1-pSPYNE or MaHIS1-pSPYCE and MaWRKY1-pSPYCE or MaWRKY1-pSPYNE, while no YFP fluorescent signal was observed either in the cells expressing MaHIS1-pSPYNE or MaHIS1-pSPYCE with only the pSPYCE or pSPYNE, or in those expressing MaWRKY1-pSPYCE or MaWRKY1-pSPYNE with only the pSPYNE or pSPYCE (Fig. 5b). The BiFC results not only demonstrated the in vivo interaction among the two proteins tested but also showed the specific localization of the interacting proteins in the nucleus, which was consistent with the subcellular localization of MaHIS1 in the nuclear compartment. It has been suggested that chromatin remodeling is an important regulator of stress-responsive genes (Chinnusamy et al. 2008; Chinnusamy and Zhu 2009; Kim et al. 2010). Histone modifications on the H3N-tail, one of the chromatin remodeling related to histone modification enzymes, altering with gene activation on the coding regions of their target genes, such as four Arabidopsis drought stress-responsive genes, RD29A, RD29B and RD20, and an AP2 domain-containing transcription factor, have been well documented (Kim et al. 2008a, b, 2010). Recently, Arabidopsis AtWRKY33 and Nicotiana benthamiana NtWRKY8 are found to be phosphorylated by MPK3/MPK6 or MAPK during abiotic defense responses, respectively (Mao et al. 2011; Ishihama et al. 2011). In addition, Arabidopsis AtWRKY53 is degraded by a HECT E3 ubiquitin ligase during leaf senescence to ensure that senescence is executed in the correct time frame (Miao and Zentgraf 2010). These results indicate that protein modifications of WRKYs play important roles in regulating their functions. Since HIS1 proteins function in gene and chromatin regulation by changing the higher-order structure of chromatin such as nucleosome packing and chromatin assembly (Izzo et al. 2008), thus, it is imperative to study whether MaHIS1 could cause such modifications on the MaWRKY1 protein in the future. Moreover, it should be pointed out that the detailed physiological significance of the interaction between MaHIS1 and MaWRKY1 requires further investigation.
Expression of MaHIS1 in different tissues
The expression pattern of MaHIS1 in different banana tissues was presented in Fig. 6. It was interesting to observe that MaHIS1 did not show fruit-specific expression. Actually, MaHIS1 mRNA could be detected in young fruit, root, petal and leaf, with higher level in root and petal, but lower level in fruit. Previous studies have showed that in tomato and maize, high levels of HIS expression were observed in buds, young fruits, and young stems than those in leaves, flowers, and roots (Razafimahatratra et al. 1991; Jayawardene and Riggs 1994). These results suggest that tissue expression pattern of HIS1 is different among different plants.
Expression of MaHIS1 in different tissues, including fruit, root, leaf, and petal. MaRPS2 gene was used as reference to normalize expression of MaHIS1 under identical condition, and expression level at each tissue was expressed as a ratio relative to the fruit, which was set to 1. Each value represents the means of three replicates, and vertical bars indicate the SE
In conclusion, a linker histone H1 gene MaHIS1 was isolated and characterized from banana fruit. MaHIS1 was a nucleus-localized protein. The expression of MaHIS1 by RT-qPCR analysis showed that MaHIS1 may be related to fruit ripening and stress responses. Moreover, MaHIS1 could interact with MaWRKY1, suggesting that they may act together to be involved in these physiological processes.
References
Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5:421–427
Ascenzi R, Gantt JS (1997) A drought-stress-inducible histone gene in Arabidopsis thaliana is a member of a distinct class of plant linker histone variants. Plant Mol Biol 34:629–641
Ascenzi R, Gantt JS (1999) Subnuclear distribution of the entire complement of linker histone variants in Arabidopsis thaliana. Chromosoma 108:345–355
Bartels S, González Besteiro MA, Lang D, Ulm R (2010) Emerging functions for plant MAP kinase phosphatases. Trends Plant Sci 15:322–329
Bassett A, Cooper S, Wu C, Travers A (2009) The folding and unfolding of eukaryotic chromatin. Curr Opin Genet Dev 19:159–165
Bharath MMS, Chandra NR, Rao MRS (2002) Prediction of an HMG-Box fold in the C-terminal domain of Histone H1: insights into its role in DNA condensation. Proteins Struct Funct Genet 49:71–81
Bray EA, Shih TY, Moses MS, Cohen A, Imai R, Plant AL (1999) Water-deficit induction of a tomato H1 histone requires abscisic acid. Plant Growth Regul 29:35–46
Chen JY, He LH, Jiang YM, Wang Y, Joyce DC, Ji ZL, Lu WJ (2008) Role of phenylalanine ammonia-lyase in heat pretreatment-induced chilling tolerance in banana fruit. Physiol Plant 132:318–328
Chen H, Lai Z, Shi J, Xiao Y, Chen Z, Xu X (2010) Roles of Arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol 10:281
Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY (2011) Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234:377–390
Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12:133–139
Chinnusamy V, Gong ZZ, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integ Plant Biol 50:1187–1195
Chung PJ, Kim YS, Jeong JS, Park SH, Nahm BH, Kim JK (2009) The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J 59:764–776
Depège-Fargeix N, Javelle M, Chambrier P, Frangne N, Gerentes D, Perez P, Rogowsky PM, Vernoud V (2011) Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. J Exp Bot 62:293–305
Fischle W, Wang Y, Allis CD (2003) Histone and chromatin cross-talk. Curr Opin Cell Biol 15:172–183
Folco HG, Freitag M, Ramón A, Temporini ED, Alvarez ME, García I, Scazzocchio C, Selker EU, Rosa AL (2003) Histone H1 is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot Cell 2:341–350
Gantt JS, Key JL (1987) Molecular cloning of a pea H1 histone cDNA. Eur J Biochem 166:119–125
Hollender C, Liu Z (2008) Histone deacetylase genes in Arabidopsis development. J Integ Plant Biol 50:875–885
Ishihama N, Yamada R, Yoshiok M, Katou S, Yoshioka H (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23:1153–1170
Ivanchenko M, Zlatanova J, van Holde K (1997) Histone H1 preferentially binds to superhelical DNA molecules of higher compaction. Biophy J 72:1388–1395
Izzo A, Kamieniarz K, Schneider R (2008) The histone H1 family: specific members, specific functions? Biol Chem 389:333–343
Jayawardene N, Riggs CD (1994) Molecular cloning, sequence analysis and differential expression of an intron-containing gene encoding tomato histone H1. Eur J Biochem 223:693–699
Jerzmanowski A, Przewloka MR, Grasser KD (2000) Linker histones and HMG1 proteins of higher plants. Plant Biol 2:586–597
Kadonaga JT (1998) Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell 92:307–313
Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M (2008a) Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 49:1580–1588
Kim KC, Lai Z, Fan B, Chen Z (2008b) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20:2357–2371
Kim JM, To TK, Nishioka T, Seki M (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33:604–611
Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130:2129–2141
Kuang JF, Chen JY, Luo M, Wu KQ, Sun W, Jiang YM, Lu WJ (2012) Histone deacetylase HD2 interacts with ERF1 and is involved in longan fruit senescence. J Exp Bot 63:441–454
Lee BH, Henderson DA, Zhu JK (2005) The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155–3175
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T) method. Methods 25:402–408
Luo M, Liu XC, Singh P, Cui YH, Zimmerli L, Wu KQ (2011) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta. doi:10.1016/j.bbagrm.2011.06.008
Mao GH, Meng XZ, Liu YD, Zheng ZY, Chen ZX, Zhang SQ (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23:1639–1653
Miao Y, Zentgraf U (2010) A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J 63:179–188
Mitsuda N, Ohme-Takagi M (2009) Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol 50:1232–1248
Neilson KA, Mariani M, Haynes PA (2011) Quantitative proteomic analysis of cold-responsive proteins in rice. Proteomics 11:1696–1706
Razafimahatratra P, Chaubet N, Philipps G, Gigot C (1991) Nucleotide sequence and expression of a maize H1 histone cDNA. Nucl Acids Res 19:1491–1496
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen XF, Finer JJ, Shen QJ, Rushton PJ (2012) WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol J 10:2–11
Scippa GS, Griffiths A, Chiatante D, Bray EA (2000) The H1 histone variant of tomato, H1-S, is targeted to the nucleus and accumulates in chromatin in response to water-deficit stress. Planta 211:173–181
Scippa GS, DiMichele M, Onelli E, Patrignani G, Chiatante D, Bray EA (2004) The histone-like protein H1-S and the response of tomato leaves to water deficit. J Exp Bot 55:99–109
Smith BJ, Harris MR, Sigournay CM, Mayes EL, Bustin M (1984) A survey of H1-and H5-like protein structure and distribution in higher and lower eukaryotes. Eur J Biochem 138:309–317
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Szekeres M, Haizel T, Adam E, Nagy F (1995) Molecular characterization and expression of a tobacco histone H1 cDNA. Plant Mol Biol 27:597–605
Tanaka I, Akahori Y, Gomi K, Suzuki T, Ueda K (1999) A novel histone variant localized in nucleoli of higher plant cells. Chromosoma 108:190–199
Usachenko SI, Gavin IM, Bavykin SG (1996) Alterations in nucleosome core structure in linker histone-depleted chromatin. J Biol Chem 271:3831–3836
Verbsky ML, Richards EJ (2001) Chromatin remodeling in plants. Curr Opin Plant Biol 4:494–500
Verk MCV, Bol JF, Linthorst HJM (2011) WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol 11:89
Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40:428–438
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wei T, O’Connell MA (1996) Structure and characterization of a putative drought-inducible H1 histone gene. Plant Mol Biol 30:255–268
Zhu Y, Dong AW, Shen WH (2011) Histone variants and chromatin assembly in plant abiotic stress responses. Biochim Biophys Acta. doi:10.1016/j.bbagrm.2011.07.012
Acknowledgments
The authors are grateful to Prof. Seiichiro Hasezawa (Department of Integrated Biosciences, The University of Tokyo) and Prof. Jörg Kudla (Institut für Biologie und Biotechnologie der Pflanzen, Universität Münster) for the generous gift of tobacco BY2 suspension cells and BiFC vectors, respectively. This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 31172007, 30972068, 30800772), China Agriculture Research System (Grant No. CARS-32-02A), and Guangdong Modern Agricultural Industry Technology System (Grant No. LNSG2010-12).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Lakshmanan.
Rights and permissions
About this article
Cite this article
Wang, Jn., Kuang, Jf., Shan, W. et al. Expression profiles of a banana fruit linker histone H1 gene MaHIS1 and its interaction with a WRKY transcription factor. Plant Cell Rep 31, 1485–1494 (2012). https://doi.org/10.1007/s00299-012-1263-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-012-1263-7