Abstract
Chromosome positioning is crucial for multiple chromosomal events, including DNA replication, repair, and recombination. The linker of nucleoskeleton and cytoskeleton (LINC) complexes, which consist of conserved nuclear membrane proteins, were shown to control chromosome positioning and facilitate various biological processes by interacting with the cytoskeleton. However, the precise functions and regulation of LINC-dependent chromosome positioning are not fully understood. During meiosis, the LINC complexes induce clustering of telomeres, forming the bouquet chromosome arrangement, which promotes homologous chromosome pairing. In fission yeast, the bouquet forms through LINC-dependent clustering of telomeres at the spindle pole body (SPB, the centrosome equivalent in fungi) and detachment of centromeres from the SPB-localized LINC. It was recently found that, in fission yeast, the bouquet contributes to formation of the spindle and meiotic centromeres, in addition to homologous chromosome pairing, and that centromere detachment is linked to telomere clustering, which is crucial for proper spindle formation. Here, we summarize these findings and show that the bouquet chromosome arrangement also contributes to nuclear fusion during karyogamy. The available evidence suggests that these functions are universal among eukaryotes. The findings demonstrate that LINC-dependent chromosome positioning performs multiple functions and controls non-chromosomal as well as chromosomal events, and that the chromosome positioning is stringently regulated for its functions. Thus, chromosome positioning plays a much broader role and is more strictly regulated than previously thought.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the nucleus of eukaryotic cells, chromosome positioning dynamically changes during development and differentiation and thereby contributes to regulation of various chromosomal events, including DNA transcription, repair, and recombination (Spector 2003; Cremer and Cremer 2010; Meister et al. 2011; Aparicio 2013; Mine-Hattab and Rothstein 2013; Rocha and Skok 2013). The linker of nucleoskeleton and cytoskeleton (LINC) complex, which consists of the conserved Sad1/Unc-84 (SUN) and Klarsicht/ANC-1/Syne homology (KASH) domain nuclear membrane proteins, connects the chromosomes to the cytoskeleton. Recent work showed that the LINC complex is a crucial factor that controls chromosome positioning and facilitates various biological processes (Fridkin et al. 2009; Hiraoka and Dernburg 2009; Oza et al. 2009; Razafsky and Hodzic 2009; Schober et al. 2009; Horigome et al. 2014).
One of the most prominent examples of LINC-dependent chromosome positioning is observed during meiosis. During meiotic prophase, homologous chromosomes pair and recombine, forming chiasmata that link homologous chromosomes and enable their segregation at meiosis I (Roeder 1997; Page and Hawley 2003; Petronczki et al. 2003). At this stage, telomeres cluster in a small area, forming the typical chromosome arrangement referred to as the “bouquet” (Zickler and Kleckner 1998; Scherthan 2001). The bouquet promotes homologous chromosome pairing by inducing homologous association and concomitantly preventing non-homologous association (Zickler and Kleckner 1998; Harper et al. 2004; Hiraoka and Dernburg 2009; Koszul and Kleckner 2009). The LINC complexes connect telomeres with the cytoskeleton and induce bouquet formation (Fridkin et al. 2009; Hiraoka and Dernburg 2009; Razafsky and Hodzic 2009; Yamamoto 2014).
Promotion of homologous chromosome pairing is a common task of the bouquet in all organisms examined to date (Zickler and Kleckner 1998; Harper et al. 2004; Hiraoka and Dernburg 2009). However, recent studies of the fission yeast Schizosaccharomyces pombe showed that the bouquet also contributes to spindle formation and assembly of meiotic centromeres (Asakawa et al. 2005; Tomita and Cooper 2007; Klutstein et al. 2015; Katsumata et al. 2016), indicating that LINC-dependent chromosome positioning performs multiple tasks and controls non-chromosomal as well as chromosomal events. Furthermore, we recently found that dissociation of centromeres from the LINC complexes is linked to telomere clustering during bouquet formation and that this connection is crucial for proper spindle formation (Katsumata et al. 2016). Thus, LINC-dependent chromosome positioning is stringently regulated, and this regulation is crucial for its functions. In this manuscript, we describe recent advances in our understanding of the functions and the regulation of the bouquet chromosome arrangement in S. pombe, including our novel finding that the bouquet also contributes to nuclear fusion during karyogamy. Finally, we discuss the universality of the multiple functions of LINC-dependent chromosome positioning.
Formation of the bouquet chromosome arrangement in S. pombe
Meiosis progression and changes in chromosome positioning in S. pombe
Before going into detail regarding the functions and regulation of LINC-dependent chromosome positioning during meiosis in S. pombe, we first briefly describe meiotic progression and changes in chromosome positioning in this organism. In S. pombe cells, mitotic centromeres are attached to the spindle pole body (SPB, the fungal centrosome), which is associated with the nuclear envelope (NE); the telomeres are also associated with the NE, but at distinct sites from the SPB. This chromosome positioning corresponds to the “Rabl” configuration observed in many organisms [Fig. 1a(i), 1c, Rabl] (Funabiki et al. 1993; Cremer and Cremer 2010). Upon nitrogen starvation, two haploid cells conjugate, forming a diploid zygote, and immediately enter meiosis [Fig. 1a(ii–x)] (Yamamoto et al. 1997). Upon entering meiosis, telomeres gather at the SPB while centromeres detach from it, forming the “bouquet” chromosome arrangement [Fig. 1a(ii, iii), c, Bouquet] (Chikashige et al. 1994). During the bouquet stage, SPB-nucleated microtubules and cytoplasmic dynein, a microtubule motor, generate back-and-forth nuclear movements between the cell ends (termed “horsetail” nuclear movements after an elongated nuclear shape) [Fig. 1a(iii)] (Chikashige et al. 1994; Ding et al. 1998; Yamamoto et al. 1999, 2001). After a couple of hours of horsetail movements, a telomere cluster resolves, and the zygote undergoes two meiotic divisions, forming four spores [Fig. 1a(v–x)].
Meiotic process and changes in chromosome positioning in S. pombe. a Meiotic process in S. pombe. S. pombe cells normally proliferate in the haploid state. During mitotic interphase, centromeres (red circles) are located beneath the SPB (black circle), which is attached to the nuclear envelope (NE; thin black lines), whereas telomeres (blue circles) are located distant from the SPB, also in association with the NE [(i) mitotic interphase]. Microtubules (green lines) extend from the SPB and the NE in parallel to the longitudinal cell axis. This chromosome arrangement corresponds to the Rabl orientation in other species. Under nitrogen-starved conditions, two cells of opposite mating types conjugate to form a diploid zygote, followed by fusion of their nuclei [(ii) Karyogamy]. During cell conjugation, telomeres gather at the SPB, whereas centromeres become detached from it, thus forming the bouquet chromosome arrangement (Bouquet stage). After nuclear fusion, the diploid nucleus becomes elongated and moves back and forth between the cell ends, led by the SPB, which radiates microtubules [referred to as “horsetail nuclear movements”; (iii) Horsetail stage]. During the horsetail nuclear movements, DNA replication, homologous chromosome pairing, and meiotic recombination take place. After the horsetail nuclear movements, the nucleus stops around the center of the zygote (iv), telomere clustering resolves, and the nucleus forms the spindle and undergoes the first division [(v) meiosis I]. At the first division, the homologous chromosomes move apart from each other with elongation of the spindle (vi). After the first division, microtubule arrays similar to that in mitotic interphase are formed (vii). Subsequently, each nucleus forms a spindle and undergoes the second division [(viii) meiosis II]. After the second division, spindles disappear (ix), and the four spores are finally formed [(x) sporulation]. S. pombe contains three chromosomes, but only one (black thick lines) is shown for simplicity. b The telomere–LINC connection and the telocentrosome. Upon entering meiosis, Bqt1 and Bqt2 connect the SUN domain inner nuclear membrane protein Sad1 with Rap1, which interacts with the telomere component Taz1. Sad1 binds with KASH domain outer nuclear membrane proteins Kms1 and Kms2, forming the LINC complexes. At the telomere-localized LINC complexes, the γ-tubulin complex and cytoplasmic dynein become localized to form the microtubule-organizing center (“telocentrosome”) and nucleate microtubules. Only representative factors or complexes are shown. ONM outer nuclear membrane, INM inner nuclear membrane. c Formation of the bouquet chromosome arrangement. During mitotic interphase, centromeres (red sphere) are attached to the SPB (green sphere), which radiates microtubules (green lines), via the LINC complexes (dark blue ellipsoid) residing in the NE (brown line), whereas telomeres (light blue spheres) are located distant from the SPB in association with the NE. This chromosome arrangement corresponds to the Rabl orientation (Rabl). Upon nitrogen starvation, the LINC complexes are localized at the telomeres by telomere–LINC connectors. The microtubule-organizing center (telocentrosome) forms at the LINC-localized telomere and extends microtubules (LINC-dependent telocentrosome formation). SPB- and telocentrosome-nucleated microtubules gather the telomeres at the SPB with the aid of microtubule motors (Microtubule-dependent telomere clustering). After telomere clustering, centromeres detach from the SPB (Centromere detachment), forming the bouquet chromosome arrangement (bouquet). For simplicity, only one chromosome (gray lines) is shown
Telomere clustering mechanism
As in other organisms, telomere clustering in S. pombe depends on the LINC complexes. LINC complexes consisting of the SUN domain protein Sad1 and the KASH domain protein Kms2 are localized at the SPB in mitosis (Hagan and Yanagida 1995; Miki et al. 2004; Walde and King 2014). Upon entering meiosis, another KASH protein, Kms1, joins the LINC complexes (Shimanuki et al. 1997), and the Kms1/Kms2-containing LINC complexes become localized at telomeres, in addition to the SPB, via the actions of the meiosis-specific factors Bqt1 and Bqt2, which connect the telomere component Rap1 with Sad1 (Fig. 1b) (Chikashige et al. 2006; Yoshida et al. 2013). The γ-tubulin complex that nucleates microtubules and cytoplasmic dynein becomes localized at LINC-localized telomeres and forms the microtubule-organizing center, which has been termed the “telocentrosome” (Fig. 1b, c, LINC-dependent telocentrosome formation) (Yoshida et al. 2013; Yamamoto 2014). Cytoplasmic microtubules extend from the telocentrosome and the SPB, and these microtubules induce clustering of telomeres at the SPB with the aid of cytoplasmic dynein and kinesin microtubule motors (Fig. 1c, microtubule-dependent telomere clustering) (Yoshida et al. 2013; Yamamoto 2014).
The pathways that initiate meiosis induce telomere clustering. Upon nitrogen starvation, the mating pheromone activates the MAP kinase, inducing cell conjugation (Yamamoto et al. 1997; Harigaya and Yamamoto 2007), and activation of MAP kinase induces telomere clustering (Chikashige et al. 1997; Yamamoto et al. 2004). However, the MAP kinase-dependent pathway is not the sole regulatory pathway that induces telomere clustering. Inactivation of Pat1 kinase, which occurs after cell conjugation and induces meiotic divisions (Yamamoto et al. 1997; Harigaya and Yamamoto 2007), also induces telomere clustering without MAP kinase activation (Chikashige et al. 2004; Yamamoto et al. 2004). Therefore, both the MAP kinase- and Pat1-dependent pathways can independently induce telomere clustering. Because MAP kinase activation precedes Pat1 inactivation, it is possible that the MAP kinase-dependent pathway primarily contributes to establishment of telomere clustering, whereas the Pat1-dependent pathway contributes to its maintenance. It is currently unknown how these pathways induce telomere clustering. However, given that one major output of these pathways is expression of meiosis-specific genes (Yamamoto et al. 1997; Harigaya and Yamamoto 2007), they likely induce production of factors required for telocentrosome formation and/or microtubule-dependent telomere clustering.
Centromere detachment mechanism
Detachment of centromeres from the SPB, another change in chromosome positioning essential for bouquet formation (Fig. 1c, centromere detachment), occurs through elimination of centromere–LINC interaction. During mitosis, centromeres are attached to the SPB via the Csi1-dependent interaction of the kinetochore components with the LINC component Sad1 (Hou et al. 2012). Upon entering meiosis, kinetochore components dissociate from the LINC complexes and the centromeres, resulting in centromere detachment from the SPB (Asakawa et al. 2005; Hayashi et al. 2006; Katsumata et al. 2016).
Like telomere clustering, centromere detachment is regulated by the MAP kinase- and Pat1-dependent pathways. However, unlike telomere clustering, centromere detachment requires both MAP kinase activation and Pat1 inactivation concomitantly. If either occurs alone, the centromeres remain attached to the SPB (Chikashige et al. 1997, 2004), whereas, when both occur, the centromeres detach (Asakawa et al. 2005). As in the case of telomere clustering, it remains unclear how the MAP kinase- and Pat1-dependent pathways induce centromere detachment.
Recent work showed that centromere detachment is linked with telomere clustering. In Bqt1- or Rap1-lacking, telomere clustering-defective cells, centromeres frequently remained attached to the SPB (Fig. 2a, impaired telomere recruitment of LINC) (Katsumata et al. 2016). Centromeres are also frequently attached to the SPB when telomere clustering is impaired by microtubule disruption (Fig. 2a, microtubule disruption). These results indicate that centromere detachment is repressed when telomere clustering is defective.
Telomere clustering–linked regulation of centromere detachment. a Inhibition of centromere detachment in bouquet formation. Impaired telocentrosome formation due to loss of a telomere–LINC connector (Impaired telomere recruitment of LINC) or disruption of SPB- and telocentrosome-nucleated microtubules (Microtubule disruption) causes inhibition of centromere detachment. b Induction of centromere detachment by a Sad1-fused Taz1 fragment. In cells lacking a telomere–LINC connector, Bqt1 or Rap1, the LINC complexes are not recruited to telomeres, and the telomeres fail to cluster (Impaired telomere recruitment of LINC). When a portion of Taz1 fused with Sad1 (Taz1∆myb–Sad1) is introduced, it is localized at the SPB and induces centromere detachment in the absence of telomere clustering (Centromere detachment without telomere clustering). c Two mechanisms promote centromere detachment. SPB recruitment of Taz1 by telomere clustering (SPB recruitment of Taz1) and SPB- and telocentrosome-nucleated microtubules (MT formation) cooperatively promote kinetochore disassembly and centromere detachment from the SPB
Telomere clustering causes recruitment of telomere components to the SPB, and one of these components, the telomere-binding protein Taz1, contributes to centromere detachment. When a portion of Taz1 that lacks the telomeric DNA-binding myb domain is artificially tethered to the SPB by fusion with Sad1, the repression of centromere detachment observed in Bqt1- or Rap1-lacking cells is alleviated (Taz1∆myb–Sad1; Fig. 2b) (Katsumata et al. 2016). This indicates that efficient centromere detachment requires SPB recruitment of Taz1 (Fig. 2c, SPB recruitment of Taz1), but it does not require SPB interaction of telomeres or formation of the telocentrosome. However, Taz1∆myb–Sad1 fails to alleviate the repression of centromere detachment when telomere clustering is impaired by disruption of cytoplasmic microtubules, indicating that intact microtubules are also required for efficient centromere detachment (Fig. 2c, MT formation) (Katsumata et al. 2016). Thus, both Taz1-dependent and microtubule-dependent mechanisms facilitate centromere detachment, linking centromere detachment with telomere clustering. Molecular mechanisms of these pathways remain to be elucidated.
Functions of the bouquet chromosome arrangement in S. pombe
The bouquet promotes homologous chromosome pairing
In S. pombe, as in other organisms, the bouquet promotes homologous chromosome pairing. Impairment of the bouquet chromosome arrangement by mutation of genes required for the telomere–LINC interaction or LINC integrity compromises pairing and segregation of homologous chromosomes and promotes non-homologous chromosome association (Shimanuki et al. 1997; Cooper et al. 1998; Nimmo et al. 1998; Niwa et al. 2000; Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001; Tuzon et al. 2004). Similarly, when horsetail nuclear movements are impaired by mutation in the dhc1 gene, which encodes a dynein motor subunit, homologous chromosome pairing is impaired (Yamamoto et al. 1997; Ding et al. 2004). These results suggested that a combination of telomere clustering and SPB movements induces alignment and contact of homologous chromosomes, thereby promoting homologous chromosome pairing (Yamamoto and Hiraoka 2001; Hiraoka and Dernburg 2009). In this model, spatial alignment of homologs requires structural integrity of chromosomes. Consistent with this, a recent study showed that impairment of chromosome structure decreases the frequency of homologous chromosome pairing (Ding et al. 2016a, b).
Centromere detachment from the SPB induces meiotic centromere formation
In addition to homologous chromosome pairing, the bouquet chromosome arrangement contributes to formation of meiosis-specific centromeres. During homologous chromosome segregation at meiosis I, kinetochores on sister centromeres fuse with one another while sister centromere cohesion persists, allowing sister chromatids to co-segregate to the same spindle pole without separating (Petronczki et al. 2003; Hauf and Watanabe 2004; Brar and Amon 2008; Miller et al. 2013). Several studies showed that when centromere detachment does not occur, the properties of meiotic centromeres are compromised. In cells lacking chiasmata, sister chromatids often become attached to both spindle poles and experience forces toward opposite poles, but they rarely undergo separation owing to the properties of meiosis-specific centromeres (Hirose et al. 2011). Importantly, when centromeres remain attached to the SPB in the absence of MAP kinase activation (Chikashige et al. 2004), sister chromatids are predominantly segregated apart from each other in chiasma-lacking cells (Yamamoto and Hiraoka 2003). Furthermore, in bouquet-defective cells, in which centromere detachment is repressed, the separation frequency of sister chromatids significantly increased when chiasmata are eliminated (Katsumata et al. 2016). All these observations indicate that centromere detachment is required for establishment of the proper meiotic centromere properties. In bouquet-defective cells, in addition, centromeric localization of the heterochromatin protein 1 homolog Swi6 and the centromere-specific histone H3 variant Cnp1 is impaired (Klutstein et al. 2015). This may mean that centromere detachment is required for proper centromere localization of these components. Because kinetochore disassembly accompanies centromere detachment, it was proposed that the properties of meiosis-specific centromeres are established through reformation of the kinetochore (Asakawa et al. 2005).
The bouquet contributes to spindle formation
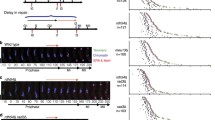
Recent studies showed that the bouquet also contributes to spindle formation. In cells defective in the LINC-dependent telomere–SPB interaction, the SPB is often missing from the spindle poles at meiosis I [Fig. 3a, bqt1∆ and rap1∆, lower graph, and c, MI (aberrant)] (Tomita and Cooper 2007; Fennell et al. 2015). This indicates that the telomere–SPB interaction is required for proper spindle formation (Tomita and Cooper 2007; Chikashige et al. 2014; Fennell et al. 2015; Katsumata et al. 2016).
Meiosis progression in diploid zygotes. a, b Changes in the population of different types of zygotes with (b) or without (a) expression of Taz1∆myb–Sad1 fusion. Cells were crossed on ME solid medium, and, at the indicated time points, the nuclear and microtubule morphology of more than 100 zygotes was examined under a microscope. (Upper graph) Populations of zygotes in various categories. Zygotes were placed into six different categories as follows: 2 nuc bi-nuclear zygotes forming an astral microtubule array with a single microtubule focus [see c, 2 nuc and 2 nuc (aberrant)], 1 nuc mono-nuclear zygotes forming an astral microtubule array with a single microtubule focus [see c, 1 nuc and 1 nuc (aberrant)], MI mono- or bi-nuclear zygotes forming a single spindle [see c, MI and MI (aberrant)] or bi-nuclear zygotes without an astral microtubule array (see Fig. 1a, vii), MII bi- or four-nuclear zygotes forming two spindles (see c, MII) or four nuclear zygotes forming short cytoplasmic microtubules [see Fig. 1a(i)]; Aberrant: zygotes forming aberrant spindles [see c, MI (aberrant)]; sporulated: zygotes forming spores (see c, sporulated). (Lower graph) Cumulative populations of zygotes forming spindles. MI zygotes forming MI spindle (see c, MI), MII zygotes forming MII spindle (see c, MII), Aberrant MI zygotes forming aberrant MI spindle [see c, MI (aberrant)]. Note that, because spindle phenotype was examined every 2 h, which is longer than the lifetime of the spindle, the population of cells with each type of spindle was undercounted. c Representative nuclear and microtubule morphology of zygotes. Blue, green, and magenta, respectively, indicate DNA, SPB (GFP-tagged Sid4), and microtubule (mCherry-tagged Atb2). White lines indicate cell shapes. The arrowhead indicates a SPB detached from the chromosome mass, and the arrow indicates a spindle pole lacking a SPB. Bar 5 µm
Interestingly, live-cell analysis of individual bouquet-defective cells revealed that deletion of a gene required for the nuclear movements, such as dhc1, eliminated the cells that formed an abnormal spindle (Chikashige et al. 2014; Fennell et al. 2015). Furthermore, in the bouquet-defective cells, the SPB is often detached from the chromosome mass during the horsetail nuclear movements [Fig. 3c, 1 nuc (aberrant)] (Tomita and Cooper 2007; Chikashige et al. 2014). Based on these observations, it was proposed that nuclear movements cause detachment of the SPB from the NE in bouquet-defective cells, impairing spindle formation (Chikashige et al. 2014).
However, several other observations argue against this idea. First, when an improper spindle forms, the SPB is associated with the nuclear periphery (Fennell et al. 2015). Second, although dhc1 deletion mostly eliminates zygotes with an aberrant meiosis I spindle, the proportion of zygotes forming a normal meiosis I spindle does not increase, and instead tends to decrease (Fig. 3a, bottom graph). Importantly, dhc1 deletion increased the frequency of bi-nuclear zygotes (Fig. 3a, top graphs). These observations raise the possibility that dhc1 deletion inhibits nuclear fusion in spindle-defective zygotes (see the next section), thereby eliminating aberrant spindle formation. This phenomenon was likely ignored in previous studies, which only examined mono-nuclear zygotes (Chikashige et al. 2014). Third, the centromere–SPB interaction can substitute for the telomere–SPB interaction to induce proper meiotic spindle formation (Fennell et al. 2015), and, when the centromere–SPB interaction is abolished during mitosis, mitotic spindle formation is impaired, as seen in bouquet-defective cells (Fernandez-Alvarez et al. 2016). Because horsetail-like nuclear movements are absent in mitosis, the mitotic spindle defects seen in cells defective in the centromere–SPB interaction cannot be attributed to such movements. Collectively, based on the observations described above, it seems unlikely that the horsetail nuclear movements cause the spindle defects seen in bouquet-defective cells.
A very recent work revealed that LINC-dependent SPB interaction of either telomeres or centromeres is required for insertion of the SPB into the NE (Fernandez-Alvarez et al. 2016). In S. pombe, the NE does not break down and continues to enclose the chromosomes during nuclear division. At the onset of nuclear division, the SPB, which is located outside of the NE, penetrates into the NE via local NE breakdown (NEBD) to form the spindle (Ding et al. 1997). Loss of telomere/centromere–SPB interaction compromises the NEBD-accompanied SPB insertion into the NE, resulting in spindle impairment (Fernandez-Alvarez et al. 2016).
It is currently not clear how the LINC-dependent telomere/centromere–SPB interaction contributes to the SPB insertion. Notably in this regard, the frequency of spindle defects is greatly reduced in haploid meiotic cells defective in the telomere–SPB interaction (Fig. 4a), in contrast to the situation in the corresponding mutant zygotes [in live-cell analysis, about half of bqt1∆ zygotes and ~80% of bqt1∆ zygotes expressing Taz1∆myb–Sad1 exhibit spindle defects (Katsumata et al. 2016)]. Because telomere/centromere–SPB interaction is also crucial for karyogamy (see the next section), a process that is missing from haploid meiosis, this may mean that defective telomere/centromere–SPB interaction often causes impaired or incomplete fusion of the NE and/or the SPBs of two nuclei during karyogamy, which in turn causes defective SPB insertion and spindle impairment during the subsequent meiosis I.
Spindle formation in haploid meiosis. a Observation frequencies of meiosis I spindle types in various types of haploid cells. Haploid cells were induced to enter meiosis in EMM-N liquid medium, and then their spindle morphology was examined every 10 min under a microscope. The spindle and SPB were visualized using mCherry-tagged Atb2 and GFP-tagged Sid4, respectively. +, no mutations otherwise depicted; +None, without Taz1∆myb–Sad1 expression; +Taz1∆myb–Sad1, with Taz1∆myb–Sad1 expression; bipolar, normal spindle with SPBs at both poles; monopolar, spindle missing the SPB at one pole; nonpolar, spindle missing SPBs at both poles; others, other types of spindles; no spindle, no spindle formation. Images show representative types of meiosis I spindles, and arrowheads indicate spindle poles lacking SPBs. Numbers in parenthesis indicate numbers of cells examined. b Dynamics of meiotic spindles in haploid cells. Numbers indicate time in min. Green shows GFP-tagged Sid4 (SPB), and magenta shows mCherry-tagged Atb2 (microtubules) and Taz1∆myb–Sad1. Arrowheads indicate mCherry dots, which were often observed in dhc1∆ cells expressing Taz1∆myb–Sad1. These dots probably represent Taz1∆myb–Sad1 that failed to localize at the SPB due to the lack of dynein activity. White lines indicate cell outlines. Bar 2 µm
The telomere clustering-linked regulation of centromere detachment is advantageous for spindle formation. Because SPB insertion requires the SPB to interact with either centromeres or telomeres, detachment of centromeres from the SPB is disastrous for spindle formation when telomere clustering is defective. Indeed, when repression of centromere detachment is alleviated by Taz1∆myb–Sad1, bouquet-defective cells mostly fail to form proper spindles and complete meiosis (Fig. 5) (Katsumata et al. 2016). Because of the catastrophic consequence of centromere detachment, repression of centromere detachment is crucial for bouquet-defective cells, although the repression causes impairment of the meiotic centromere properties, which is relatively subtle compared to the spindle impairment caused by a loss of the repression (see the previous section). In nature, various environmental factors such as low temperature or osmotic stress perhaps often disrupt cytoplasmic microtubules and compromise telomere clustering. Accordingly, cells likely evolved the telomere clustering–linked centromere regulatory system to ensure spindle formation and accomplish meiotic division under such suboptimal conditions.
The role of telomere clustering-linked centromere detachment in spindle formation and nuclear fusion. In normal meiosis, telomeres cluster at the SPB (telomere clustering), and the centromeres subsequently detach (centromere detachment), resulting in formation of the bouquet arrangement. When telomere clustering is compromised (Impaired telomere clustering), Taz1- and microtubule-dependent regulatory mechanisms (Taz1- and MT-dependent regulation) inhibit centromere detachment from the SPB. In both cases, the spindle forms properly and nuclear fusion occurs efficiently due to the interaction of the SPB with telomeres or centromeres (Proper spindle formation, Efficient nuclear fusion). However, when both telomeres and centromeres concurrently become detached from the SPB (Concurrent telomere and centromere detachment), spindle formation and nuclear fusion are compromised (Defective spindle formation, Inefficient nuclear fusion)
The bouquet contributes to nuclear fusion during karyogamy
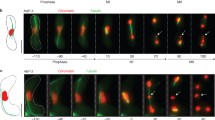
We further discovered that the bouquet contributes to nuclear fusion during karyogamy. This is demonstrated by the frequent observation of bi-nuclear bqt1∆ or rap1∆ zygotes with typical karyogamy or a horsetail microtubule array (an astral microtubule array with a single microtubule focus) (Fig. 3a, bqt1∆ and rap1∆, upper graph, and c, 2 nuc). In addition, the telomere clustering-linked regulation of centromere detachment contributes to nuclear fusion. In bqt1∆ or rap1∆ cells, Taz1∆myb–Sad1 further compromises nuclear fusion and increases the proportion of bi-nuclear zygotes (Fig. 3b, bqt1∆ + Taz1∆myb and rap1∆ + Taz1∆myb, upper graph), indicating that centromere detachment further compromises nuclear fusion in telomere clustering-defective cells. Thus, the centromere–SPB interaction can probably substitute for the telomere–SPB interaction to facilitate nuclear fusion, as in the case of spindle formation, and repression of centromere detachment ensures nuclear fusion in telomere clustering–defective cells (Fig. 5).
How does telomere/centromere–SPB interaction contribute to nuclear fusion? During nuclear fusion, SPB-bridging microtubules and minus end-directed microtubule motors, Klp2 kinesin and dynein, move the SPBs and drive nuclear fusion (Fig. 6a, Interaction-dependent mechanism) (Troxell et al. 2001; Scheffler et al. 2015). Because the SPBs often dissociate from chromosome masses during karyogamy in bouquet-defective cells [Fig. 3c, 2 nuc (aberrant)], the telomere/centromere–SPB interaction is probably required for the SPBs to move two nuclei (Fig. 6b). However, even in the absence of the telomere/centromere–SPB interaction, nuclear fusion still occurs, albeit inefficiently (Fig. 3b, bqt1∆ + Taz1∆myb and rap1∆ + Taz1∆myb, upper graph). Remarkably, the residual nuclear fusion depends exclusively on dynein, as demonstrated by the observation that introduction of the dhc1∆ mutation almost completely eliminates nuclear fusion (Fig. 3b, bqt1∆ dhc1∆ + Taz1∆myb–Sad1 and rap1∆ dhc1∆ + Taz1∆myb–Sad1). Because dynein mediates association of the centrosome with the nucleus and nuclear migration in many different organisms (Gonczy et al. 1999; Robinson et al. 1999; Salina et al. 2002; Malone et al. 2003; Tanaka et al. 2004; Burke and Roux 2009; Zhang et al. 2009; Fridolfsson and Starr 2010; Splinter et al. 2010; Bolhy et al. 2011; Jodoin et al. 2012; Sitaram et al. 2012), dynein-dependent association of the SPBs with the NE likely drives nuclear fusion independently of the telomere/centromere–SPB interaction (Fig. 6a, Interaction-independent mechanism, c). We speculate that cooperation of the mechanisms dependent on and independent of the telomere/centromere–SPB interaction drives efficient nuclear fusion (Fig. 6a).
Two mechanisms driving nuclear fusion during karyogamy in S. pombe. a Nuclear fusion mechanisms dependent on and independent of telomere/centromere–SPB interaction (interaction-dependent mechanism and interaction-independent mechanism, respectively). In the interaction-dependent mechanism, nuclear fusion is driven by microtubules (green lines) bridging the SPBs (blue spheres) of two nuclei. Klp2 (purple spheres) localized at microtubules and cytoplasmic dynein (red spheres) localized at the SPB move on the microtubules and generate forces on the SPBs that promote their approach and drive nuclear fusion, as proposed previously (Scheffler et al. 2015). In the interaction-independent mechanism, NE-localized dynein interacts with SPB-nucleated microtubules and drives nuclear fusion by generating forces that promote approach. Blue arrows show the forces generated by microtubule motors. b Effects of the loss of telomere/centromere–SPB interaction on interaction-dependent nuclear fusion. When the interaction-dependent mechanism drives nuclear fusion, only SPBs approach and fuse in the absence of the telomere/centromere–SPB interaction, and chromosome masses are left behind. c Effects of the loss of telomere/centromere–SPB interaction on the interaction-independent nuclear fusion. When the interaction-independent mechanism drives nuclear fusion, even in the absence of the telomere/centromere–SPB interaction, chromosome masses are able to approach and fuse, albeit inefficiently
Universality of the bouquet functions
As described in the “Introduction”, bouquet-dependent homologous chromosome pairing is a highly conserved event in eukaryotic organisms (Zickler and Kleckner 1998; Scherthan 2001; Yamamoto and Hiraoka 2001; Hiraoka and Dernburg 2009; Koszul and Kleckner 2009). However, some variation in chromosome behavior exists among species. In most organisms, telomere clustering is transient, and telomeres repeatedly cluster and disperse, whereas, in S. pombe, telomeres remain clustered during most of the meiotic prophase (Trelles-Sticken et al. 2005; Conrad et al. 2008; Koszul et al. 2008; Penkner et al. 2009; Baudrimont et al. 2010; Morimoto et al. 2012; Wynne et al. 2012; Shibuya et al. 2014). In C. elegans, rather than telomeres, chromosomal domains, called pairing centers, cluster (MacQueen et al. 2005; Phillips et al. 2005). In S. cerevisiae, actin filaments rather than microtubules drive telomere clustering (Trelles-Sticken et al. 2005; Koszul et al. 2008). Despite this mechanistic diversity, it is clear that the bouquet promotes homologous chromosome pairing in all organisms.
By contrast, bouquet-dependent meiotic centromere formation is observed only in yeast species. As in S. pombe, in S. cerevisiae centromeres are attached to the SPB during mitotic interphase and detach from it concomitant with dissociation of kinetochores from centromeres upon entry into meiosis (Miller et al. 2012; Kim et al. 2013; Meyer et al. 2013). Centromere detachment is probably also required for meiotic centromere formation in S. cerevisiae, because, in mutants defective in centromere detachment, sister chromatids frequently undergo equational segregation (Miller et al. 2012). However, unlike the situation in S. pombe, in which Csi1 connects the centromeres with the SPB (Hou et al. 2012), microtubule disruption in S. cerevisiae induces centromere detachment and proper meiotic centromere formation (Miller et al. 2012). Therefore, the mechanism of centromere detachment may not be identical between the two species. In multicellular eukaryotes, although centromeres are not associated with the centrosome before meiosis, kinetochore disassembly probably occurs, as evidenced by delocalization of outer kinetochore components from the centromeres during meiotic prophase (Parra et al. 2009); therefore, in multicellular species, kinetochore disassembly may induce meiotic centromere formation, as in S. pombe.
The contribution of the bouquet to spindle formation or NEBD-dependent SPB insertion seen in S. pombe has not been reported in other organisms. However, in S. cerevisiae, the SUN domain protein Mps3 contributes to SPB insertion into the NE during mitosis (Jaspersen et al. 2006; Friederichs et al. 2011; Chen et al. 2014), and, in mammals, SUN domain proteins contribute to spindle formation and the associated NEBD (Turgay et al. 2014). These facts indicate that the LINC complexes also contribute to spindle formation and NEBD in other organisms. Furthermore, in S. cerevisiae, Mps3 contributes to nuclear fusion during karyogamy (Nishikawa et al. 2003; Jaspersen et al. 2006), and, in multicellular organisms, the LINC complexes contribute to attachment of the centrosome to the nucleus and nuclear migration (Gonczy et al. 1999; Robinson et al. 1999; Salina et al. 2002; Malone et al. 2003; Tanaka et al. 2004; Burke and Roux 2009; Zhang et al. 2009; Fridolfsson and Starr 2010; Splinter et al. 2010; Bolhy et al. 2011; Jodoin et al. 2012; Sitaram et al. 2012). Thus, contribution of the LINC complexes to nuclear migration is also common among eukaryotes. It remains unknown whether chromosome–LINC interactions contribute to these events; however, given that Mps3 tethers telomeres to the NE in S. cerevisiae (Bupp et al. 2007), and that SUN domain proteins interact with chromosomes via nuclear lamins in mammals (Crisp et al. 2006; Haque et al. 2006), it is quite possible that the LINC–chromosome interaction is crucial for these events, as in the bouquet-dependent case in S. pombe.
Conclusion
It has long been known that chromosome positioning contributes to chromosomal events, including DNA transcription, replication, and recombination. However, the finding in S. pombe that the LINC-dependent bouquet chromosome arrangement contributes to non-chromosomal events, such as spindle formation and nuclear fusion, indicates that chromosome positioning has more diverse functions than previously thought. In addition, the findings that centromere detachment is linked with telomere clustering and that this connection is critical for spindle formation and nuclear fusion indicate that chromosome positioning is stringently regulated for its functions. The molecular mechanisms underlying these relatively novel functions and regulation of the bouquet remain to be determined. More importantly, it remains to be determined why chromosomes participate in regulation of non-chromosomal events.
The available evidence suggests that the functions of LINC-dependent chromosome positioning are common in other organisms. However, most previous studies of LINC-dependent chromosome positioning focused solely on chromosomal events. Conversely, studies on LINC-dependent non-chromosomal events rarely took chromosomal positioning into account. Therefore, many exciting chromosome positioning-dependent activities may remain unnoticed. Therefore, to comprehensively elucidate the functions of chromosome positioning, future studies should adopt a much broader perspective. A wider view would also be clinically important, given that LINC complexes are involved in lamin-related deceases such as Emery-Dreifuss muscular dystrophy (Fridkin et al. 2009; Razafsky and Hodzic 2009; Mejat and Misteli 2010). It is clear that chromosome positioning is not strictly a chromosomal phenomenon but instead a phenomenon that is relevant to a wide range of biological fields.
Materials and methods
Yeast strains, media, and basic genetic methods
The fission yeast strains used in this study are shown in Table 1. Genes fused with the GFP or mCherry gene and deletion alleles of the genes used in this study were described previously (Katsumata et al. 2016). Integration of the mCherry-atb2 + fusion gene at the atb2 + locus was carried out as follows. First, an integration plasmid, pEN5, was constructed. A DNA fragment carrying the mCherry-atb2 + fusion gene along with the nda3 + promoter was amplified from pMY53 using two synthetic oligonucleotide primers, TCCGAGCTCCATATATGCCGTATTCTTGAATGT and CCGCTCGAGTCAATCCGACATTTTTGCCTCG (Yoshida et al. 2013). The PCR product was digested with XhoI and SacI, and then inserted between the corresponding sites of pRS306, a URA3-bearing plasmid (Sikorski and Hieter 1989). The resultant plasmid pEN5 was linearized by digestion with EcoRI at a site in the atb2 + gene, and then transformed into ura4 – cells. Transformants bearing the mCherry-atb2 + fusion gene integrated at the atb2 + locus were selected by the ura + phenotype and confirmed by colony PCR. Media and basic genetic manipulation methods used in this study were described by Moreno et al. (1991).
Analysis of chromosome and microtubule morphology in zygotic meiosis
Cells grown on YES solid medium at 30 °C were transferred to ME solid medium and induced to enter meiosis by incubation at 25 °C. Nuclear DNA in meiotic zygotes was stained with DNA-specific Hoechst 33342 dye, as described previously (Ding et al. 1998). Images of the cells at seven focal planes spaced at 0.4 µm intervals were taken through a 60×/1.42 NA Plan Apo oil immersion objective lens on an Olympus IX71 inverted microscope (Olympus Corp., Tokyo, Japan) equipped with a cooled charge-coupled device camera (CoolSNAP-HQ2; Nippon Roper Co., Ltd., Tokyo, Japan). The resultant images were processed by deconvolution and analyzed using the MetaMorph (version 7) software (Molecular Devices Japan, Inc., Tokyo, Japan).
Live-cell analysis of spindle dynamics in haploid meiotic cells
Haploid cells bearing both mating-type genes were grown in liquid YES medium and induced to enter meiosis by incubation at 30 °C in liquid EMM-N medium, as described previously (Yoshida et al. 2013). For analysis of spindle dynamics, a drop of the cell suspension was placed on the bottom of 35 mm glass-bottom dishes (Matsunami Glass Ind., Ltd., Tokyo, Japan) coated with 5 mg/ml lectin (Sigma-Aldrich Japan, Inc., Tokyo, Japan). The cells were observed through a 60×/1.42 NA Plan Apo oil immersion objective lens (Olympus Corp., Tokyo, Japan) on an IX71 inverted microscope operated by the MetaMorph software. Using a cooled CCD camera, time-lapse images of the cells were collected every 10 min at nine focal planes spaced at 0.5 µm intervals. During collection of time-lapse images, the cells were kept at 25 °C. All obtained images were processed as for analysis of zygotic cells.
References
Aparicio OM (2013) Location, location, location: it’s all in the timing for replication origins. Genes Dev 27:117–128
Asakawa H, Hayashi A, Haraguchi T, Hiraoka Y (2005) Dissociation of the Nuf2-Ndc80 complex releases centromeres from the spindle-pole body during meiotic prophase in fission yeast. Mol Biol Cell 16:2325–2338
Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, Jantsch V (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6:e1001219
Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V (2011) A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192:855–871
Brar GA, Amon A (2008) Emerging roles for centromeres in meiosis I chromosome segregation. Nat Rev Genet 9:899–910
Bupp JM, Martin AE, Stensrud ES, Jaspersen SL (2007) Telomere anchoring at the nuclear periphery requires the budding yeast Sad1-UNC-84 domain protein Mps3. J Cell Biol 179:845–854
Burke B, Roux KJ (2009) Nuclei take a position: managing nuclear location. Dev Cell 17:587–597
Chen J, Smoyer CJ, Slaughter BD, Unruh JR, Jaspersen SL (2014) The SUN protein Mps3 controls Ndc1 distribution and function on the nuclear membrane. J Cell Biol 204:523–539
Chikashige Y, Hiraoka Y (2001) Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11:1618–1623
Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science 264:270–273
Chikashige Y, Ding DQ, Imai Y, Yamamoto M, Haraguchi T, Hiraoka Y (1997) Meiotic nuclear reorganization: switching the position of centromeres and telomeres in the fission yeast Schizosaccharomyces pombe. EMBO J 16:193–202
Chikashige Y, Kurokawa R, Haraguchi T, Hiraoka Y (2004) Meiosis induced by inactivation of Pat1 kinase proceeds with aberrant nuclear positioning of centromeres in the fission yeast Schizosaccharomyces pombe. Genes Cells 9:671–684
Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y (2006) Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125:59–69
Chikashige Y, Yamane M, Okamasa K, Mori C, Fukuta N, Matsuda A, Haraguchi T, Hiraoka Y (2014) Chromosomes rein back the spindle pole body during horsetail movement in fission yeast meiosis. Cell Struct Funct 39:93–100
Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME (2008) Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133:1175–1187
Cooper JP, Watanabe Y, Nurse P (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392:828–831
Cremer T, Cremer M (2010) Chromosome territories. Cold Spring Harb Perspect Biol 2:a003889
Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006) Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172:41–53
Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR (1997) The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell 8:1461–1479
Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci 111(Pt 6):701–712
Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y (2004) Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6:329–341
Ding DQ, Haraguchi T, Hiraoka Y (2016a) A cohesin-based structural platform supporting homologous chromosome pairing in meiosis. Curr Genet 62:499–502
Ding DQ, Matsuda A, Okamasa K, Nagahama Y, Haraguchi T, Hiraoka Y (2016b) Meiotic cohesin-based chromosome structure is essential for homologous chromosome pairing in Schizosaccharomyces pombe. Chromosoma 125:205–214
Fennell A, Fernandez-Alvarez A, Tomita K, Cooper JP (2015) Telomeres and centromeres have interchangeable roles in promoting meiotic spindle formation. J Cell Biol 208:415–428
Fernandez-Alvarez A, Bez C, O’Toole ET, Morphew M, Cooper JP (2016) Mitotic nuclear envelope breakdown and spindle nucleation are controlled by interphase contacts between centromeres and the nuclear envelope. Dev Cell 39:544–559
Fridkin A, Penkner A, Jantsch V, Gruenbaum Y (2009) SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci 66:1518–1533
Fridolfsson HN, Starr DA (2010) Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol 191:115–128
Friederichs JM, Ghosh S, Smoyer CJ, McCroskey S, Miller BD, Weaver KJ, Delventhal KM, Unruh J, Slaughter BD, Jaspersen SL (2011) The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet 7:e1002365
Funabiki H, Hagan I, Uzawa S, Yanagida M (1993) Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121:961–976
Gonczy P, Pichler S, Kirkham M, Hyman AA (1999) Cytoplasmic dynein is required for distinct aspects of MTOC positioning, including centrosome separation, in the one cell stage Caenorhabditis elegans embryo. J Cell Biol 147:135–150
Hagan I, Yanagida M (1995) The product of the spindle formation gene sad1 + associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129:1033–1047
Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S (2006) SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 26:3738–3751
Harigaya Y, Yamamoto M (2007) Molecular mechanisms underlying the mitosis-meiosis decision. Chromosome Res 15:523–537
Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117:4025–4032
Hauf S, Watanabe Y (2004) Kinetochore orientation in mitosis and meiosis. Cell 119:317–327
Hayashi A, Asakawa H, Haraguchi T, Hiraoka Y (2006) Reconstruction of the kinetochore during meiosis in fission yeast Schizosaccharomyces pombe. Mol Biol Cell 17:5173–5184
Hiraoka Y, Dernburg AF (2009) The SUN rises on meiotic chromosome dynamics. Dev Cell 17:598–605
Hirose Y, Suzuki R, Ohba T, Hinohara Y, Matsuhara H, Yoshida M, Itabashi Y, Murakami H, Yamamoto A (2011) Chiasmata promote monopolar attachment of sister chromatids and their co-segregation toward the proper pole during meiosis I. PLoS Genet 7:e1001329
Horigome C, Oma Y, Konishi T, Schmid R, Marcomini I, Hauer MH, Dion V, Harata M, Gasser SM (2014) SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol Cell 55:626–639
Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S (2012) Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol 199:735–744
Jaspersen SL, Martin AE, Glazko G, Giddings TH Jr, Morgan G, Mushegian A, Winey M (2006) The Sad1-UNC-84 homology domain in Mps3 interacts with Mps2 to connect the spindle pole body with the nuclear envelope. J Cell Biol 174:665–675
Jodoin JN, Shboul M, Sitaram P, Zein-Sabatto H, Reversade B, Lee E, Lee LA (2012) Human Asunder promotes dynein recruitment and centrosomal tethering to the nucleus at mitotic entry. Mol Biol Cell 23:4713–4724
Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11:1624–1630
Katsumata K, Hirayasu A, Miyoshi J, Nishi E, Ichikawa K, Tateho K, Wakuda A, Matsuhara H, Yamamoto A (2016) A Taz1- and microtubule-dependent regulatory relationship between telomere and centromere positions in bouquet formation secures proper meiotic divisions. PLoS Genet 12:e1006304
Kim S, Meyer R, Chuong H, Dawson DS (2013) Dual mechanisms prevent premature chromosome segregation during meiosis. Genes Dev 27:2139–2146
Klutstein M, Fennell A, Fernandez-Alvarez A, Cooper JP (2015) The telomere bouquet regulates meiotic centromere assembly. Nat Cell Biol 17:458–469
Koszul R, Kleckner N (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19:716–724
Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133:1188–1201
MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF (2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123:1037–1050
Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115:825–836
Meister P, Mango SE, Gasser SM (2011) Locking the genome: nuclear organization and cell fate. Curr Opin Genet Dev 21:167–174
Mejat A, Misteli T (2010) LINC complexes in health and disease. Nucleus Austin 1:40–52
Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS (2013) Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science 339:1071–1074
Miki F, Kurabayashi A, Tange Y, Okazaki K, Shimanuki M, Niwa O (2004) Two-hybrid search for proteins that interact with Sad1 and Kms1, two membrane-bound components of the spindle pole body in fission yeast. Mol Genet Genomics 270:449–461
Miller MP, Unal E, Brar GA, Amon A (2012) Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife 1:e00117
Miller MP, Amon A, Unal E (2013) Meiosis I: when chromosomes undergo extreme makeover. Curr Opin Cell Biol 25:687–696
Mine-Hattab J, Rothstein R (2013) DNA in motion during double-strand break repair. Trends Cell Biol 23:529–536
Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194:795–823
Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198:165–172
Nimmo ER, Pidoux AL, Perry PE, Allshire RC (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392:825–828
Nishikawa S, Terazawa Y, Nakayama T, Hirata A, Makio T, Endo T (2003) Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J Biol Chem 278:9938–9943
Niwa O, Shimanuki M, Miki F (2000) Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J 19:3831–3840
Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL (2009) Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev 23:912–927
Page SL, Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785–789
Parra MT, Gomez R, Viera A, Llano E, Pendas AM, Rufas JS, Suja JA (2009) Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet 5:e1000417
Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, Jantsch V (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139:920–933
Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423–440
Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF (2005) HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123:1051–1063
Razafsky D, Hodzic D (2009) Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol 186:461–472
Robinson JT, Wojcik EJ, Sanders MA, McGrail M, Hays TS (1999) Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J Cell Biol 146:597–608
Rocha PP, Skok JA (2013) The origin of recurrent translocations in recombining lymphocytes: a balance between break frequency and nuclear proximity. Curr Opin Cell Biol 25:365–371
Roeder GS (1997) Meiotic chromosomes: it takes two to tango. Genes Dev 11:2600–2621
Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B (2002) Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108:97–107
Scheffler K, Minnes R, Fraisier V, Paoletti A, Tran PT (2015) Microtubule minus end motors kinesin-14 and dynein drive nuclear congression in parallel pathways. J Cell Biol 209:47–58
Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2:621–627
Schober H, Ferreira H, Kalck V, Gehlen LR, Gasser SM (2009) Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev 23:928–938
Shibuya H, Morimoto A, Watanabe Y (2014) The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet 10:e1004821
Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O (1997) A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet 254:238–249
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27
Sitaram P, Anderson MA, Jodoin JN, Lee E, Lee LA (2012) Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development 139:2945–2954
Spector DL (2003) The dynamics of chromosome organization and gene regulation. Annu Rev Biochem 72:573–608
Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, Akhmanova A (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8:e1000350
Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG (2004) Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol 165:709–721
Tomita K, Cooper JP (2007) The telomere bouquet controls the meiotic spindle. Cell 130:113–126
Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H (2005) Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol 170:213–223
Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR (2001) pkl1 + and klp2 +: two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell 12:3476–3488
Turgay Y, Champion L, Balazs C, Held M, Toso A, Gerlich DW, Meraldi P, Kutay U (2014) SUN proteins facilitate the removal of membranes from chromatin during nuclear envelope breakdown. J Cell Biol 204:1099–1109
Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O (2004) The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165:759–765
Walde S, King MC (2014) The KASH protein Kms2 coordinates mitotic remodeling of the spindle pole body. J Cell Sci 127:3625–3640
Wynne DJ, Rog O, Carlton PM, Dernburg AF (2012) Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196:47–64
Yamamoto A (2014) Gathering up meiotic telomeres: a novel function of the microtubule-organizing center. Cell Mol Life Sci 71:2119–2134
Yamamoto A, Hiraoka Y (2001) How do meiotic chromosomes meet their homologous partners? Lessons from fission yeast. BioEssays 23:526–533
Yamamoto A, Hiraoka Y (2003) Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J 22:2284–2296
Yamamoto M, Imai Y, Watanabe Y (1997) Mating and sporulation in Schizosaccharomyces pombe. In: Pringle JR, Broach JR, Jones EW (eds) The molecular and cellular biology of the yeast Saccharomyces: cell cycle and cell biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, pp 1037–1106
Yamamoto A, West RR, McIntosh JR, Hiraoka Y (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol 145:1233–1249
Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y (2001) Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell 12:3933–3946
Yamamoto TG, Chikashige Y, Ozoe F, Kawamukai M, Hiraoka Y (2004) Activation of the pheromone-responsive MAP kinase drives haploid cells to undergo ectopic meiosis with normal telomere clustering and sister chromatid segregation in fission yeast. J Cell Sci 117:3875–3886
Yoshida M, Katsuyama S, Tateho K, Nakamura H, Miyoshi J, Ohba T, Matsuhara H, Miki F, Okazaki K, Haraguchi T, Niwa O, Hiraoka Y, Yamamoto A (2013) Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J Cell Biol 200:385–395
Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M (2009) SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64:173–187
Zickler D, Kleckner N (1998) The leptotene-zygotene transition of meiosis. Annu Rev Genet 32:619–697
Acknowledgements
We thank Akira Shinohara for reagents used for this study, and Takashi Ushimaru and Motoaki Hiraoka for critical reading of the manuscript and helpful comments. This work was supported by JSPS KAKENHI Grant Number 16K07248 (Grant-in-Aid for Scientific Research C) and performed under the Cooperative Research Program of the Institute for Protein Research, Osaka University (CR-13/14/15/16-03).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Kupiec.
Rights and permissions
About this article
Cite this article
Katsumata, K., Nishi, E., Afrin, S. et al. Position matters: multiple functions of LINC-dependent chromosome positioning during meiosis. Curr Genet 63, 1037–1052 (2017). https://doi.org/10.1007/s00294-017-0699-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-017-0699-2