Abstract
Somatic embryogenesis (SE) is a propagation tool of particular interest for accelerating the deployment of new high-performance planting stock in multivarietal forestry. However, genetic conformity in in vitro propagated plants should be assessed as early as possible, especially in long-living trees such as conifers. The main objective of this work was to study such conformity based on genetic stability at simple sequence repeat (SSR) loci during somatic embryogenesis in maritime pine (Pinus pinaster Ait.). Embryogenic cell lines (ECLs) subjected to tissue proliferation during 6, 14 or 22 months, as well as emblings regenerated from several ECLs, were analyzed. Genetic variation at seven SSR loci was detected in ECLs under proliferation conditions for all time points, and in 5 out of 52 emblings recovered from somatic embryos. Three of these five emblings showed an abnormal phenotype consisting mainly of plagiotropism and loss of apical dominance. Despite the variation found in somatic embryogenesis-derived plant material, no correlation was established between genetic stability at the analyzed loci and abnormal embling phenotype, present in 64% of the emblings. The use of microsatellites in this work was efficient for monitoring mutation events during the somatic embryogenesis in P. pinaster. These molecular markers should be useful in the implementation of new breeding and deployment strategies for improved trees using SE.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant propagation through tissue culture should result, theoretically, in production of clonal individuals identical to the starting material. However, changes at several levels, including morphological, biochemical, genetic and/or epigenetic, have been observed during in vitro plant propagation (Rahman and Rajora 2001; Etienne and Bertrand 2003; Peredo et al. 2006; El-Dougdoug et al. 2007).
Somaclonal variation can have a major effect in determining quality of cloned individuals, especially in long-living forest trees like pine. Thus, early detection of variants is important so that deleterious effects that might be expressed later in growth do not result in having a severe economic impact (Fourré 2000). In in vitro culture, several factors, such as exposure to high concentrations of growth regulators and long-term maintenance under artificial conditions, can be potential inducers of somaclonal variation (DeVerno 1995).
In vitro clonal propagation of conifer species by somatic embryogenesis (SE) is considered a valuable process in breeding programs and commercial forestry. The maritime pine, Pinus pinaster (Ait.), is the most important industrial tree species in Southwestern Europe. It is one of the highly abundant forest species in Portugal, following cork oak, occupying an area of approximately 711,000 ha (DGRF 2008). During the last few years, several factors, including extensive fires, have contributed to a shortage of raw material for the forestry industry. Additionally, this decrease in forest biomass presents a concomitant concern with regard to carbon sequestration.
Somatic embryogenesis in Portuguese genotypes of maritime pine was previously established from zygotic embryos of half-sib families (Miguel et al. 2004). Following the established procedure, the SE process takes a minimum of 9 months to complete. This process involves the initiation of embryogenic cell lines (ECL) and their proliferation. This is followed by maturation of somatic embryos and germination, up to the stage where somatic embryo plants (emblings) are transferred to ex vitro conditions. However, this process may be longer in duration depending on how long the cultures must be maintained under proliferation conditions in order to obtain sufficient amounts of tissue for regeneration of emblings. Several cases of somaclonal variation have been reported in embryogenic cultures of conifers including Larix decidua (Von Aderkas et al. 2003), Picea abies (Burg et al. 1999), Picea glauca (DeVerno et al. 1999) and Pinus sylvestris (Burg et al. 2007). Therefore, it appears that routine monitoring of somaclonal variation in SE-derived conifers would be prudent in order to maintain conformity and quality of cloned plants.
Variation can be assessed based on changes in phenotypic traits, changes in isozyme and protein patterns, changes in chromosome structure and number, and DNA alterations, or epigenetic characteristics (Fourré 2000). Some genetic molecular markers, such as RFLP (DeVerno et al. 1994), RAPD (Isabel et al. 1993; Fourré et al. 1997) and AFLP (Hornero et al. 2001), have been used for analysis of true-to-typeness. Microsatellites can serve as highly sensitive markers for monitoring genetic variation that may signal potential deleterious mutations during in vitro culture, because they reflect a relatively high rate of mutation and corresponding degree of genetic variability (Burg et al. 1999; Leroy et al. 2000; Wilhelm et al. 2005; Lopes et al. 2006; Burg et al. 2007). Microsatellites consist of tandem repeats of short sequence motifs (2–8). Observed variation, or instability, in microsatellites sometimes corresponds to an alteration in the size of simple repeat sequences (Leroy et al. 2000). In humans, the average overall mutation rate for 28 di- and tetranucleotide microsatellites was estimated at about 0.001, with tetranucleotide repeats significantly more mutable than dinucleotide repeats (Goldstein and Pollock 1997). The primary mutational mechanism in microsatellites is believed to be replication slippage (adding or subtracting of repeat units) (Whittaker et al. 2003).
Despite several published studies on SE of P. pinaster (Lelu et al. 1999; Miguel et al. 2004; Harvengt 2005; Lelu-Walter et al. 2006) and the high level of commercial interest in application of this propagation tool, there is no information currently available regarding genetic conformity of cloned plants from in vitro culture.
The report presented here is the first on analysis of true-to-typeness in P. pinaster propagated by SE. For this study, we analyzed genetic stability of maritime pine based upon microsatellite variability at several simple sequence repeat (SSR) loci during SE. Plant material was collected for analysis at several points during SE up to the final stage of regenerated emblings. The effects of prolonged culture under proliferation conditions (exposure to auxin and cytokinin) were specifically targeted for analysis as these treatments may potentially contribute to variation.
Material and methods
Somatic embryogenesis
Embryogenic cell lines (ECLs) of P. pinaster were established from immature zygotic embryos, as previously described (Miguel et al. 2004). The zygotic embryos were excised from seeds collected from open-pollinated trees growing in Mata do Escaroupim, Portugal. The ECLs were numbered as x/y, where x is the number of the seed family (or mother tree from which the seeds were collected) and y the genotype number corresponding to each zygotic embryo.
The initiated ECLs with at least 1 g fresh weight of embryogenic tissue were considered established. Establishment is accomplished approximately 3 months after inoculation of the zygotic embryos. All the somatic embryogenesis-derived material used in this study was obtained from ECLs established in years 2000, 2001 and 2002. Established ECLs were maintained on proliferation medium consisting of DCR salts and vitamins (Gupta and Durzan 1985) supplemented with 4.4 μM BAP, 14 μM 2,4-d, casein hydrolysate (500 mg/l), l-glutamine (1,460 mg/l) and 2% (w/v) sucrose, solidified with 0.2% (w/v) Phytagel (Sigma), in the dark at 22 ± 2°C (Miguel et al. 2004).
Somatic embryo maturation was performed as described by Tereso et al. (2006), except that plant growth regulator-free DCR medium supplemented with 3% of sucrose and 1% activated charcoal, was used for resuspension of embryogenic tissues. Embryogenic suspensions were then spread over filter papers and placed on maturation medium. To evaluate maturation ability, the average number of cotyledonary embryos obtained per gram of FW tissue was recorded in five replicate Petri dishes of ECLs 13/100, 16/385, 32/4 and 82/73.
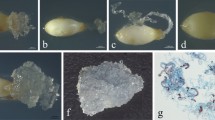
Conversion of somatic embryos to plants growing in the greenhouse (emblings) was obtained following the protocol described by Tereso et al. (2006). Emblings with a well-developed epicotyle were transferred to a mixture of peat:perlite:pine tree bark (1:1:1) and acclimatized in a growth chamber over a 10-day period (Miguel et al. 2004). After 4 and 12 months under ex vitro conditions (nursery and outdoors) morphology of the emblings was examined, and phenotypes were characterized. Abnormal phenotypes were identified based on plagiotropism, loss of apical dominance and bifurcation traits. The different tissues derived from SE used for SSR analysis are described below.
Plant material used in SSR analysis
Experiment 1. Effect of prolonged culture under proliferation conditions (E1)
In this experiment, 17 ECLs from 6 different families, established in 2001 (13/148; 13/154; 13/161; 16/404; 31/472; 49/420; 49/423; 49/432; 49/433; 49/439; 68/334; 68/345; 68/357; 68/363; 68/367; 82/248; 82/281) were used for SSR analysis. The beginning of the experiment (T0) corresponds to the time of establishment of the ECLs. After establishment, these lines were further subcultured every 2 weeks for 6 (T6), 14 (T14) and 22 (T22) months before samples were collected for DNA extraction. Each sample, consisting of approximately 100 mg of FW tissue, was immediately frozen in liquid nitrogen and stored at −80°C until DNA extraction.
We have considered the SSR pattern of each ECL at establishment (T0) as our reference pattern to which the T6, T12 and T22 SSR patterns were compared. However, to check for any possible mosaicism in the embryogenic cultures, three tissue sectors of each ECL in T0 stage were sampled and analyzed separately.
Experiment 2. Genetic stability in emblings (E2)
Needles were collected from a total of 52 emblings regenerated from four ECLs. The ECLs were 32/4 (22 emblings), 82/73 (19 emblings) and 13/100 (8 emblings) established in 2002, and 16/385 (3 emblings) established in 2001. Emblings were derived from biological material under in vitro conditions for 22 months and needles were collected following 12 months of ex vitro growth.
Approximately 100 mg FW needles were collected from each embling and immediately frozen in liquid nitrogen and stored at −80°C until DNA extraction.
To establish a reference SSR profile, 100 mg FW tissue of each ECL, from which the emblings were regenerated, was collected at the time of ECL establishment (T0).
DNA isolation and quantification
Plant tissues from ECLs and needles (100 mg) were ground to a fine powder using a mortar and a pestle in liquid nitrogen. DNA extraction was performed using the DNeasy Plant Mini Kit (QIAGEN) following instructions provided by the manufacturer. DNA concentration was quantified spectrophotometrically (GeneQuant, Pharmacia Biotech) at 260 nm. DNA purity, integrity and concentration were assessed using electrophoresis on 1% agarose gels after staining with ethidium bromide. DNA concentrations were normalized to 100 ng/μl in all samples.
SSR amplification and fragment analysis
Seven primer pairs, from available nuclear microsatellite primers already tested in P. pinaster, were chosen for this study. These primers were selected based on the relatively high quality of PCR products produced for fragment analysis. The pine nuclear microsatellite loci analyzed were FRPP91, FRPP94, ITPH4516, PtTX3107, NZPR114, NZPR544 and RPtEST11, as described in Mariette et al. (2001), González-Martínez et al. (2004) and Chagné et al. (2004). The sequences of the primers used are shown in Table 1. Each forward primer was labeled with CEQ dye blue (D4, Beckman Coulter).
PCR reactions for analysis of FRPP91, FRPP94 and ITPH4516 loci were performed in a final volume of 10 μl containing 0.5 U Taq DNA polymerase (Invitrogen Life Technologies), 1.0 μl 10 × reaction buffer, 200 μM of each dNTP, 0.1 μM of each primer, 2 mM MgCl2 for ITPH4516 (or 2.25 mM for FRPP91 and 2.50 mM for FRPP94), and 0.6 ng/μl of genomic DNA (or 0.3 ng/μl for FRPP94). The amplification conditions followed the protocol described by Mariette et al. (2001) for FRPP94 and by González-Martínez et al. (2002) for FRPP91 and ITPH4516 loci.
PCR reactions for analysis of the PtTX3107 locus were performed in the same volume and levels of reagents, as above, but with 100 μM of each dNTP, 4 mM of MgCl2, and 0.15 ng/μl of genomic DNA. The amplification conditions followed the protocol described by Auckland et al. (2002) as modified by González-Martínez et al. (2004).
PCR reactions for analysis of NZPR114, NZPR544 and RPtEST11 loci were performed using the same volumes and levels of reagents as the other reactions, but with 200 μM of each dNTP, 0.1 μM for NZPR114 and RPtEST11 primers (or 0.25 μM for NZPR544), 2.00 mM of MgCl2 and 0.3 ng/μl of genomic DNA for NZPR114 and RPtEST11 (or 0.6 ng/μl for NZPR544). Amplification conditions followed the protocol described by (Chagné et al. 2004). All PCR amplifications were performed in a Uno-Thermoblock thermal cycler (Biometra).
An individual aliquot from each PCR reaction was initially assessed by electrophoresis on 1.0% agarose gels to check amplification. Samples were then separated by capillary electrophoresis using a Beckman Coulter automatic DNA Sequencer (CEQ™ 8000), with linear polyacrylamide (LPA) denaturing gel and CEQ separation buffer. Depending on the sample, 0.5 μl (FRPP94) or 0.1 μl (FRPP91, ITPH4516, PtTX3107, NZPR114, NZPR544, RPtEST11) of PCR product were mixed with 24 μl of deionized formamide (SLS, sample loading solution), 0.5 μl of CEQ™ DNA Size Standard 400 and one drop of mineral oil for fragment analysis. Fragment lengths of PCR products were evaluated by CEQ™ 8000 Genetic Analysis System Software.
For DNA samples in which fragment size variations could be attributed to mutations, PCR amplification and subsequent fragment analysis were repeated at least three times for validation. An allele was considered mutated when more than a 2 bp mismatch was observed in comparison with the size of the original allele.
Statistical analysis
Significant genotype effects on average number of somatic embryos obtained per gram of FW ECL tissue were analyzed by ANOVA. Means were compared using a Tukey’s HSD post hoc test.
Effects of loci, allele, time in culture under proliferation conditions, and genotype on occurrence of SSR mutations were evaluated by logistic regression analysis.
Results
The aim of this research was to assess genetic stability in P. pinaster at several SSR loci along the process of SE. Experiments were designed to analyze the effects of prolonged ECL culture under proliferation conditions. In addition, the effect of the overall SE process was also assessed through the analysis of SSR loci stability in the regenerated emblings which can be considered the end product of SE.
PCR amplification of SSR loci
A total of 158 DNA samples from tissues of ECLs under proliferation conditions, and from needles of regenerated emblings were analyzed. This analysis was based upon PCR amplification products from P. pinaster using primers of seven nuclear microsatellite loci selected based on unambiguous and reproducible amplification patterns using described protocols (data not shown).
A total of 41 alleles were obtained for the tested loci. The number of alleles ranged from 3 to 11 per locus (Table 1). The FRPP91 locus was the most polymorphic marker with 11 different alleles. The estimated average number of alleles per locus was 5.9. Representative electropherogram profiles of the FRPP94 locus are shown in Fig. 1.
Effect of prolonged culture under proliferation conditions (E1)
The proliferation of ECLs is one step of the SE process that starts at the time the cell lines are established. If desired, this step can be prolonged for indefinite periods until the somatic embryos are transferred to maturation conditions, where further development occurs. We analyzed the genetic stability of SSR loci in several ECLs maintained for 6 (T6), 14 (T14) and 22 (T22) months under proliferation conditions. From a total of 17 ECLs, 2 ECLs (11.8%) showed variation in amplification profiles at one or more time points in comparison with T0 (Table 2). Variation in allele sizes was observed for three SSR loci (FRPP91, ITPH4516, PtTX3107). However, logistic regression analysis revealed there were no significant effects on mutation rate related to genotype (Wald(16) = 0.213; P = 1.000), duration in culture under proliferation conditions (Wald(3) = 0.415; P = 0.937) and loci (Wald(1)0.826; P = 0.801).
Allele size variation was detected at the FRPP91 locus in ECL 49/432 subcultured for 6 months (T6) on proliferation medium. However, this mutation (from 170/186 to 170/170) was not observed in subsequent time points (T14 and T22) (Table 2). For the same ECL (49/432), a variation in allele size was detected at T14 and T22 (from 150/150 to 150/164 bp) for the PtTX3107 locus (Table 2).
In ECL 13/148, subcultured for 14 and 22 months (T14 and T22), a slippage mutation (from 152/164 to 152/162) at ITPH4516 locus was detected. The new allele with 162 bp identified in T14 and T22 versus 164 bp in T0, could result from loss of a repeat unit at this locus.
Genetic stability in emblings (E2)
One of the main objectives of this research was to evaluate reliability of SE methodology established for P. pinaster as a means of producing clonal individuals. Since emblings can be considered end products of the SE process, we analyzed allelic profiles of the seven SSR loci in emblings in comparison to starting material (established ECL). In addition, we analyzed allelic profiles among emblings regenerated from the same ECL in order to detect possible mosaicism. Furthermore, in order to identify any correlation between maturation ability and genetic stability, we collected morphological data from the 52 emblings analyzed and recorded maturation rates of the four ECLs from which the emblings were established.
With regard to maturation, the four ECLs developed cotyledonary somatic embryos after abscisic acid treatment. However, there was a highly significant effect of genotype on maturation (F(3.16) = 17.361; P < 0.001), in terms of number of cotyledonary embryos produced per gram FW. ECL 32/4 showed highest maturation rate (63 embryos/g FW). This rate was significantly different from rates obtained for ECLs 16/385 (16 embryos/g FW) and 82/73 (4 embryos/g FW), according to Tukey’s HSD post hoc test. Significant differences in maturation rates were also found between ECLs 13/100 (39 embryos/g FW) and 82/73, and ECLs 82/73 and 32/4.
Thirty-three plants (64%) of the total emblings used in the SSR analysis showed an abnormal phenotype after 4 months under ex vitro conditions (Fig. 2). Morphology of the abnormal phenotypes was related to plagiotropism (10/33), loss of apical dominance (25/33) and bifurcation traits (3/33). However, 8 months later, 50% of plagiotropism initially detected had reverted.
With regard to SSR profiles, 5 (9.6%) of the 52 emblings showed variation in amplification profiles for tested loci compared with the ECL of origin (Table 3). The mean variation frequency per locus was 2.7%. However, no significant effects of loci (Wald(6) = 4.659; P = 0.588) and allele (Wald(1) = 0.104; P = 0.588) on occurrence of mutations were detected. Nevertheless, the highest mutation rate (7.7%) was observed in the ITPH4516 locus (Table 4), where genetic variation was found in four individuals, from ECLs 82/73, 32/4, and 16/385. This locus was one of the most polymorphic of those tested (Table 1). Moreover, a new allele was also obtained at this locus. No SSR allele size variation was detected in locus PtTX3107 where polymorphism was relatively low.
There was a significant effect of genotype on occurrence of mutation (Wald(3) = 21.745; P < 0.001), with significant differences obtained between emblings from ECL 16/385 and from ECLs 13/100, 32/4 and 82/73 (Wald(1) = 13.885; P < 0.001). However, the highest mutation rate observed in emblings from ECL 16/385 may reflect the low number of plants analyzed in comparison to other ECLs tested. Low maturation (16 embryos/g FW) and germination rates (data not shown) in ECL 16/385 resulted in a low number of regenerated emblings for SSR analysis.
From the emblings showing an abnormal phenotype (33 out of the 52) only three revealed variation in analysis of SSR patterns. Anomalous phenotypes associated with detection of SSR variation were loss of apical dominance in all three cases. In the two emblings (D and E, Table 3) where mutations were found in more than one allele, an abnormal phenotype was also observed.
In three individual emblings (A, B and C, Table 3), regenerated from ECL 82/73, allele size variation was detected. However, only one of these individuals had an abnormal phenotype. There was no apparent relationship between SSR mutations detected and phenotype in the above emblings. However, in the mutated individuals D and E, from ECLs 32/4 and 16/385, respectively, allelic size variations were detected in more than one locus. The SSR analysis of individual E showed mutations in FRPP94, ITPH4516, RPtEST11, NZPR114 and NZPR544 loci. This individual showed a pronounced loss of apical dominance.
Discussion
Studies of genetic stability during SE of P. pinaster may provide useful tools to monitor and improve the SE process. This improvement can occur through selection of protocols conducive to maintaining genetic stability. Polyembryony is a common phenomenon in gymnosperms, whereby multiple embryos develop in a seed as a result of fertilization of several archegonia (simple polyembryony) or by embryo cleavage (cleavage polyembryony). In order to assure that detected variations in a single ECL resulted from in vitro culture, and not from mosaicism created by co-culturing more than one immature embryo, individual ECLs should be obtained from a single immature zygotic embryo. In this work, each embryo was isolated at the pre-cotyledonary stage, the most responsive to SE initiation (Miguel et al. 2004). Therefore, any variations detected in SSR loci in ECLs under prolonged proliferation conditions resulted from genetic instability that occurred during this stage. It should be noted that the variation at several SSR loci found in this work may result from mutation at the locus itself (or primer site) or from other genomic changes including ploidy level. For instance, the variation detected for ECL 49/432 at T14 and T22 (from 150/150 bp to 150/164 bp) for the PtTX3107 locus, may result from a mutation in the second allele yielding a new allelic profile compatible with 150/164. Alternatively, an addition of a third allele might have occurred as a result of a duplication of a chromosome fragment or addition of a single extra chromosome during in vitro culture. In this case, an allelic profile of 150/150/164 in the PtTX3107 locus would correctly describe the detected mutation.
Somatic mutations can result in the production of genetic chimeras, where unchanged alleles are retained by some cells (López et al. 2004; Burg et al. 2007). The variation at locus FRPP91, obtained in ECL 49/432 under proliferation for 6 months, was not detected in the subsequent time points of analysis. The inability to detect this variation at later time points suggests that only non-mutated cells survived and continued to proliferate. DeVerno et al. (1999) suggested somatic embryogenic tissue may be composed of a mixture of genetically altered and unaltered cells with different populations of altered cells occurring at various stages during in vitro culture. Etienne and Bertrand (2003) described the effects of age of embryogenic cell suspensions on frequency and phenotype of variants of Coffea arabica, where somaclonal variation increased 25% in plants produced from 12-month-old cell suspensions. Other authors have noted increasing mutation in relation to prolonged culture in the presence of synthetic growth regulators, such as 2,4 dichlorophenoxyacetic acid (Lee and Phillips 1988; DeVerno 1995; Bordallo et al. 2004; Lakshmanan et al. 2007).
In pine species, it is widely accepted that the maturation ability tends to decrease with ECL age. The same observation is true for P. pinaster (Klimaszewska et al. 2009). It has been hypothesized that the reduced ability to regenerate from such embryogenic tissues is related to genetic instability. One explanation may be that during proliferation, cells are rapidly dividing and there is an increased probability of genetic errors accumulating during this stage. The loss of ability to regenerate could be explained by either loss or mutation of genes essential for regeneration, or by changes in ploidy level (Deverno 1995). The activation and insertion of transposable elements is another mechanism that can cause variation in tissue culture during repeated subcultures (Fourré 2000).
In our study, a mutation was detected in an ECL as soon as 6 months of culture under proliferation conditions. This results clearly leads to the conclusion that embryogenic tissues should be cryopreserved as soon as possible after the establishment of ECLs in order to avoid genetic instability that may arise during the proliferation stage, with potential consequences on the embling regeneration ability or quality.
Until now, there were no reported studies concerning genetic stability of Pinus clonal plants propagated by SE. The assessment of genetic stability in the end product of somatic embryogenesis, the emblings, is essential before this technology can be considered for large-scale clonal propagation. In our study, about 9.6% of the analyzed emblings of P. pinaster showed changes in the genomic regions analysed. Despite the high frequency of abnormal phenotypes in plants derived from SE, these morphological traits could not be directly related to the genetic variation detected. In Pinus sylvestris, high mutation rates in SSR sequences were also found in embryogenic cells lines and cotyledonary embryos (Burg et al. 2007). These authors suggest that the high mutation rates found might reflect the plasticity to adapt to stress, which also includes adaptation to in vitro conditions. However, no information was presented on SSR analysis of emblings. In contrast, stability of nuclear microsatellite sequences was reported in Picea abies, at successive stages of SE, including plants regenerated from SE (Harvengt et al. 2001; Helmersson et al. 2004).
The emblings of P. pinaster analyzed in our study were regenerated from ECLs cultured on proliferation medium for over 12 months. It is clear from our study that there is a risk of mutations accumulating in embryogenic tissues during the SE process. This risk is particularly high during the proliferation stage, which can lead to regeneration of mutant plants. Furthermore, our results show that plants with different mutation patterns were regenerated from the same ECL (e.g., ECL 82/73). This can be explained by the presence in the same ECL of a mixture of cell populations carrying different mutations resulting in the regeneration of plants which are mutant and not truly clonal. Genetic variation detected in the loci we analyzed could not, however, be related to somaclonal variation expressed at morphological level. It may be possible that phenotypic changes in these emblings are correlated with changes in other regions of the genome. Thus, we cannot discount the possibility that somaclonal variation expressed at morphological level is not related to genetic mutations. A wider screening of genomic regions may be required to prove or disprove this relationship. Another possibility is that epigenetic factors, or a combination of genetic and epigenetic alterations, are on the basis of the observed somaclonal variation seen at the morphological level. Epigenetic factors could include reversible changes in the methylation of DNA or acetylation of histones that occur during somatic embryogenesis, as already suggested for Medicago truncatula (Santos and Fevereiro 2002). In our study with P. pinaster, after 12 months of ex vitro conditions, 50% of the plants in which plagiotropism was initially detected and started to revert to orthotropism. Similarly, plagiotropic growth was also initially detected in plants of Larix (Ewald 1999) and in Picea (Tremblay et al. 1999) propagated by SE, but after approximately 5 years a normal orthotropic growth was completely restored in these plants.
The optimization of SE protocols for P. pinaster including a reduction of the in vitro culture period and/or the use of lower levels of growth regulators may be required. The microsatellites analyzed in our study proved to be useful for monitoring mutational changes related to the SE process in P. pinaster. The implementation of new breeding and deployment strategies for improved pine trees using SE may profit from integrating the use of these molecular markers in the monitorization of intra-clonal variation.
Abbreviations
- ECL:
-
Embryogenic cell lines
- SE:
-
Somatic embryogenesis
- SSR:
-
Simple sequence repeat
References
Auckland LD, Bui T, Zhou Y, Shepherd M, Williams CG (2002) Conifer microsatellite handbook. Corporate Press, Raleigh NC, p 57
Bordallo PN, Silva DH, Maria J, Cruz CD, Fontes EP (2004) Somaclonal variation on in vitro callus culture potato cultivars. Hortic Bras 22:300–304
Burg K, Hristoforoglu K, Fluch S, Hohl K, Burg A, Schmidt J (1999) Analysis of the fidelity of DNA replication in embryogenic cultures of Norway spruce. In: Biofor 99—“Applications of biotechnology to forest genetics”, Vitoria-Gasteiz, Spain, pp 231–235
Burg K, Helmersson A, Bozhkov P, von Arnold S (2007) Developmental and genetic variation in nuclear microsatellite stability during somatic embryogenesis in pine. J Exp Bot 58:687–698. doi:610.1093/jxb/erl1241
Chagné D, Chaumeil P, Ramboer A, Collada C, Guevara A, Cervera MT, Vendramin GG, Garcia V, Frigerio J-M, Echt C, Richardson T, Plomion C (2004) Cross species transferability and mapping of genomic and cDNA SSRs in pines. Theor Appl Genet 109:1204–1214. doi:1210.1007/s00122-00004-01683-z
DeVerno LL (1995) An evaluation of somaclonal variation during somatic embryogenesis. In: Jain SM, Gupta PK, Newton RJ (eds) Somatic embryogenesis in woody plants. Kluwer, Dordrecht, pp 361–377
DeVerno LL, Charest PJ, Bonen L (1994) Mitochondrial DNA variation in somatic embryogenic cultures of Larix. Theor Appl Genet 88:727–732. doi:710.1007/BF01253977
DeVerno LL, Park YS, Bonga JM, Barret JD (1999) Somaclonal variation in cryopreserved clones of white spruce (Picea glauca (Moench) Voss.). Plant Cell Rep 18:948–953. doi:910.1007/s002990050689
DGRF (2008) Inventário Florestal National, Direcção Geral de Recursos Florestais. In: http://www.dgrf.min-agricultura.pt
El-Dougdoug KA, El-Harthi HMS, Korkar HM, Taha RM (2007) Detection of somaclonal variations in banana tissue culture using isozyme and DNA fingerprint analysis. J Appl Sci Res 3:622–627
Etienne H, Bertrand B (2003) Somaclonal variation in Coffea arabica: effects of genotype and embryogenic cell suspension age on frequency and phenotype of variants. Tree Physiol 23:419–426
Ewald D (1999) Is plagiotropic growth in micropropagated larch a marker for ageing? In: International Congress “Applications of biotechnology to forest genetics”—BIOFOR 99, Vitoria-Gasteiz, pp 479–486
Fourré J-L (2000) Somaclonal variation and genetic molecular markers in woody plants. In: Jain SM, Minocha SC (eds) Molecular biology of woody plants. Kluwer, The Netherlands, pp 425–449
Fourré J-L, Berger P, Niquet L, André P (1997) Somatic embryogenesis and somaclonal variation in Norway spruce: morphogenic, cytogenetic and molecular approaches. Theor Appl Genet 94:159–169. doi:110.1007/s001220050395
Goldstein DB, Pollock DD (1997) Launching microsatellites: a review of mutation processes and methods of phylogenetic inference. J Hered 88:335–342
González-Martínez SC, Gerber S, Cervera MT, Martínez-Zapater JM, Gil L, Alía R (2002) Seed gene flow and fine-scale structure in a Mediterranean pine (Pinus pinaster Ait.) using nuclear microsatellite markers. Theor Appl Genet 104:1290–1297. doi:1210.1007/s00122-00002-00894-00124
González-Martínez SC, Robledo-Arnuncio JJ, Collada C, Díaz A, Williams CG, Alía R, Cervera MT (2004) Cross-amplification and sequence variation of microsatellite loci in Eurasian hard pines. Theor Appl Genet 109:103–111
Gupta PK, Durzan DJ (1985) Shoot multiplication from mature trees of Douglas fir (Pseudotsuga menziesii) and sugar pine (Pinus lambertina). Plant Cell Rep 4:177–179. doi:110.1007/BF00269282
Harvengt L (2005) Somatic embryogenesis in maritime pine (Pinus pinaster Ait.). In: Jain SM, Gupta PK (eds) “Protocol for somatic embryogenesis in woody plants”, Series: forestry sciences. Springer, The Netherlands, pp 107–120
Harvengt L, Trontin J-F, Reymond I, Canlet F, Pâques M (2001) Molecular evidence of true-to-type propagation of a 3-year-old Norway spruce through somatic embryogenesis. Planta 213:828–832. doi:810.1007/s004250100628
Helmersson A, von Arnold S, Burg K, Bozhkov PV (2004) High stability of nuclear microsatellite loci during the early stages of somatic embryogenesis in Norway spruce. Tree Physiol 24:1181–1186
Hornero J, Martínez I, Celestino C, Gallego FJ, Torres V, Toribio M (2001) Early checking of genetic stability of cork oak somatic embryos by AFLP analysis. Int J Plant Sci 162:827–833. doi:810.1086/320784
Isabel N, Tremblay L, Michaud M, Tremblay FM, Bousquet J (1993) RAPDs as an aid to evaluate the genetic integrity of somatic embryogenesis-derived populations of Picea mariana (Mill.) B.S.P. Theor Appl Genet 86:81–87. doi:10.1007/BF00223811
Klimaszewska K, Noceda C, Pelletier G, Label P, Rodriguez R, Lelu-Walter M-A (2009) Biological characterization of young and aged embryogenic cultures of Pinus pinaster (Ait.). In Vitro Cell Dev Biol Plant. doi:10.1007/s11627-008-9158-6
Lakshmanan V, Venkataramareddy SR, Neelwarne B (2007) Molecular analysis of genetic stability in long-term micropropagated shoots of banana using RAPD and ISSR markers. Electron J Biotechnol 10:106–113
Lee M, Phillips RL (1988) The chromosomal basis of somaclonal variation. Ann Rev Plant Physiol Plant Mol Biol 39:413–437. doi:410.1146/annurev.pp.1139.060188.002213
Lelu M-A, Bastien C, Drugeault A, Gouez M-L, Klimaszewska K (1999) Somatic embryogenesis and plantlet development in Pinus sylvestris and Pinus pinaster on medium with and without growth regulators. Physiol Plantarum 105:719–728. doi:710.1034/j.1399-3054.1999.105417.x
Lelu-Walter M-A, Bernier-Cardou M, Klimaszewska K (2006) Simplified and improved somatic embryogenesis for clonal propagation of Pinus pinaster (Ait.). Plant Cell Rep 25:767–776
Leroy XJ, Leon K, Branchard M (2000) Plant genomic instability detected by microsatellite-primers. Electron J Biotechnol 3:5–10
Lopes T, Pinto G, Loureiro J, Costa A, Santos C (2006) Determination of genetic stability in long-term somatic embryogenic cultures and derived plantlets of cork oak using microsatellite markers. Tree Physiol 26:1145–1152
López CMR, Wetten AC, Wilkinson MJ (2004) Detection and quantification of in vitro-culture induced chimerism using simple sequence repeat (SSR) analysis in Theobroma cacao (L). Theor Appl Genet 110:157–166. doi:110.1007/s00122-00004-01823-00125
Mariette S, Chagné D, Decroocq S, Vendramin GG, Lalanne C, Madur D, Plomion C (2001) Microsatellite markers for Pinus pinaster Ait. Ann For Sci 58:203–206
Miguel C, Gonçalves S, Tereso S, Marum L, Maroco J, Oliveira MM (2004) Somatic embryogenesis from 20 open-pollinated families of Portuguese plus trees of maritime pine. Plant Cell Tissue Organ Cult 76:121–130
Peredo EL, Revilla MÁ, Arroyo-García R (2006) Assessment of genetic and epigenetic variation in hop plants regenerated from sequential subcultures of organogenic calli. J Plant Physiol 163:1071–1079. doi:1010.1016/j.jplph.2005.1009.1010
Rahman MH, Rajora OP (2001) Microsatelite DNA somaclonal variation in micropropagated trembling aspen (Populus tremuloides). Plant Cell Rep 20:531–536. doi:510.1007/s002990100365
Santos D, Fevereiro P (2002) Loss of DNA methylation affects somatic embryogenesis in Medicago truncatula. Plant Cell Tissue Organ Cult 70:155–161
Tereso S, Miguel C, Zoglauer K, Valle-Piquera C, Oliveira MM (2006) Stable Agrobacterium-mediated transformation of embryogenic tissues from Pinus pinaster Portuguese genotypes. Plant Growth Regul 6:24–33. doi:10.1007/s10725-10006-19126-10722
Tremblay L, Levasseur C, Tremblay FM (1999) Frequency of somaclonal variation in plants of black spruce (Picea mariana, Pinaceae) and white spruce (P. glauca, Pinaceae) derived from somatic embryogenesis and identification of some factors involved in genetic instability. Am J Bot 86:1373–1381
Von Aderkas PRP, Hristoforoglu K, Ma Y (2003) Embryogenesis and genetic stability in long term megagametophyte-derived cultures of larch. Plant Cell Tissue Organ Cult 75:27–34
Whittaker JC, Harbord RM, Boxall N, Mackay I, Dawson G, Sibly RM (2003) Likelihood-based estimation of microsatellite mutation rates. Genetics 164:781–787
Wilhelm E, Hristoforoglu K, Fluch S, Burg K (2005) Detection of microsatellite instability during somatic embryogenesis of oak (Quercus robur L.). Plant Cell Rep 23:790–795. doi:710.1007/s00299-00004-00891-y
Acknowledgements
The authors are grateful to Dr. Manuela Veloso from Estação Agronómica Nacional, Portugal for technical support and to Dr. Bruce Campbell for the critical reading of the manuscript and English language revising. This research was supported by Fundação Ciência e Tecnologia (FCT) and POCI 2010, through a PhD Grant SFRH/BD/17906/2004/28UE to Liliana Marum. Estação Florestal Nacional (EFN) is acknowledged for making plant material available.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Puigdomenech.
Rights and permissions
About this article
Cite this article
Marum, L., Rocheta, M., Maroco, J. et al. Analysis of genetic stability at SSR loci during somatic embryogenesis in maritime pine (Pinus pinaster). Plant Cell Rep 28, 673–682 (2009). https://doi.org/10.1007/s00299-008-0668-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-008-0668-9