Abstract
Changes in ascorbate peroxidase (APX) enzyme activity in response to nematode (Heterodera avenae) attack were studied in roots of three hexaploid wheat lines carrying Cre2, Cre5, or Cre7 nematode resistance genes and the susceptible Triticum aestivum cv. Anza. A spectrophotometric analysis was carried out with root extracts of infected plants 4, 7, 11, and 14 days after nematode inoculation using uninfected plant as control. APX induction in infected resistant genotypes was similar and higher than in the susceptible control. The introgression wheat/Aegilops ventricosa H-93-8 line, carrying the Cre2 gene, and its parental line H-10-15 as susceptible control were used to analyze whether this increase of activity was correlated with the induction of APX gene expression. Genes encoding cytosolic forms of APX were induced in roots of both lines in response to nematode infection. This induction took place both earlier and with greater intensity in the resistant line than in the susceptible one, and it was also higher in the root area at the site of nematode attachment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cereal cyst nematode (CCN) (Heterodera avenae Woll.) is the most widely distributed and damaging species on cereals cultivated in less temperate regions. This nematode species has been detected in many countries and it is responsible for yield losses in wheat of up to 30%. The CCN induces syncytial feeding sites in the roots of its hosts. Infective second-stage juveniles (J2) enter the plant roots at the level of the differentiation zone and penetrate intracellularly towards the vascular cylinder. Once there, they select and pierce with their stylet a single cell where they release esophageal secretions. In the following hours, the affected plant cells start to develop feeding structures (Das et al. 2008). Approximately 2 weeks after infection, the nematode reaches maturity and it is associated with a syncytium, typically including a great number of cells (Golinowski et al. 1996). Many different methods are available to control nematodes, but one of the most effective is the use of host resistance, which is defined as the reduction in nematode multiplication in the presence of resistant plants. Globally, this is acknowledged as one of the most cost-effective, environmentally sustainable, and accessible methods to control CCN. There has been considerable progress in identifying resistance to H. avenae, with all reported genes exhibiting a major effect (i.e., controlled by a single dominant gene), making their use in a breeding program very amenable (Nicol and Rivoal 2007). Sources of H. avenae resistance (named Cre genes) have been found predominantly in wild relatives of wheat, with six of the eight coming from four Aegilops spp. Most of these genes have already been introgressed into hexaploid wheat backgrounds for bread wheat breeding purposes (Delibes et al. 1993; Eastwood et al. 1994; Romero et al. 1998; Seah et al. 2000; Ogbonnaya et al. 2001). However, the effectiveness of Cre resistance genes toward H. avenae depends on the nematode pathotype in conferring total or partial resistance to different CCN populations. For example, it has been demonstrated that Cre3 has the greatest impact on reducing the Ha13 Australian population, followed by Cre1 and Cre8 (Safari et al. 2005), but it is ineffective against European pathotypes (Montes et al. 2008; Rivoal et al. 2001). In contrast, Cre2, which confers a high level of resistance to several European CCN populations, including the preponderant Spanish pathotype, Ha71, used in this study (Delibes et al. 1993), has no effect on the Australian pathotype (Ogbonnaya et al. 2001). In a previous study, the efficacy of Cre genes against Spanish pathotype Ha71 was rank-ordered as Cre1 ≥ Cre4 ≥ Cre5 ≥ Cre2 ≥ Cre7 ≫ Cre8 > Cre3 > Cre6 (Montes et al. 2008).
Plants defend themselves from nematodes using a wide variety of mechanisms, including rapid induction of localized necrosis at the site of infection (the hypersensitive response: HR), increased expression of defense-related proteins, production of antimicrobial compounds, lignin formation, and the oxidative burst. The nature of the plant genes switched on or off during the nematode–host interaction is a subject of great interest as some of the products of these genes are directly responsible for promoting nematode parasitism or for fending-off the nematode attack in the resistant genotypes. Moreover, the timing in the surveillance system of receptor molecules that can recognize elicitor molecules of pathogen origin is fundamental for the host response and it is functional against a wide spectrum of pathogens (Nürnberger et al. 2004). Following this initial recognition, multiple biochemical pathways are activated and lead to a cascade of reactions, ultimately resulting in the formation and accumulation of secondary metabolites which help in the plant to overcome the parasite (Zhao et al. 2005). Among the altered biochemical pathways are those involving peroxidases, which comprise a large group of enzymes that use different peroxides (ROOH) as electron acceptors. According to Welinder (1992), these enzymes are classified into three classes (I, II, and III). Within class I enzymes, the ascorbate peroxidases (APX, EC 1.11.1.11) are intracellular and characterized to show a high affinity for ascorbate (AsA) as electron donor to reduce H2O2. This reaction gives rise to monohydroascorbate and H2O, being highly unstable in the absence of AsA (Chen and Asada 1989). Through a wide series of reactions, AsA plays an important role in the neutralization of hydrogen peroxide (H2O2) in photosynthetic organisms. AsA is responsible for H2O2 detoxification in green leaves (Chaudie`re and Ferrari-Iliou 1999) and APX is considered as a key antioxidant enzyme in plants (Orvar and Ellis 1997). Thus, the reactions catalyzed by APX and the coupled reaction AsA–GSH prevents the accumulation of toxic levels of H2O2, part of which is generated by class III peroxidases in photosynthetic organisms. APX activities are located in chloroplasts (chAPX), cytosol (cAPX), peroxisomes, or microbodies (pAPX) and mitochondria, each cellular compartment possessing one or several APX isoforms. In Arabidopsis, the same protein is dually targeted to mitochondria and chloroplast stroma (Chew et al. 2003). The different APX isozymes are very similar sharing 50–70% of sequence homology between them, so that a common evolutionary route for different APX genes has been proposed (Teixeira et al. 2004). Recent studies have focused on changes in activity and gene expression for APX isozymes in plants subjected to environmental and pathogen stress (Shigeoka et al. 2002; Chen et al. 2006).
Class II peroxidases are secreted by fungi while class III peroxidases are plant enzymes that target the secretory pathway via their signal peptide. Class III peroxidases (PRX) have been proposed as key regulators of the level of extracellular H2O2 and as producers of the extremely reactive oxygen species (ROS), hydroxyl (OH·), and hydroperoxyl (OOH·) radicals (Bolwell et al. 2002; Kawano 2003; Passardi et al. 2005; Bindschedler et al. 2006). The three described classes of peroxidases, in combination with other systems, regulate H2O2 levels that act as a systemic signal (Shigeoka et al. 2002).
Previous studies from our group (Andrés et al. 2001; Montes et al. 2003, 2004) demonstrated that cyst nematode infection enhances PRX, esterase (EST), and superoxide dismutase (SOD) activity in wheat roots carrying Cre2, Cre5, or Cre7 resistance genes. The highest response was found in the PRX system. More recently, we have identified three groups of PRX genes greatly induced by the CCN H. avenae in a resistant wheat line carrying the Cre2 gene (Simonetti et al. 2009a), which was accompanied by an increase in H2O2 synthesis (Simonetti et al. 2009b). All these findings suggest that apoplastic PRX from wheat plays a role in triggering some defense-related responses regulating H2O2 levels, a process in which APX activity would also be involved through H2O2 detoxification.
Therefore, to understand the precise role of APX in the wheat response to H. avenae infection, it is imperative to study its activity and regulation following nematode attack. The aim of the present study was to assess the involvement of APX in the defense response against nematode infection. Here, we describe the changes produced in APX enzyme activity and in its gene expression due to infection by the Ha71 pathotype of H. avenae in wheat lines carrying Cre resistance genes. The accumulation of cytosolic APX gene transcripts in wheat roots challenged by CCN suggests that the different ROS neutralization systems are co-regulated.
Materials and methods
Plant material and nematodes
The wheat introgression hexaploid lines, H-93-8 and TR-3531, carriers of Cre2 and Cre7 CCN resistance genes transferred from Aegilops ventricosa and Aegilops triuncialis, respectively, have been previously described (Delibes et al. 1993; Mena et al. 1993; Romero et al. 1998; Ogbonnaya et al. 2001; Montes et al. 2003). The substitution line 6Nv/6D, carrying Cre5 gene from Ae. ventricosa #10, was provided by Dr. Jahier from INRA, France (Jahier et al. 2001). Triticum aestivum cvs Anza and Almatense, H-10-15 (parent of H-93-8 and TR-3531) were used as susceptible controls (compatible interaction). Hereafter, abbreviations H-93-8, TR-3531, 6Nv/6D, Anza and H-10-15 will be used.
The CCN (H. avenae) population from a naturally infested plot at “La Poveda” Experimental Station in Arganda (Madrid) has been previously characterized as belonging to Ha71 pathotype (López-Braña et al. 1996). Seeds of resistant and susceptible wheat lines were germinated on wet paper after surface sterilization with 5% sodium hypochlorite and sowed in 5-cm diameter plates containing 10 ml of wet sterilized sand and maintained at 15 ± 2°C in a growth chamber. At 48 h, seedlings were divided into two groups: one was inoculated with 100 J2s of Ha71 pathotype per plant and the second one (uninfected seedlings) was used as a control as described by Andrés et al. (2001). RNA was extracted from root sections, with and without nematode, and leaves of infected plants, harvested 4 and 7 days after inoculation (dai), as well as from the uninoculated tissues used as control. Whole infected roots 4, 7, 11 and 14 dai and an equal part of uninfected material were used for enzyme analysis. Root and leaf strips were flash frozen in liquid nitrogen and then stored at −80°C for further use.
APX enzyme activity
To analyze quantitative changes in APX enzyme after wheat inoculation with pathotype Ha71 of H. avenae, global enzyme activity of root extracts was measured spectrophotometrically in resistant (introgression lines carrying Cre2, Cre5, or Cre7 genes) and susceptible (T. aestivum cv. Anza) genotypes. The supernatant used for enzyme essays was obtained by grinding 100 mg of root tissue with 200 µl of extraction buffer (25 mM potassium phosphate pH 7.0, 1 mM EDTA and 5 mM ascorbic acid) in a small mortar in the presence of glass beads, followed by centrifugation at 13,000 rpm for 15 min at 4°C. Global APX activity was determined spectrophotometrically measuring the decrease in absorbance at 290 nm wavelength of ascorbate in 0.5 ml of a reaction mixture, containing 25 mM potassium phosphate buffer pH 7.0, 0.5 mM ascorbic acid and enzyme extracts (100 µl), as previously described (Nakano and Asada 1981). Then, the peroxide reaction was started by the addition of 100 µl of 30% H2O2 (v/v). Enzyme activity was calculated as the concentration of ascorbic acid oxidized min−1 mg−1 protein using the molar extinction coefficient (ε = 2.8 mM−1 cm−1). Concentration of total protein in samples was determined using the Bradford method (1976). Each activity value was the average of three essays.
RNA purification
Total RNA was isolated according to Båga et al. (1995). Concentration of each RNA sample was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). Only the RNA samples with a 260/280 ratio between 1.9 and 2.1 and with a 260/230 ratio greater than 2.0 were used. Validation of RNA quality was performed using the RNA 6000 Nano Assay on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA).
Primers design and real-time PCR analysis
First-strand cDNA was synthesized by reverse transcribing 2 µg of total RNA in a final reaction volume of 20 µl using a high-capacity cDNA reverse transcription kit (Applied Biosystems) following the manufacturer’s instructions and using a poli-T oligonucleotide.
Specific primers for each APX group were designed from 3′ low homology region sequences of different groups of APX available in the databases of the NCBI website (National Center for Biotechnology Information; Altschul et al. 1997). The APX sequences of the different cereal species were aligned according to their subcellular location (cytosol, chloroplast and peroxisome) by MULTALIN program (Corpet 1988) and then manually adjusted using the GeneDoc program version 2.7.000 (Nicol and Rivoal 2007). Primers were designed using Primer Express program version 2.0 (Applied Biosystems). The primers selected from a preliminary list are shown in Table 1. This selection was carried out by picking primer sequences that were present in all APX of the same location (cytosol, chloroplast, or peroxisome), but absent in those of different location. The sizes of the amplified fragments (amplicons) obtained by PCR were in all cases 101 base pairs in length (except actin that was 151 base pairs).
The qPCR mixture contained 1 µl of fivefold diluted cDNA, 10 µl of Power SYBR Green PCR Master Mix (Applied Biosystems), and 300 nM each gene-specific primer in a final volume of 20 µl. PCR without template was also performed for each primer pair as controls. Data acquisitions were carried out with an ABI Prism 7300 Sequence Detection System and Software (Applied Biosystems). Cycling conditions for all reactions were 10 min at 95°C (denaturation), 40 cycles of 15 s at 95°C, and 1 min at 60–68°C in 96-well optical reaction plates (Applied Biosystems). The specificity of amplicons was verified by melting curve analysis (60–95°C) after 40 cycles and agarose gel electrophoresis. All genes were analyzed in triplicate from two independently isolated RNA samples. The cycle threshold (Ct) values were normalized to a conventional reference gene (actin). Two different approaches were used to process qRT-PCR data: (1) the relative quantification method which assumes that amplification efficiencies are equal to 2 for all reactions (Pfaffl 2001) and (2) the method based on the kinetics of individual PCR reactions. In the latter case, raw fluorescence values, as a function of cycles, were exported into the real-time PCR Miner program (Zhao and Fernald 2005) to calculate the efficiency and Ct values for individual PCR reactions.
Results
APX activity
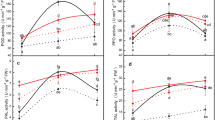
APX specific activity showed a significant increase in roots of wheat lines following the inoculation with juveniles of the Ha71 H. avenae pathotype (Fig. 1). The maximum activity level was seen 11 dai in all assayed genotypes. Enzyme levels were quite similar in the resistant lines and, in all cases, higher than those in the susceptible one. The pre-infection levels of APX were not significantly different between resistant lines carrying Cre2, Cre5, or Cre7 and the susceptible control (data not shown) and no differences were detected 4 dai. There was a significant increase in the activity in resistant lines as compared to the susceptible control 7 dai, reaching the maximum activity approximately 11 dai. This increase was 1.8–2.0 fold in resistant genotypes, over the uninfected controls. The activity of all resistant lines 14 dai declined significantly to levels observed in the susceptible control.
Ascorbate peroxidase activity at different time intervals post-infection (4, 7, 11 and 14 days) in roots of cereal cyst nematode. (1) Resistant (H-93-8, 6Nv/6D and TR-3531) or (2) susceptible (T. aestivum cv Anza) genotypes, infected with juveniles of H. avenae (filled square). Open triangle indicate their respective uninfected controls. Values represent the mean ± standard errors of three assays
TaAPx genes expression
To evaluate the expression of the different groups of TaAPx genes during the process of wheat infection by H. avenae, transcript levels were monitored by quantitative reverse transcription polymerase chain reaction (qRT-PCR) 4 and 7 dai in both susceptible H-10-15 and resistant H-93-8 lines. Different sets of TaAPx genes analyzed in this study are named according to the nomenclature proposed in the PeroxiBase database (Passardi et al. 2007) with the letters “Ta” for Triticum aestivum and “APx” for APX, preceded by different letters according to their subcellular location (cTaAPx for cytosol, chTaAPx for chloroplast, and pTaAPx for peroxisome or microbody). In Figs. 2, 3, 4 we have differentiated attacked plants (+) from control plants without attack (−) and in Fig. 2 also the root zone in which the nematodes have attached (+N) from the areas without nematodes (+).
Expression of cTaAPrx genes in wheat roots following Heterodera avenae infection. Steady-state mRNA levels of resistant (H-93-8) and susceptible (H-10-15) wheat were normalized with respect to actin. All data are mean ± standard error of three replicates from two biological samples. – Uninfected root section, + infected root section without nematode, +N infected root section with nematode 4 and 7 days, respectively, after nematode infection
Expression of cTaAPrx genes in wheat leaves following Heterodera avenae infection. Steady-state mRNA levels of resistant (H-93-8) and susceptible (H-10-15) wheat were normalized with respect to actin. All data are mean ± standard error of three replicates from two biological samples. – Leaves of uninfected plants, + leaves of infected plants 4 and 7 days, respectively, after nematode infection
Expression of chTaAPrx genes in wheat leaves following Heterodera avenae infection. Steady-state mRNA levels of resistant (H-93-8) and susceptible (H-10-15) wheat were normalized with respect to actin. All data are mean ± standard error of three replicates from two biological samples. – Leaves of uninfected plants, + leaves of infected plants 4 and 7 days, respectively, after nematode infection
There were no significant differences between the two methods used for data analysis (relative quantification assuming efficiencies of two and real-time PCR Miner). The results obtained using the second method are reported in the corresponding figures.
The largest increase in cTaAPx expression levels (Fig. 2) took place 4 dai in roots of line H-93-8, especially near the area where the nematode had established its feeding site (+N). The changes in expression of cTaAPx after infection in the susceptible line H-10-15 were much less pronounced, although, there was also a slight increase at 4 dai. The expression levels, at this time, were found to be three times higher in the resistant line than in the susceptible under all conditions. However, higher values were detected 7 dai in the susceptible wheat, in the region where the nematodes were attached (+N). Differences between regions of infected plants in the susceptible wheat disappeared 7 dai, but showed an increase in the levels in infected root areas without nematode (+). It can be seen in resistant wheat that the mRNA levels 7 dai were similar in all conditions regardless of infection (Fig. 2).
The expression levels were lower in leaves than in roots of both lines (Fig. 3), the differences being 8–tenfold in H-10-15 and 20-50 fold in the H-93-8 line. Moreover, cTaAPx mRNA levels in leaves were higher in the susceptible line, H-10-15, than in the resistant one, in all conditions, with and without attack, and both 4 and 7 dai. The H-10-15 line showed a slight induction in leaves in response to infection, both 4 and 7 dai, the levels being higher in the latter. The H-93-8 line exhibited very low levels in all cases.
The expression levels of chTaAPx in the H-10-15 line were higher than in the H-93-8 line, except in leaves of resistant plants 7 dai, which exhibited a light induction in response to the infection (Fig. 4). The H-10-15 line did not show significant differences in expression levels at 4 and 7 dai, regardless of whether they were attacked or not. The expression levels in roots (data not shown) were, in all the cases, 20–30 times lower than in leaves and also lower than those of cTaAPx (≈40 times) and the changes observed could not be considered significant.
Relative expression levels of pTaAPx in leaves were lower than those of chTaAPx (≈20–50 times), in both the H-10-15 and H-93-8 lines. Furthermore, the expression levels did not show differences that can be considered significant at 4 and 7 dai, except in attacked leaves of resistant plants 7 dai, which exhibited a very slight induction (Fig. 5). It is worth mentioning that levels were lower in roots (≈4 times) than in leaves (data not shown) and could be considered negligible both in the resistant and the susceptible lines.
Expression of pTaAPrx genes in wheat leaves following Heterodera avenae infection. Steady-state mRNA levels of resistant (H-93-8) and susceptible (H-10-15) wheat were normalized with respect to actin. All data are mean ± standard error of three replicates from two biological samples. – Leaves of uninfected plants, + leaves of infected plants 4 and 7 days, respectively, after nematode infection
Discussion
The results reported here are consistent with our previous work in which changes in SOD, EST and PRX were detected in roots of the same biological material (H-93-8, 6Nv/6D and TR-3531), inoculated with juveniles from the same H. avenae pathotype (Montes et al. 2003, 2004). Increase in the activity of these enzymes was detected 4 dai in lines carrying Cre2, Cre5, or Cre7 genes, being yet more evident 7 dai. We have previously observed that induction in PRX activity was accompanied by an increase in levels of some mRNAs belonging to this enzyme system (Simonetti et al. 2009a).
High levels of H2O2 are produced as a consequence of PRX action (Simonetti et al. 2009b), which may have several important effects on the host–pathogen interaction. APX and CAT (catalase) are some of the key enzymes of the plant antioxidant system, playing a central role in H2O2 scavenging and therefore its action is considered to be later than PRX. On the other hand, Alkharouf et al. (2004) noticed an early response of APX genes in a 12 h library made from mRNA extracted from roots of a resistant soybean inoculated with H. glycine, but the expression of these genes was not so abundant at 6–8 days. The induction of APX activity as a result of subjecting various plant species to different types of stress such as drought (Mittler and Zilinskas 1994), presence of salts (Seckin et al. 2009), high temperatures (Shi et al. 2001), excess light (Mullineaux and Karpinski 2000) and infection by pathogens (Mittler et al. 1998; Chen et al. 2006) has also been described.
Given this background, we were interested to study the changes in APX enzyme activity as a result of nematode infection in wheat lines carrying Cre resistance genes. In our first approach, we tried to detect changes in APX activity by means of conventional electrophoresis or isoelectrofocusing in wheat roots infected with H. avenae, following the methods described by Mittler and Zilinskas (1994) and Bai et al. (2009). However, these techniques were not sensitive enough to detect APX activity in roots. For this reason, we decided to carry out the study by spectrophotometry, which detects activity at lower concentrations.
The nematode infection produced an increase in APX activity (Fig. 1) in extracts from roots of the resistant genotypes as well as in the susceptible control T. aestivum cv. Anza. The activity was always higher 11 dai and with a greater increase (×2) in the infected lines carrying resistance genes. This finding was similar to that previously obtained by isoelectrofocusing for SOD, PRX and EST enzyme systems in roots of the same biological material (H-93-8, 6Nv/6D and TR-3531) inoculated with juveniles of the Ha71 H. avenae pathotype (Andrés et al. 2001; Montes et al. 2003, 2004).
The induction of APX activity as a result of nematode attack led us to study the accumulation of messenger RNA (transcripts) for TaAPx genes. This enzyme group, as it has been previously mentioned, is located in cytosol, chloroplasts and peroxisomes. Accumulation of cTaAPx transcripts over their normal basal level was produced by nematode infection in roots of both resistant and susceptible genotypes (Fig. 2). In contrast, changes in chTaPAx and pTaAPx groups were lower and not significant, suggesting that these two gene groups might not be directly involved in the early defense response to H. avenae. Furthermore, the cTaAPx genes were induced in roots of the resistant line in the zone in which the nematode establishes its feeding site (+N, Fig. 2) and this induction was greater than in the same area (+N) of the susceptible line. Unlike the spectrophotometric data, this value was higher 4 dai and then it fell 7 dai. These results are coherent if we keep in mind that mRNA expression precedes the enzymatic activity of the product that it encodes. The susceptible line showed a lower response to infection in cTaAPx, constant over time, in agreement with the results obtained by Sobczak et al. (2005). Transcription cTaAPX levels in leaves were lower than those registered in roots in both lines studied (Fig. 3). In addition, cTaAPx genes were induced in leaves of susceptible line 7 dai (Fig. 3). The expression of chTaAPx was much lower (about 102 times, data not shown) in roots, in both lines, and was not affected by nematode attack, suggesting that these genes would be not directly involved in the plant defense response against H. avenae. As predicted, the levels of chTaAPx were higher in leaves than in roots, in both lines. Moreover, pTaAPx expression was very low in leaves and roots as to be insignificant. The higher levels of cTaAPx in roots and chTaAPx in leaves agree with previous findings in Arabidopsis (Pnuelli et al. 2003), rice (Teixeira et al. 2006) and potato (Pereira et al. 2005). Nevertheless, it has been observed in Arabidopsis that only one of the cytosolic APX (AtAPx1) increases in response to diverse types of stress (Pnuelli et al. 2003; Rizhsky et al. 2004). Induction of another cytosolic APX (APx2) has been detected in roots of resistant soybean, during H. glycine infection, but this increase took place prior, at 12 h post-inoculation (Alkharouf et al. 2004). In contrast, Chen et al. (2006) described induction of pTaAPx in leaves of a resistant wheat cultivar in response to infection by Blumeria graminis. In our case, the levels of pTaAPx were very low and without significant differences in response to nematode attack. This may reflect changes in gene expression mediated by the two different ways of pathogen penetration, intracellularly in CCN and appressorium-mediated in B. graminis, with and without wounding. Nevertheless, our results are consistent with that obtained by analyzing transcriptomes of syncytia in Arabidopsis infected by H. schachtii (Szakasits et al. 2009) and also with studies carried out on whole galls and giant cells in tomato and Arabidopsis attacked by Meloydogyne javanica (Portillo et al. 2009; Barcala et al. 2010) that are more similar systems to that used in this work.
We have previously described an accumulation of TaPrx111, TaPrx112, and TaPrx113 peroxidase transcripts over their normal basal level in response to nematode infection in both resistant, H-93-8, and susceptible, H-10-15, genotypes (Simonetti et al. 2009a), but the induction was not higher where the nematode was established. This was accompanied by an increase in H2O2 levels and PRX activity around nematode feeding site cells (Simonetti et al. 2009b), which again was higher on the resistant H-93-8 line than in the susceptible H-10-15. It could be suggested that TaAPx and TaPrx genes respond differently to signals from the nematode and, as a whole, they cooperate to limit root damage and to regulate hydrogen peroxide levels and such a defense mechanism in the resistant genotypes is activated immediately after infection by H. avenae. Probably the same mechanism could operate in the susceptible wheat cultivar, but it may function quite slowly and could reach lower levels. Such a mechanism has also been proposed for the wheat–Hessian fly interaction (Liu et al. 2010). Because two APX genes of cytoplasmic location have been described, and the existence of a functional specialization of each member has been suggested (Teixeira et al. 2006), it would be useful to study the response of the different wheat cytoplasmic APX genes to the infection by the nematode. Evaluating the function of each peroxidase isoform is quite difficult because of the redundancy of PRX and APX genes in plants (Cosio and Dunand 2009). Therefore, it would be necessary to carry out further studies such as the generation of transgenic plants overexpressing (Schweizer 2008) or silencing (Bindschedler et al. 2006) different peroxidase genes to analyze their specific involvement in the plant resistance response. Silenced transgenic plants exhibited a reduction in mRNA levels of two PRX genes accompanied by a greater susceptibility to different pathogens than that showed by wild plants (Bindschedler et al. 2006). Moreover, Johrde and Schweizer (2008) induced transient overexpression of the HvPrx40 gene in epidermal cells of wheat and barley leaves and detected, in both cases, induction of resistance to Bl. graminis. These findings were complemented by transiently induced HvPrx40 gene silencing showing hypersusceptibility of barley leaf epidermal cells to this fungus.
It would also be advisable to identify the tissue location in planta of each APX and PRX isoform by fusing the upstream region of each of the genes to a reporter gene such as β-glucuronidase (GUS) or green fluorescent protein (GFP). Using this method, in transgenic plants of Arabidopsis thaliana, one zucchini isoperoxidase was localized to the zone of elongation of the hypocotyl (Cosio et al. 2009).
In summary, TaPrx could be responsible for the HR as a result of increased levels of hydrogen peroxide, which are higher in the resistant genotype. The nematode could reduce its exposure to peroxide by inducing an increase in cTaAPx expression, particularly in the syncytium. Increased hydrogen peroxide would not affect catalase levels, because it is only involved when the amount of H2O2 is not very high (Ioannidi et al. 2009). A key role in the detoxification and maintenance of the ROS levels may be played by AsA, necessary for APX activity which is quickly inactivated in its absence (Chen and Asada 1989). The observed increased cTaAPx levels 7 dai in leaves of the susceptible line attacked by the nematode (Fig. 3) could be the result of systemic increase of H2O2 in the whole plant that would occur as a result of lower APX activity in roots of this line. Finally, our results suggest that APX present in wheat roots could play a role in Cre-mediated resistance to H. avenae, either directly or indirectly and that the biochemical basis of defense in hexaploid wheat against the Ha71 pathotype of H. avenae could be the same in all the resistant genotypes tested.
Abbreviations
- APX:
-
Ascorbate peroxidase
- AsA:
-
Ascorbic acid
- Cre:
-
Cereal root eelworm
- CCN:
-
Cereal cyst nematode
- dai:
-
Days after inoculation
- EST:
-
Esterase
- GSH:
-
Reduced form of glutathione
- PRX:
-
Plant class III peroxidases
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
References
Alkharouf N, Khan R, Matthews BF (2004) Analysis of expressed sequence tags from roots of resistant soybean infected by the soybean cyst nematode. Genome 47:380–388
Altschul SF, Madden TL, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped blast and psiblast, a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Andrés MF, Melillo MT, Delibes A, Romero MD, Bleve-Zacheo T (2001) Changes in wheat root enzymes correlated with resistance to cereal cyst nematodes. New Phytol 152:343–354
Båga M, Chibbar RN, Kartha KK (1995) Molecular cloning and expression analysis of peroxidase genes from wheat. Plant Mol Biol 29:647–662
Bai R, Ma F, Liang D, Zhao X (2009) Phthalic acid induces oxidative stress and alters the activity of some antioxidant enzymes in roots of Malus prunifolia. J Chem Ecol 35:488–494
Barcala M, García A, Cabrera J, Casson S, Lindsey K, Favery B, García-Casado G, Solano R, Fenoll C, Escobar C (2010) Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J 61:698–712
Bindschedler LV, Dewdney J, Blee KA, Stone JM, Asai T, Plotnikov J, Denoux C, Hayes T, Gerrish C, Davies DR, Ausubel FM, Bolwell GP (2006) Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J 47:851–863
Bolwell GP, Bindschedler LV, Blee KA, Butt VS, Davies DR, Gardner SL, Gerrish C, Minibayeva F (2002) The apoplastic oxidative burst in response to biotic stress in plants: a three component system. J Exp Bot 53:1367–1376
Bradford MM (1976) A rapid and sensitive method for 1 the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chaudie`re J, Ferrari-Iliou R (1999) Intracellular antioxidants: from chemical to biochemical mechanisms. Food Chem Toxicol 37:949–962
Chen GX, Asada K (1989) Ascorbate peroxidase in tea leaves: occurrence of two isoenzymes and the differences in their enzymatic and molecular properties. Plant Cell Physiol 30:987–998
Chen YP, Wang HZ, Cao AZ, Wang CM, Chen PD (2006) Cloning of a resistance gene analog from wheat and development of a codominant PCR marker for Pm21. J Integr Plant Biol 48:715–721
Chew O, Whelan J, Millar AH (2003) Molecular definition of the ascorbate glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278:46869–46877
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Cosio C, Dunand C (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60:391–408
Cosio C, Vuillemin L, De Meyer M, Kevers C, Penel C, Dunand C (2009) An anionic class III peroxidase from zucchini may regulate hypocotyl elongation through auxin oxidase activity. Planta 229:823–836
Das S, DeMason DA, Ehlers JD, Close TJ, Roberts PA (2008) Histological characterization of root-knot nematode resistance in cowpea and its relation to reactive oxygen species modulation. J Exp Bot 59:1305–1313
Delibes A, Romero D, Aguaded S, Duce A, Mena M, Lopez-Braña I, Andrés MF, Martin-Sanchez JA, García-Olmedo F (1993) Resistance to the cereal cyst nematode (Heterodera avenae Woll.) transferred from the wild grass Aegilops ventricosa to hexaploid wheat by a “stepping-stone procedure. Theor Appl Genet 87:402–408
Eastwood RF, Lagudah ES, Appels R (1994) A directed search for DNA sequence tightly linked to cereal cyst nematode resistance genes in Triticum tauschii. Genome 37:311–319
Golinowski W, Grundler FMW, Sobczak M (1996) Changes in the structure of Arabidopsis thaliana during female development of the plant parasitic nematode Heterodera schachtii. Protoplasma 194:103–116
Ioannidi E, Kalamaki MS, Engineer C, Pateraki I, Alexandrou D, Mellidou I, Giovannonni J, Kanellis AK (2009) Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. J Exp Bot 60:663–678
Jahier J, Abelard P, Tanguy M, Dedryver F, Rivoal R, Khatkar S, Bariana HS (2001) The Aegilops ventricosa segment on chromosome 2AS of the wheat cultivar ‘VPM1’ carries the cereal cyst nematode resistance gene Cre5. Plant Breed 120:125–128
Johrde A, Schweizer P (2008) A class III peroxidase specifically expressed in pathogen-attacked barley epidermis contributes to basal resistance. Mol Plant Pathol 9:687–696
Kawano T (2003) Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep 21:829–837
Liu XM, Williams CE, Nemacheck JA, Wang H, Subramanyam S, Zheng C, Chen MS (2010) Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol 152:985–999
López-Braña I, Romero MD, Delibes A (1996) Application of the random amplified polymorphic DNA (RAPD) technique for analysis of Heterodera avenae complex populations. Genome 39:118–122
Mena M, Orellana J, López-Braña I, García-Olmedo F, Delibes A (1993) Characterization of wheat/Aegilops ventricosa introgression and addition lines with respect to the Mv genome. Theor Appl Genet 86:197–204
Mittler M, Zilinskas BA (1994) Regulation of pea cytosolic ascorbate peroxidase and other antioxidant enzymes during the progression of drought stress and following recovery from drought. Plant J 5:397–405
Mittler R, Feng X, Cohen M (1998) Post-transcriptional suppression of cytosolic ascorbate peroxidase expression during pathogen-induced programmed cell death in tobacco. Plant Cell 10:461–474
Montes MJ, Lopez-Braña I, Romero MD, Sin E, Andres MF, Martin-Sánchez JA, Delibes A (2003) Biochemical and genetic studies of two Heterodera avenae resistance genes transferred from Aegilops ventricosa to wheat. Theor Appl Genet 107:611–618
Montes MJ, López-Braña I, Delibes A (2004) Root enzyme activities associated with resistance to Heterodera avenae conferred by gene Cre7 in a wheat/Aegilops triuncialis introgression line. J Plant Physiol 161:1135–1140
Montes MJ, Andrés MF, Sin E, López-Braña I, Martín-Sánchez JA, Romero MD, Delibes A (2008) Cereal cyst nematode resistance conferred by the Cre7 gene from Aegilops triuncialis and its relationship with Cre genes from Australian wheat cultivars. Genome 51:315–319
Mullineaux P, Karpinski S (2000) Signal transduction in response to excess light: getting out of the chloroplast. Curr Opin Plant Biol 5:43–48
Nakano Y, Asada K (1981) Hydrogen-peroxide is scavenged by ascorbate specific peroxidase in spinach-chloroplasts. Plant Cell Physiol 22:867–880
Nicol JM, Rivoal R (2007) Global knowledge and its application for the integrated control and management of nematodes on wheat. In: Ciancio A, Mukerji KG (eds) Integrated management and biocontrol of vegetable and grain crops nematodes. Springer, The Netherlands, pp 243–287
Nürnberger T, Brunner F, Kemmerling B, Piater L (2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev 198:249–266
Ogbonnaya FC, Seah S, Delibes A, Jahier J, López-Braña I, Eastwood RF, Lagudah ES (2001) Molecular–genetic characterisation of a new nematode resistance gene in wheat. Theor Appl Genet 102:623–629
Orvar BL, Ellis BE (1997) Transgenic tobacco plants expressing antisense RNA for cytosolic ascorbate peroxidase show increased susceptibility to ozone injury. Plant J 11:1297–1305
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265
Passardi F, Theiler G, Zamocky M, Cosio C, Rouhier N, Teixeira F, Margis-Pinheiro M, Ioannidis V, Penel C, Falquet L, Dunand C (2007) Peroxibase: the peroxidase database. Phytochemistry 68:1605–1611
Pereira CS, Soares da Costa D, Teixeira J, Pereira S (2005) Organ-specific distribution and subcellular localisation of ascorbate peroxidase isoenzymes in potato (Solanum tuberosum L.) plants. Protoplasma 226:223–230
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Pnuelli L, Liang H, Rozenberg M, Mittler R (2003) Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (APX1)-deficient Arabidopsis plants. Plant J 34:187–203
Portillo M, Lindsey K, Casson S, García-Casado G, Solano R, Fenoll C, Escobar C (2009) Isolation of RNA from laser-capture microdissected giant cells at early differentiation stages suitable for differential transcriptome análisis. Mol Plant Pathol 10:523–535
Rivoal R, Bekal S, Valette S, Gauthier JP, Bel Hadj Fradj M, Mokabli A, Jahier J, Nicol J, Yahyaoui A (2001) Variation in reproductive capacity and virulence on different genotypes and resistance genes of Triticeae in the cereal cyst nematode species complex. Nematology 3:581–592
Rizhsky L, Davletova S, Liang H, Mittler R (2004) The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem 279:11736–11743
Romero D, Montes MJ, Sin E, López-Braña I, Duce A, Martín-Sánchez JA, Andrés MF, Delibes A (1998) A cereal cyst nematode (Heterodera avenae Woll.) resistance gene transferred from Aegilops triuncialis to hexaploid wheat. Theor App Genet 96:1135–1140
Safari E, Gororo NN, Eastwood RF, Lewis J, Eagles HA, Ogbonnaya FC (2005) Impact of Cre1, Cre8 and Cre3 genes on cereal cyst nematode resistance in wheat. Theor Appl Genet 110:567–572
Schweizer P (2008) Tissue-specific expression of a defense-related peroxidase in transgenic wheat potentiates cell death in pathogen-attacked leaf epidermis. Mol Plant Pathol 9:45–57
Seah S, Miller C, Sivasithamparam K, Lagudah ES (2000) Root responses to cereal cyst nematode (Heterodera avenae) in hosts with different resistance genes. New Phytol 146:527–533
Seckin B, Sekmen AH, Türkan I (2009) An enhancing effect of exogenous mannitol on the antioxidant enzyme activities in roots of wheat under salt stress. J Plant Growth Regul 28:12–20
Shi WM, Muramoto Y, Ueda A, Takabe T (2001) Cloning of peroxisomal ascorbate peroxidase gene from barley and enhanced thermotolerance by overexpressing in Arabidopsis thaliana. Gene 273:23–27
Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K (2002) Regulation and function of ascorbate peroxidase isoenzymes. J Exp Bot 372:1305–1319
Simonetti E, Veronico P, Melillo MT, Delibes A, Andrés MF, López-Braña I (2009a) Analysis of class III peroxidase genes expressed in roots of resistant and susceptible wheat lines infected by Heterodera avenae. Mol Plant–Microbe Interact 22:1081–1092
Simonetti E, Veronico P, Melillo MT, Bleve-Zacheo T, Delibes A, Andrés MF, López-Braña I (2009b) Class III peroxidase are involved in wheat resistance to Heterodera avenae. NEMAGENICS. Exploiting genomics to understand plant–nematode interactions. Cost 872 Workshop and Meeting. Toledo, Spain, p p 60 (abstracts book)
Sobczak M, Avrova A, Jupowicz J, Phillips MS, Ernst K, Kumar A (2005) Characterization of susceptibility and resistance responses of potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad spectrum nematode resistance Hero gene. Mol Plant–Microbe Interact 18:158–168
Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FMW, Bohlmann H (2009) The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J 57:771–784
Teixeira FK, Menezes-Benavente L, Margis R, Margis-Pinheiro M (2004) Analysis of the molecular evolutionary history of the ascorbate peroxidase gene family: inferences from the rice genome. J Mol Evol 59:761–770
Teixeira FK, Menezes-Benavente L, Costa Galvão V, Margis R, Margis-Pinheiro M (2006) Rice ascorbate peroxidase gene family encodes functionally diverse isoforms localized in different subcellular compartments. Planta 224:300–314
Welinder KG (1992) Plant peroxidases: structure-function relationships. In: Penel C et al. (eds) Plant peroxidases. Geneve University, Switzerland, pp 1–24
Zhao S, Fernald RD (2005) Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comp Biol 12:1047–1064
Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnol Adv 23:283–333
Acknowledgments
This research was supported by Grant 083/Q060210-031 from the Comunidad Autonoma de Madrid (Spain). We thank M. López, C. Martínez-Belinchón and R. Rodriguez-Ríos for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. Zaitlin.
Rights and permissions
About this article
Cite this article
Simonetti, E., Alba, E., Montes, M.J. et al. Analysis of ascorbate peroxidase genes expressed in resistant and susceptible wheat lines infected by the cereal cyst nematode, Heterodera avenae . Plant Cell Rep 29, 1169–1178 (2010). https://doi.org/10.1007/s00299-010-0903-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0903-z