Abstract
During meiosis, telomeres cluster and promote homologous chromosome pairing. Telomere clustering depends on conserved SUN and KASH domain nuclear membrane proteins, which form a complex called the linker of nucleoskeleton and cytoskeleton (LINC) and connect telomeres with the cytoskeleton. It has been thought that LINC-mediated cytoskeletal forces induce telomere clustering. However, how cytoskeletal forces induce telomere clustering is not fully understood. Recent study of fission yeast has shown that the LINC complex forms the microtubule-organizing center (MTOC) at the telomere, which has been designated as the “telocentrosome”, and that microtubule motors gather telomeres via telocentrosome-nucleated microtubules. This MTOC-dependent telomere clustering might be conserved in other eukaryotes. Furthermore, the MTOC-dependent clustering mechanism appears to function in various other biological events. This review presents an overview of the current understanding of the mechanism of meiotic telomere clustering and discusses the universality of the MTOC-dependent clustering mechanism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During meiosis, eukaryotic organisms recombine homologous chromosomes to generate chromosomes that harbor new sets of the genes, and partition the recombined homologous chromosomes to halve the chromosome number in the gametes. Both recombination and segregation of the homologous chromosomes depend on physical interaction of the chromosomes along their entire length, which is termed “homologous chromosome pairing” (it is also known as “synapsis”, but “pairing” is used in this review). How the homologous chromosomes approach each other and undergo pairing has been one of the major questions in the field of meiosis.
During the period of homologous chromosome pairing, telomeres become clustered at the nuclear periphery [1, 2]. This telomere clustering was noted more than 100 years ago, and has been observed in various types of meiotic cells. The chromosome arrangement with clustered telomeres is called a “bouquet,” because it resembles a bouquet of flowers. It has long been predicted that the bouquet arrangement of the chromosomes contributes to homologous chromosome pairing, because the formation of the bouquet arrangement coincides with homologous chromosome pairing. Recent studies have shown that this is indeed the case.
The molecular mechanism of telomere clustering has been recently revealed. Telomere clustering has been shown to depend on two different types of nuclear membrane proteins, which respectively contain the conserved Sad1/Unc-84 (SUN) and Klarsicht/ANC-1/Syne homology (KASH) domains [3–5]. The SUN and KASH domain proteins form linker of nucleoskeleton and cytoskeleton (LINC) complexes [6] and connect the telomere with the cytoskeleton (Fig. 1). It has been speculated that LINC-mediated cytoskeletal forces move and gather the telomeres, and a recent study of fission yeast has demonstrated that the LINC complex induces telomere clustering by forming a microtubule-organizing center (MTOC) at the telomere [7]. In this review, I present an overview of the current understanding of the telomere clustering mechanism. I also describe the effects of the finding of the telomeric MTOC on MTOC studies, and discuss the universality of the MTOC-dependent clustering mechanism.
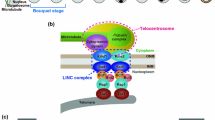
Schematic structure of the LINC complex. A SUN domain protein (blue) has a coiled-coil region (CC) that extends into the nuclear lumen and a transmembrane region (TM) that resides in the inner nuclear envelope (NE), while a KASH domain protein (brown) has a TM that resides in the outer NE. Both proteins form trimers, and SUN and KASH trimers interact with each other via their SUN and KASH domains in the nuclear lumen. The SUN trimer interacts with nuclear lamins or chromosomes with its domains plunged into the nucleoplasm, while the KASH trimer interacts with cytoskeleton with its domain exposed to the cytoplasm
The role of telomere clustering in homologous chromosome pairing
Before addressing the telomere clustering mechanism in detail, I will briefly discuss the role of telomere clustering in homologous chromosome pairing. The role of telomere clustering is described in greater detail elsewhere [2, 4, 8–10].
The first implication of the significance of telomere clustering in homologous chromosome pairing was probably brought about through studies of fission yeast, Schizosaccharomyces pombe. This organism normally propagates in the haploid state [8, 11]. Under nitrogen-starved conditions, S. pombe cells with opposite mating types fuse to form a diploid cell and immediately enter meiosis. The diploid cells undergo two meiotic divisions and eventually form four spores. During the majority of the period that precedes meiotic division (thereafter, this period is comprehensively called “meiotic prophase”), telomeres remain clustered at the spindle pole body (SPB; a fungal centrosome) while centromeres are located away from it, resulting in the typical bouquet arrangement of chromosomes (Fig. 2a) [12]. During this stage, the nucleus becomes elongated and moves back and forth between the cell ends (Fig. 2b) [12]. This nuclear oscillation is called “horsetail nuclear movement” because of the horsetail-like nuclear shape. The horsetail nuclear movements are driven by cytoplasmic microtubules that extend from the SPB located at the nuclear membrane [13]. The cytoplasmic microtubules interact with the cell cortex, and the minus end-directed microtubule motor, cytoplasmic dynein, accumulates at the cortical interaction sites and generates pulling forces via the microtubules that drive nuclear movements (Fig. 2b) [14–18]. A combination of nuclear movements and telomere clustering leads to chromosome movements led by the bundled telomeres [12]. It has been proposed that the telomere-bundled chromosome movements bring about the alignment of homologous chromosomes from the telomeres and the frequent contact of homologous regions, promoting homologous chromosome pairing [8, 12, 14, 19]. This view is supported by the impairment of homologous chromosome pairing in mutants that are defective in nuclear movements or telomere clustering. Because nuclear movements are dependent on cytoplasmic dynein, the loss of dynein function leads to defective nuclear movement. Cytoplasmic dynein is a large complex, and dynein heavy and light chains (DHC and DLC) act respectively as motor and regulatory subunits of dynein. The depletion of either Dhc1 (S. pombe DHC) or Dlc1 [S. pombe Tctex-1 (t-complex testis-expressed-1)-type DLC] eliminates or severely compromises nuclear movements in S. pombe [14, 20]. In these cells, homologous chromosome pairing is severely impaired [14, 19, 20]. Telomere clustering, on the other hand, depends on factors that are required for telomere integrity or SUN/KASH nuclear membrane proteins (see below), and the loss of any of these factors leads to defective telomere clustering [21–26]. Similar to the nuclear movement-defective mutants, telomere clustering mutants fail to establish proper homologous chromosome pairing [19, 27].
Meiotic chromosome arrangement and dynamics in S. pombe and C. elegans. a Telomere clustering and chromosome arrangement during meiotic prophase in S. pombe. Telomeres are clustered at the SPB while centromeres are located away from it, resulting in a bouquet-like chromosome arrangement. b Chromosome and nuclear dynamics during meiotic prophase in S. pombe. Microtubules extending from the SPB interact with the cell cortex, and pull the nucleus, causing back-and-forth nuclear movements between the cell ends. Telomere clustering and nuclear movements promote side-by-side alignment of homologous chromosomes from the bundled telomeres and contact of homologous regions. c Dynamics of PCs in C. elegans. PCs move along the nuclear periphery by interacting with cytoplasmic microtubules via the LINC complexes. The PCs repeatedly associate and dissociate, and eventually interact with their homologous partners. Black arrows indicate PC movements. In a and c, blue or red lines indicate respective pairs of homologous chromosomes. NE nuclear envelope
The importance of telomere clustering is also recognized in budding yeast, Saccharomyces cerevisiae. As in S. pombe, telomeres gather during meiotic prophase [28]. However, the telomere cluster is not stable in S. cerevisiae: telomeres frequently form small aggregates of various sizes, and these aggregates associate and dissociate repetitively [29–31]. In addition, the whole nucleus does not move around inside the cell, and telomeres move around solely at the nuclear periphery, driving chromosome movements inside the nucleus. Despite these differences, telomere clustering and chromosome movements appear to promote homologous chromosome pairing in S. cerevisiae, as in S. pombe. Depletion of the meiosis-specific telomere-binding factor Ndj1 impairs telomere clustering and chromosome movements, and homologous chromosome pairing is compromised in Ndj1-depleted cells [28, 30–35]. However, because telomere clustering is not stable and impairment of the clustering and/or chromosome movements leads to the association of non-homologous regions [36, 37], it has also been proposed that repeating association and dissociation of the telomeres is required in order to resolve the non-homologous interaction and/or chromosome entanglement [10, 30, 31].
A similar kind of story has also been observed in the nematode Caenorhabditis elegans. In C. elegans, instead of telomeres, special chromosomal regions called “pairing centers (PCs)” play a critical role in homologous chromosome pairing (Fig. 2c) [38, 39]. During the period of homologous chromosome pairing, the PCs are attached to the nuclear membrane and gather as meiotic telomeres do. As in S. cerevisiae, the PCs do not form a stable single cluster; small aggregations of PCs associate and dissociate repetitively, and move around at the nuclear periphery without movements of the whole nucleus (Fig. 2c) [40–42]. The PCs are essential for homologous chromosome pairing, as demonstrated by the fact that homologous chromosomes that lack the PCs fail to pair properly [38]. Based on these observations, it has been proposed that PC clustering and PC-led chromosome movements induce homologous chromosome pairing and eliminate the entanglement or improper association of chromosomes, like telomeres do in S. cerevisiae [40, 43].
Telomere clustering and telomere-led chromosome movements are also observed in mammalian cells. In mouse and human spermatocytes, telomeres become clustered at the nuclear periphery during the period of homologous chromosome pairing [44]. In addition, telomeres move around at the nuclear periphery in mouse spermatocytes, much like telomeres/PCs do in S. cerevisiae or C. elegans [45]. Telomere clustering also occurs in maize cells [46–48]. Collectively, these observations show that telomere clustering and telomere-led chromosome movements are conserved biological events that are essential for proper homologous chromosome pairing.
Telomere clustering and the LINC complex
SUN and KASH domain proteins are essential proteins for telomere clustering [3–5]. X-ray analysis of a crystal structure of the LINC complex revealed that three SUN domains firmly interact with three KASH domains (Fig. 1) [49, 50]. By forming a firm complex, the LINC complexes connect the nucleus to various types of cytoskeleton, such as microtubules, actin filaments, and intermediate filaments. The LINC complexes originally attracted attention owing to their essential roles in the migration of the nucleus during the development of various tissues, including the muscle and the brain [3–5, 51–53]. SUN proteins have also been shown to interact with nuclear lamins [54–59], whose defects lead to a type of cardiac and skeletal muscle dysfunction called Emery-Dreifuss muscular dystrophy [60, 61]. Furthermore, it was very recently found that the LINC complexes drive biased sister chromatid segregation during stem cell division [62, 63].
The significance of the LINC complexes in telomere clustering has been well recognized in S. pombe (Table 1). In this organism, a SUN domain-containing protein, Sad1, is localized at the SPB and plays a pivotal role in spindle formation during mitosis [64]. However, when telomere clustering occurs, Sad1 also becomes localized at telomeres, which are tethered to the nuclear membrane by Bqt3 and Bqt4 [65, 66] (Fig. 3). Sad1 telomere localization is dependent on the meiosis-specific proteins Bqt1 and Bqt2 [65]. Bqt1 localizes to the telomeres by forming a complex with Bqt2 and the telomere-binding protein Rap1, and tethers Sad1 to the telomeres by interacting directly with it. When either Bqt1 or Bqt2 is depleted, Sad1 fails to accumulate at telomeres, and the telomeres do not form a cluster [65–67]. Like Sad1, S. pombe KASH proteins Kms1 and Kms2 (which are localized at the SPB during mitosis) also become co-localized with telomeres during meiosis [7, 21]. In addition, Kms1 depletion compromises telomere clustering. These results indicate that recruitment of the LINC complex to telomeres is an essential step to induce telomere clustering.
A model for molecular organization of the telocentrosome. Telomere-recruited SUN/KASH recruits the γ-TuC to form the telocentrosome (dashed purple circle). It also recruits cytoplasmic dynein together with dynactin, which aids dynein function. Recruited subunits of dynein [dynein heavy chain (DHC) and dynein light chain (DLC)] and dynactin (Ssm4) are shown in green, and the SUN and KASH LINC components are shown in orange
Similar stories have also emerged in other organisms. In S. cerevisiae, a SUN protein, Mps3, is localized at the SPB, and additionally becomes co-localized with telomeres when telomere clustering occurs, as seen for Sad1 in S. pombe (Table 1) [30, 68, 69]. Mps3 interacts with the telomere-binding protein Ndj1, and depletion of an Ndj1-interacting domain of Mps3 causes defects in telomere clustering. In C. elegans, SUN protein Metafin/SUN-1 and KASH protein ZYG-12 are distributed throughout the nuclear membrane in germ cells; however, when the PCs aggregate, these proteins concomitantly accumulate at the sites where the PCs are located (Table 1) [40, 43, 70]. Mutations in the genes encoding Metafin/SUN-1 and Zyg12 or RNAi depletion of Metafin/SUN-1 lead to defective aggregation of the PCs [43, 70]. Metafin/SUN-1 becomes phosphorylated during meiotic prophase in a manner that is dependent on the meiosis-specific CHK-2 kinase and Polo-like kinases, PLK-1 and PLK-2; the loss of Metafin/SUN-1 phosphorylation impairs aggregation [40, 71–73]. In mouse spermatocytes, SUN proteins SUN1 and SUN2 and KASH protein KASH5 become co-localized with telomeres during meiotic prophase; the loss of SUN1 or KASH5 impairs homologous chromosome pairing (Table 1) [45, 74–76]. All of these observations made in different organisms indicate that the clustering process depends on the LINC complexes.
Consistent with an essential task of the LINC complexes (that is, connecting the nuclear structure and/or chromosomes to the cytoskeleton), accumulating lines of evidence demonstrate that the LINC complexes induce telomere clustering via the cytoskeleton. In S. pombe, C. elegans, and mice, aggregation of the telomeres or the PCs is abolished by the disruption of microtubules [7, 43, 45]. In contrast, in S. cerevisiae an actin-depolymerizing drug inhibits telomere clustering, and telomeres become co-localized with actin filaments and move with the filaments [29, 31]. These results provoked the idea that the LINC complexes connect telomeres/PCs with the cytoskeleton, enabling LINC-mediated cytoskeletal forces to drive telomere/PC clustering.
Cytoskeleton-dependent telomere clustering mechanism
How the LINC-mediated cytoskeletal forces drive telomere clustering is not fully understood. Cytoplasmic dynein is a cytoskeletal motor protein that generates LINC-mediated cytoskeletal forces. In S. pombe, cytoplasmic dynein is co-localized with telomeres, and simultaneous depletion of a motor subunit Dhc1 and a regulatory subunit Dlc1 impairs telomere clustering [7, 20]. In C. elegans, PC movements are dependent on cytoplasmic dynein, which is co-localized with the PCs [42, 43]. In mice, dynein-associated factors become co-localized with telomeres during meiotic prophase [45, 76]. Based on these observations, it has been suggested that cytoplasmic dynein tethered to telomeres/PCs directly transports those telomeres/PCs along the cytoplasmic microtubules towards the minus ends, inducing their clustering. This model seems to fit reasonably well with the case of S. pombe, in which telomeres remain clustered at the SPB that is associated with the microtubule minus ends [12]. However, the following two facts do not fit with the model. First, dynein is unnecessary for the clustering process. A loss of dynein motor subunit Dhc1 alone does not compromise telomere clustering in S. pombe [14]. Similarly, RNAi depletion of DLC together with the temperature-sensitive allele of the DHC-encoding gene does not eliminate the association of the pairing centers in C. elegans, although it significantly reduces their movements [42]. Second, in C. elegans, PC aggregates form independent of the centrosome, and repetitively dissociate and associate [40–42]. Apparently, dynein motor-dependent transport of telomeres/PCs on microtubules alone is not sufficient to support the observed telomere/PC clustering.
A very recent study of S. pombe has provided a breakthrough in understanding of the cytoskeleton-dependent telomere clustering mechanism. The study showed that cytoplasmic dynein and dynactin, which aids dynein functions, are tethered to telomeres and contribute to telomere clustering [7]. It also demonstrated that, in addition to cytoplasmic dynein, several different kinesin motors (including those that move in the same direction as dynein) contribute to telomere clustering. In addition, it was observed that when cells were treated with a microtubule depolymerizer and subsequently allowed to reform microtubules by removal of the inhibitor, telomeres moved along the microtubules directly towards the nucleation sites. This observation supports the dynein-dependent transport of telomeres along microtubules. More importantly, however, it was observed that microtubules nucleated from the dispersed telomeres in the cells, and that telomeres drifted inside the cell and gathered once they were connected with the telomere-nucleated microtubules. This observation indicates that the MTOC is formed at the telomere, and that telomere clustering is driven by the telomere-nucleated microtubules. This novel telomeric MTOC has been named the “telocentrosome” after the telomere and the centrosome. Consistent with the microtubule nucleation activity, a component of the γ-tubulin complex (γ-TuC) that is responsible for microtubule nucleation is co-localized with the telomeres. Telocentrosome formation depends on Kms1, a KASH protein. The telocentrosome is essential for telomere clustering, because the loss of Kms1 leads to severe telomere clustering defects in addition to defective telocentrosome formation [21].
Based on these findings, the following model has been proposed. Upon entering meiosis, the LINC complex recruits the γ-TuC to telomeres to form the telocentrosome (Fig. 3). Next, oligomerized, minus end-directed microtubule motors crosslink the telocentrosome- and the SPB-nucleated microtubules and gather the telomeres by moving along the microtubules towards the nucleation sites (Fig. 4a). The kinesin and dynein motors may also cooperate to regulate the polymerization and/or bundling of microtubules to promote connection between telomeres and the SPB and to promote drift of the telomeres inside the cell to facilitate the encounter of telocentrosome- and SPB-nucleated microtubules. In addition, cytoplasmic dynein becomes tethered to telomeres and directly transports the telomeres towards the nucleation sites to aid telomere clustering.
Models for telomere clustering mechanisms. a The telomere clustering mechanism in S. pombe. During mitotic interphase, telomeres (blue spheres) are located away from the SPB, which consists of γ-TuC (green sphere) and LINC-containing structure (purple sphere), whereas the centromere (yellow sphere) is located near the SPB (mitotic interphase). Upon entering meiosis, the LINC complexes become localized at telomeres and form telocentrosomes (purple and green spheres at telomeres), and microtubules extend from the telocentrosomes (telocentrosome formation). After telocentrosome formation, microtubule motors (MT motor) gather telomeres at the SPB by crosslinking SPB- and/or telocentrosome-nucleated microtubules and moving along those microtubules towards the nucleation sites (motor-dependent telomere aggregation, top). Telocentrosome- and SPB-localized microtubule motors also facilitate telomere clustering by directly transporting the telomeres or the SPB along microtubules towards the microtubule nucleation sites (motor-dependent telomere aggregation, bottom). As a consequence of telomere clustering together with centromere dissociation from the SPB, a bouquet chromosome arrangement in meiotic prophase is established (meiotic prophase). b MTOC-dependent clustering and dispersal of telomeres/PCs. MTOCs are formed at telomeres/PCs by telomere-localized LINC complexes. The minus-end-directed microtubule motors that are tethered to the MTOCs gather telomeres/PCs by moving along the microtubules (i). Oligomerized minus-end-directed microtubule motors also gather telomeres/PCs by crosslinking MTOC-nucleated microtubules and moving along them towards nucleation sites (ii). Similarly, telomeres/PCs are separated by telomere/PC-tethered plus-end-directed microtubule motors (iii) and oligomerized ones (iv). MT microtubule motor; NE nuclear envelope. Yellow arrows indicate movements of telomeres/PCs. Blue and red arrows indicate movements of minus- and plus-end-directed MT motors, respectively
The telocentrosome-dependent model solves a spatial problem that has previously been unexplained. If the SPB were the sole MTOC, then the SPB-telomere connection would not be easily established, because it is difficult for microtubules, which extend straight from the SPB, to reach the telomeres, which are attached to the spherical nuclear membrane. For instance, if telomeres are located on the opposite side of the nucleus from the SPB, then the SPB-nucleated microtubules probably never reach them. However, the situation is different if telomeres simultaneously nucleate microtubules. Both SPB-nucleated and telomere-nucleated microtubules can easily interact with each other and establish SPB-telomere connections, even when the SPB and telomeres are on opposite sides of the nucleus (Fig. 4a). It is also apparent that mutual microtubule nucleation is more efficient for establishing a connection than nucleation from the SPB alone. By nucleating microtubules, the SPB and the telomere mutually search for each other and efficiently establish their connection.
The telocentrosome-dependent clustering mechanism does not appear to function only during the telomere-clustering stage. During nuclear movements, a small fraction of wild-type cells exhibit the dissociation of some telomeres from the SPB during meiotic prophase [7]. It appears that telomeres sometimes fall off of the SPB, but move back to it during nuclear movements. It is likely that the telomere-clustering mechanism also functions during nuclear movements and maintains telomere clustering at the SPB.
These new findings in S. pombe revealed that the LINC complexes induce MTOC formation in addition to tethering cytoplasmic dynein. Once the MTOC is formed, several different microtubule motors cooperate to move and gather the telomeres by acting on the microtubules that nucleated from the LINC-induced MTOC. This mechanism can account for microtubule-dependent telomere clustering in other organisms. In particular, it can account for the centrosome-independent formation of small aggregates of the PCs in C. elegans (Fig. 4b). By nucleating microtubules, the PCs gather by themselves independently of the centrosome. Furthermore, the opposing motile activities of kinesin and dynein motors might be able to drive the repeated association and dissociation of the small aggregates. Indeed, LINC-dependent antagonistic participation of dynein and kinesin has been observed in the nuclear migration of various organisms. During the development of C. elegans, a distinct type of the LINC complex (UNC-84/UNC-83) tethers the nucleus to microtubules via both cytoplasmic dynein and kinesin, and the tethered dynein and kinesin drive bidirectional nuclear migrations by generating forces in opposite directions [77–79]. A similarly antagonistic type of motor participation has also been observed in LINC-dependent nuclear migration during eye development in Drosophila melanogaster [80] and muscle development in mice [81]. During meiosis in C. elegans, the LINC complex (SUN-1/ZYG-12) may tether kinesin in addition to dynein, and the tethered kinesin and dynein may induce bidirectional movements of the PCs (Fig. 4b). It is also possible that oligomerized kinesin and dynein contribute to bi-directional movements by linking and moving along the PC-nucleated microtubules. Because telomere movements are similar in mouse spermatocytes [45], a similar mechanism might also drive telomere movements in mammals.
Although the telomeric MTOC-dependent mechanism accounts for the telomere clustering that has been observed in different organisms, it is not the only mechanism that induces meiotic telomere clustering. In S. cerevisiae, actin filaments drive telomere clustering instead of cytoplasmic microtubules [29, 31]. Telomere clustering also appears to be independent of cytoplasmic microtubules in plant cells, because depolymerization of cytoplasmic microtubules does not inhibit telomere clustering [82]. Therefore, at least in these organisms, telomere clustering is driven by a different mechanism(s). In S. cerevisiae, because the telomere appears to be associated with the same point on the lateral side of the actin filament during its movements, it is unlikely that telomere movements are driven by actin motor-dependent transport along the actin filaments [31]. It is also unlikely that actin filaments nucleate from the telomere, as observed regarding cytoplasmic microtubules in S. pombe. A “piggy-backing” mechanism has been proposed for telomere movements, in which telomeres are hooked to the actin filaments and moved by the elongation or shortening of the filaments. Although the major mechanism that drives telomere clustering is different in S. cerevisiae, a microtubule-dependent mechanism might still be involved in telomere clustering, because telomere clustering tends to occur around the SPB [31] and the depletion of microtubule motor Kar3 affects meiotic telomere dynamics [83].
Additional effects of the finding of the telocentrosome
MTOC regulation
The study of the telocentrosome has provided new information about MTOC regulation. During interphase of the vegetative cell cycle in S. pombe, the MTOCs are formed at the nuclear surface, and these MTOCs form parallel arrays of cytoplasmic microtubules together with the SPB [84–86]. MTOCs are also formed at the equatorial region where the septum is assembled during telophase [87]. During meiotic prophase, in addition, the SPB forms radial microtubules [13]. These MTOCs are respectively termed iMTOC, eMTOC, and rMTOC [88, 89], and their formation is dependent on Mto1, which interacts with the γ-TuC and is a potential conserved constituent of the centrosome [7, 90–93].
Telocentrosome formation also depends on Mto1, and Mto1 depletion severely compromises both telocentrosome formation and telomere clustering [7]. However, regulatory mechanisms of the telocentrosome and mitotic MTOCs are different. Dlc1 contributes to telocentrosome formation independently of a dynein motor [7], while it does not contribute to iMTOCs or eMTOCs, as shown by the lack of detectable defects on mitotic division in Dlc1-lacking cells [20]. The dynein-independent involvement of Dlc1 in telocentrosome formation is probably a reason for the severe telomere clustering defects that have been observed in cells lacking both Dhc1 and Dlc1.
In contrast, the telocentrosome and the rMTOC likely share the same mechanism for their formation, because the rMTOC is formed by gathering the telocentrosomes and the SPB. Indeed, simultaneous depletion of Dhc1 and Dlc1 compromises rMTOC formation as well as telocentrosome formation; in cells that lack both Dhc1 and Dlc1, a radial microtubule array is frequently dissociated from the SPB during meiotic prophase [7]. Because dissociation has not been observed in cells lacking either Dhc1 or Dlc1 [14, 20], a reasonable interpretation is that Dhc1 and Dlc1 independently contribute to the anchoring of the radial microtubule array to the SPB to form the rMTOC. The fact that the radial microtubule array can be formed without attaching to the SPB also implies that oligomerization of γ-TuC and tethering of the γ-TuC to the SPB are distinctly regulated, and Dhc1 and Dlc1 are likely dispensable for γ-TuC oligomerization. Dlc1 appears to be required only during the later stage of telocentrosome formation, and dispensable in the early stage [7]. Because the telocentrosome is formed in cells that contain an interphase-like microtubule array, the same mechanism that generates the iMTOC might induce telocentrosome formation at the beginning; subsequently, the meiosis-specific, Dlc1-dependent MTOC-forming mechanism (perhaps the rMTOC-forming mechanism) might take over to induce MTOC maturation and/or perform MTOC maintenance.
MTOC aggregation and spindle formation
In addition to MTOC formation, the study of MTOC-dependent telomere clustering is informative for the study of other MTOC-dependent events. The gathering of multiple MTOCs is not a process unique to telomere clustering and has been observed in various other biological events. Although centrosomes are absent during oogenesis of animal cells, a bipolar spindle is still formed in a centrosome-independent manner [94, 95]. Studies of the formation of the acentrosomal spindle in mouse oocytes showed that multiple MTOCs are formed in the cytoplasm, and these MTOCs gather to form spindle poles, like telocentrosomes do in S. pombe [96]. The centrosome-independent spindle formation mechanism also functions in mitosis [97]. Interestingly, in Xenopus egg extracts, acentrosomal spindle pole formation depends on cytoplasmic dynein, like telomere clustering [98, 99]. These similarities suggest that acentrosomal spindle pole formation is driven by a similar mechanism that drives meiotic telomere clustering. In support of this view, SUN domain proteins (Sad1 in S. pombe and Mps3 in S. cerevisiae) are required for spindle integrity and telomere clustering in yeasts [64, 100], although the present available evidence denies involvement of the SUN/KASH proteins in spindle formation in higher eukaryotes. Furthermore, in S. pombe cells that are defective in meiotic telomere clustering, meiotic spindle integrity is compromised in addition to homologous chromosome pairing [101]. This fact shows the presence of a link between telomere clustering and spindle formation, and further supports the idea that telomere clustering and spindle formation share a common mechanism.
An MTOC-dependent connection between the centrosome and cellular organelles
The MTOC-dependent clustering mechanism appears to function in other biological events. Various organelles are connected with the centrosome via microtubules, and some connections are dependent on MTOCs formed on the organelles, as observed in telocentrosome-dependent telomere clustering at the SPB. Microtubules extend from the organelle MTOCs, as well as the centrosome (Fig. 5a). The organelles and the centrosome are connected with each other through centrosome-nucleated and organelle-nucleated microtubules, and are subsequently brought into proximity by those microtubules.
MTOC-dependent centrosome-organelle/structure interactions. a MTOCs are formed on the organelle and nucleate microtubules (i). Microtubules nucleated from the organelle MTOC and the centrosome elongate and shorten repeatedly (black arrows). After the organelle-centrosome connection is established by interaction of the microtubules (ii), the organelle and the centrosome are brought into close proximity via the connecting microtubules [iii and iv, magenta arrows]. b Comparison of various centrosome-organelle/structure interactions is shown. NE nuclear envelope, H human, Y budding yeast, M mouse, W worm, F fly
An example of the MTOC-dependent centrosome-organelle connection is observed in the interaction of chromosomes with the centrosome during mitotic division (Fig. 5b, Kinetochores in human and budding yeast). During chromosome segregation, chromosomes interact with microtubules extending from the centrosome via the kinetochore and are pulled towards the centrosome by the microtubules. Recent studies have shown that the kinetochore itself nucleates microtubules, and kinetochore-nucleated microtubules interact with centrosome-nucleated microtubules to establish the centrosome-kinetochore connection [102–106]. It has been shown in human cells that the Nup107-160 nuclear pore subcomplex is recruited to the kinetochore and induces microtubule nucleation by forming a complex with γ-tubulin; the Ran GTPase activator RanGAP1-RanBP2 regulates Nup107-160-dependent microtubule nucleation and the spindle attachment of chromosomes [106–109]. The Nup107-160 complex also interacts with CENP-F [110], which binds to the cytoplasmic dynein regulators NudE/NdeI and NudEL/NdelI, as well as microtubules [111, 112]. CENP-F may also contribute to the nucleation of kinetochore microtubules, because it has been shown to regulate microtubule nucleation at the centrosome in mouse cells [113]. In S. cerevisiae, the kinetochore has also been shown to nucleate microtubules with the plus ends distal to the kinetochore, although microtubule nucleation is dependent not on γ-tubulin, but on the microtubule-plus-end-tracking protein Stu2 (an yeast ortholog of vertebrate XMAP215/TOG) [105]. Detailed analysis of kinetochore and microtubule dynamics in this organism provided evidence that the kinetochore-nucleated microtubules facilitate the establishment of the centrosome-kinetochore connection. Nucleation of kinetochore microtubules has not been detected thus far in S. pombe; however, this process may also be involved in the centrosome-kinetochore interaction, because Dlc1 (which is involved in telocentrosome formation) contributes to the kinetochore-spindle interaction [114, 115].
The MTOC-dependent mechanism is also likely to facilitate the connection between the Golgi apparatus and the centrosome (Fig. 5b, Golgi in human). The Golgi, which is composed of stacks of membrane cisternae, is required for the modification and sorting of various proteins synthesized in the endoplasmic reticulum. It is located near the centrosome during interphase and disperses to form small vesicles during mitotic division [116, 117]. After mitotic division, the scattered Golgi vesicles gather to reform the membrane cisternae structure near the centrosome. The interphase centrosomal location of the Golgi cisternae and its reconstitution after mitotic division are thought to depend on microtubules and cytoplasmic dynein, because microtubule disruption or dynein depletion cause fragmentation and dispersal of the Golgi cisternae [117–121]. The Golgi vesicles attracted attention as cargoes that were transported on the microtubules to the centrosome via cytoplasmic dynein [116]. However, it has been demonstrated that the Golgi vesicle itself accumulates γ-TuC at its surface in a manner that is dependent on the microtubule-associated protein GMAP-210 and microtubule-plus-end-tracking proteins, CLASPs; in addition, the Golgi has microtubule nucleation activity [122–124]. The centrosomal protein myomegalin, which is likely responsible for microtubule nucleation, is also co-localized with the Golgi apparatus [125]. Because of the similarity to the kinetochore, it has been proposed that the Golgi-nucleated microtubules facilitate establishment of the connection between the centrosome and the Golgi [123]. Furthermore, because the fragmented Golgi cisternae that are formed by microtubule disruption gather by themselves independently of the centrosome after microtubule reformation, it has also been proposed that the Golgi-nucleated microtubules induce self-gathering of the fragmented Golgi, as has been proposed for meiotic telomere clustering [123, 126].
The MTOC-dependent mechanism may also contribute to the centrosome-nucleus attachment (Fig. 5b, Nucleus in human, mouse, worm, and fly). In eukaryotic organisms, the centrosome interacts with the nuclear surface [127]. This interaction is essential for the development of various tissues, including brain, muscle, and eye [127], as well as for efficient breakdown of the nuclear envelope during mitotic division [128, 129]. A centrosome-nucleus connection depends on the LINC complexes, microtubules, and cytoplasmic dynein [3–5, 52, 53, 127, 130–133]. In the C. elegans embryo, the SUN domain protein SUN-1 and the KASH domain protein ZYG-12 are required for centrosome-nucleus attachment [134]. ZYG-12 contributes to centrosome-nucleus attachment by tethering cytoplasmic dynein to the nuclear envelope. During Drosophila eye development, the SUN domain protein Klaroid and the KASH domain protein Klarsicht are required for centrosome-nucleus attachment and nuclear migration [54, 135]. During Drosophila spermatogenesis, cytoplasmic dynein likely contributes to centrosome-nucleus attachment, because Asunder, which tethers cytoplasmic dynein to the nuclear envelope, is essential for centrosome-nucleus attachment [136, 137]. Similarly, human Asunder recruits dynein to the nuclear envelope and tethers the centrosome to the nucleus [138]. In mice, the SUN domain proteins SUN1 and SUN2 and KASH domain proteins Syne/Nesprin-1 and Syne/Nesprin-2 form complexes and connect the centrosome to the nucleus during neurogenesis and neuronal migration, and Syne/Nesprin-2 interacts with cytoplasmic dynein together with dynactin [139]. The kinetochore proteins responsible for microtubule nucleation also contribute to centrosome-nucleus attachment. In human cells, the components of the nuclear pore complex (Nup133 and RanBP2) and the centromere component (CENP-F) are required in order to tether the centrosome to the nucleus [140, 141]. Similar to other centrosome-nucleus attachments, the dynein regulators NudE, NudEL, and Lis1, are also involved [140–142].
Because SUN/KASH proteins, the nuclear components Nup133 and RanBP2, and CENP-F potentially contribute to microtubule nucleation, it is possible that these factors induce microtubule nucleation from the nuclear surface to facilitate interaction between the nucleus and the centrosome. Indeed, the nucleus has a γ-tubulin-dependent, microtubule nucleation activity at its surface in plant cells [143]. One might think that nuclear microtubule nucleation may not be necessary for centrosome-nucleus attachment, because the nucleus itself is a large organelle and can be easily captured by centrosomal microtubules. However, because many centrosomal microtubules probably must attach to the nucleus to move such a large organelle, it is reasonable to think that cells might utilize the MTOC-dependent mechanism to efficiently establish and/or maintain numerous attachments.
Conclusions and perspectives
It is currently evident that telomere clustering is required for meiotic homologous chromosome pairing. The LINC complex induces telomere clustering by forming the telocentrosome in S. pombe. However, the molecular mechanism that induces telocentrosome formation remains poorly understood. What molecules induce telocentrosome formation together with the LINC complex and how telocentrosome formation is regulated are the next challenging questions in the field. Similarly, how clustered telomeres are released from the SPB before meiotic division is a future question to be solved. In addition, it is of great interest to examine whether the MTOC-dependent telomere clustering observed in S. pombe is common to other organisms. Molecular mechanisms of microtubule-independent telomere clustering in budding yeast and plant cells also must be elucidated. Clarifying these points is critical to an integrated and comprehensive understanding of meiosis and of various LINC-dependent activities, such as nuclear migration, nuclear positioning, and biased sister chromatid segregation during stem-cell division. It would also contribute to a general understanding of the regulatory mechanisms of MTOC, MTOC-dependent centrosome-organelle connection, and DLC functions. Finally, it is clinically important to understand these mechanisms because the impairment of LINC complexes in human cells is involved in Emery-Dreifuss muscular dystrophy, and defective telomere clustering causes improper homologous chromosome segregation that might be a cause of miscarriage or Down’s syndrome. The mechanism of meiotic telomere clustering is undoubtedly one of the most exciting biological subjects to be studied in greater depth in the future.
Abbreviations
- DHC:
-
Dynein heavy chain
- DLC:
-
Dynein light chain
- LINC:
-
Linker of nucleoskeleton and cytoskeleton
- MTOC:
-
Microtubule-organizing center
- PC:
-
Pairing center
- SPB:
-
Spindle pole body
- γ-TuC:
-
γ-Tubulin complex
References
Scherthan H (2001) A bouquet makes ends meet. Nat Rev Mol Cell Biol 2(8):621–627
Zickler D, Kleckner N (1998) The leptotene-zygotene transition of meiosis. Annu Rev Genet 32:619–697. doi:10.1146/annurev.genet.32.1.619
Fridkin A, Penkner A, Jantsch V, Gruenbaum Y (2009) SUN-domain and KASH-domain proteins during development, meiosis and disease. Cellular Mol Life Sci: CMLS 66(9):1518–1533. doi:10.1007/s00018-008-8713-y
Hiraoka Y, Dernburg AF (2009) The SUN rises on meiotic chromosome dynamics. Dev Cell 17(5):598–605. doi:10.1016/j.devcel.2009.10.014
Razafsky D, Hodzic D (2009) Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol 186(4):461–472. doi:10.1083/jcb.200906068
Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006) Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 172(1):41–53. doi:10.1083/jcb.200509124
Yoshida M, Katsuyama S, Tateho K, Nakamura H, Miyoshi J, Ohba T, Matsuhara H, Miki F, Okazaki K, Haraguchi T, Niwa O, Hiraoka Y, Yamamoto A (2013) Microtubule-organizing center formation at telomeres induces meiotic telomere clustering. J Cell Biol 200(4):385–395. doi:10.1083/jcb.201207168
Yamamoto A, Hiraoka Y (2001) How do meiotic chromosomes meet their homologous partners? Lessons from fission yeast. Bioessays 23(6):526–533. doi:10.1002/bies.1072
Harper L, Golubovskaya I, Cande WZ (2004) A bouquet of chromosomes. J Cell Sci 117(Pt 18):4025–4032. doi:10.1242/jcs.01363
Koszul R, Kleckner N (2009) Dynamic chromosome movements during meiosis: a way to eliminate unwanted connections? Trends Cell Biol 19(12):716–724. doi:10.1016/j.tcb.2009.09.007
Yamamoto M, Imai Y, Watanabe Y (1997) Mating and sporulation in Schizosaccharomyces pombe. In: Pringle JR, Broach JR, Jones EW (eds) The molecular and cellular biology of the yeast Saccharomyces; cell cycle and cell biology, vol 3. Cold Spring Harbor Laboratory Press, New York, pp 1037–1106
Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y (1994) Telomere-led premeiotic chromosome movement in fission yeast. Science 264(5156):270–273
Ding DQ, Chikashige Y, Haraguchi T, Hiraoka Y (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J Cell Sci 111(Pt 6):701–712
Yamamoto A, West RR, McIntosh JR, Hiraoka Y (1999) A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol 145(6):1233–1249
Yamamoto A, Tsutsumi C, Kojima H, Oiwa K, Hiraoka Y (2001) Dynamic behavior of microtubules during dynein-dependent nuclear migrations of meiotic prophase in fission yeast. Mol Biol Cell 12(12):3933–3946
Yamamoto A, Hiraoka Y (2003) Cytoplasmic dynein in fungi: insights from nuclear migration. J Cell Sci 116(Pt 22):4501–4512. doi:10.1242/jcs.00835
Ananthanarayanan V, Schattat M, Vogel SK, Krull A, Pavin N, Tolic-Norrelykke IM (2013) Dynein motion switches from diffusive to directed upon cortical anchoring. Cell 153(7):1526–1536. doi:10.1016/j.cell.2013.05.020
Vogel SK, Pavin N, Maghelli N, Julicher F, Tolic-Norrelykke IM (2009) Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol 7(4):e1000087. doi:10.1371/journal.pbio.1000087
Ding DQ, Yamamoto A, Haraguchi T, Hiraoka Y (2004) Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev Cell 6(3):329–341
Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O (2002) The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell 13(3):930–946. doi:10.1091/mbc.01-11-0543
Shimanuki M, Miki F, Ding DQ, Chikashige Y, Hiraoka Y, Horio T, Niwa O (1997) A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet: MGG 254(3):238–249
Nimmo ER, Pidoux AL, Perry PE, Allshire RC (1998) Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392(6678):825–828. doi:10.1038/33941
Cooper JP, Watanabe Y, Nurse P (1998) Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392(6678):828–831. doi:10.1038/33947
Tuzon CT, Borgstrom B, Weilguny D, Egel R, Cooper JP, Nielsen O (2004) The fission yeast heterochromatin protein Rik1 is required for telomere clustering during meiosis. J Cell Biol 165(6):759–765. doi:10.1083/jcb.200312061
Chikashige Y, Hiraoka Y (2001) Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol 11(20):1618–1623
Kanoh J, Ishikawa F (2001) spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol 11(20):1624–1630
Niwa O, Shimanuki M, Miki F (2000) Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J 19(14):3831–3840. doi:10.1093/emboj/19.14.3831
Trelles-Sticken E, Dresser ME, Scherthan H (2000) Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J Cell Biol 151(1):95–106
Trelles-Sticken E, Adelfalk C, Loidl J, Scherthan H (2005) Meiotic telomere clustering requires actin for its formation and cohesin for its resolution. J Cell Biol 170(2):213–223
Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME (2008) Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133(7):1175–1187
Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S (2008) Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133(7):1188–1201
Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, Kaback DB (2007) Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104(43):16934–16939
Conrad MN, Dominguez AM, Dresser ME (1997) Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276(5316):1252–1255
Chua PR, Roeder GS (1997) Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev 11(14):1786–1800
Rockmill B, Roeder GS (1998) Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev 12(16):2574–2586
Goldman AS, Lichten M (2000) Restriction of ectopic recombination by interhomolog interactions during Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci USA 97(17):9537–9542
Schlecht HB, Lichten M, Goldman AS (2004) Compartmentalization of the yeast meiotic nucleus revealed by analysis of ectopic recombination. Genetics 168(3):1189–1203. doi:10.1534/genetics.104.029157
MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF (2005) Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell 123(6):1037–1050
Phillips CM, Wong C, Bhalla N, Carlton PM, Weiser P, Meneely PM, Dernburg AF (2005) HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123(6):1051–1063
Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, Woglar A, Csaszar E, Pasierbek P, Ammerer G, Gruenbaum Y, Jantsch V (2009) Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell 139(5):920–933. doi:10.1016/j.cell.2009.10.045
Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, Jantsch V (2010) Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6(11):e1001219. doi:10.1371/journal.pgen.1001219
Wynne DJ, Rog O, Carlton PM, Dernburg AF (2012) Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196(1):47–64. doi:10.1083/jcb.201106022
Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF (2009) Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139(5):907–919. doi:10.1016/j.cell.2009.10.039
Scherthan H, Weich S, Schwegler H, Heyting C, Harle M, Cremer T (1996) Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134(5):1109–1125
Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198(2):165–172. doi:10.1083/jcb.201204085
Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ (1997) Telomeres cluster de novo before the initiation of synapsis: a three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol 137(1):5–18
Carlton PM, Cande WZ (2002) Telomeres act autonomously in maize to organize the meiotic bouquet from a semipolarized chromosome orientation. J Cell Biol 157(2):231–242. doi:10.1083/jcb.200110126
Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, Cande WZ (2000) Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J Cell Sci 113(Pt 6):1033–1042
Sosa BA, Rothballer A, Kutay U, Schwartz TU (2012) LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149(5):1035–1047. doi:10.1016/j.cell.2012.03.046
Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, Wu SL, Karger BL, Greene MI, Wang Q (2012) Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem 287(8):5317–5326. doi:10.1074/jbc.M111.304543
Starr DA, Han M (2003) ANChors away: an actin based mechanism of nuclear positioning. J Cell Sci 116(Pt 2):211–216
Starr DA, Fischer JA (2005) KASH ‘n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays 27(11):1136–1146. doi:10.1002/bies.20312
Wilhelmsen K, Ketema M, Truong H, Sonnenberg A (2006) KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci 119(Pt 24):5021–5029
Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA (2004) The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell 15(2):600–610. doi:10.1091/mbc.E03-06-0374
Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S (2006) SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 26(10):3738–3751. doi:10.1128/MCB.26.10.3738-3751.2006
Libotte T, Zaim H, Abraham S, Padmakumar VC, Schneider M, Lu W, Munck M, Hutchison C, Wehnert M, Fahrenkrog B, Sauder U, Aebi U, Noegel AA, Karakesisoglou I (2005) Lamin A/C-dependent localization of Nesprin-2, a giant scaffolder at the nuclear envelope. Mol Biol Cell 16(7):3411–3424. doi:10.1091/mbc.E04-11-1009
Mislow JM, Kim MS, Davis DB, McNally EM (2002) Myne-1, a spectrin repeat transmembrane protein of the myocyte inner nuclear membrane, interacts with lamin A/C. J Cell Sci 115(Pt 1):61–70
Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM (2005) Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci 118(Pt 4):673–687. doi:10.1242/jcs.01642
Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y (2002) Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell 13(3):892–901. doi:10.1091/mbc.01-06-0294
Emery AE (1989) Emery-Dreifuss syndrome. J Med Genet 26(10):637–641
Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D (1994) Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet 8(4):323–327. doi:10.1038/ng1294-323
Yadlapalli S, Yamashita YM (2013) Chromosome-specific nonrandom sister chromatid segregation during stem-cell division. Nature 498(7453):251–254. doi:10.1038/Nature12106
Yamashita YM (2013) Nonrandom sister chromatid segregation of sex chromosomes in Drosophila male germline stem cells. Chromosome Res 21(3):243–254. doi:10.1007/S10577-013-9353-0
Hagan I, Yanagida M (1995) The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol 129(4):1033–1047
Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y (2006) Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125(1):59–69
Chikashige Y, Yamane M, Okamasa K, Tsutsumi C, Kojidani T, Sato M, Haraguchi T, Hiraoka Y (2009) Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J Cell Biol 187(3):413–427. doi:10.1083/jcb.200902122
Tang X, Jin Y, Cande WZ (2006) Bqt2p is essential for initiating telomere clustering upon pheromone sensing in fission yeast. J Cell Biol 173(6):845–851. doi:10.1083/jcb.200602152
Conrad MN, Lee CY, Wilkerson JL, Dresser ME (2007) MPS3 mediates meiotic bouquet formation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 104(21):8863–8868
Rao HB, Shinohara M, Shinohara A (2011) Mps3 SUN domain is important for chromosome motion and juxtaposition of homologous chromosomes during meiosis. Genes Cells 16(11):1081–1096. doi:10.1111/j.1365-2443.2011.01554.x
Penkner A, Tang L, Novatchkova M, Ladurner M, Fridkin A, Gruenbaum Y, Schweizer D, Loidl J, Jantsch V (2007) The nuclear envelope protein Matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev Cell 12(6):873–885
Labella S, Woglar A, Jantsch V, Zetka M (2011) Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Dev Cell 21(5):948–958. doi:10.1016/j.devcel.2011.07.011
Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF (2011) Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev Cell 21(5):934–947. doi:10.1016/j.devcel.2011.09.001
Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, La Volpe A, Jantsch V (2013) Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLoS Genet 9(3):e1003335. doi:10.1371/journal.pgen.1003335
Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M (2007) SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell 12(6):863–872
Schmitt J, Benavente R, Hodzic D, Hoog C, Stewart CL, Alsheimer M (2007) Transmembrane protein Sun2 is involved in tethering mammalian meiotic telomeres to the nuclear envelope. Proc Natl Acad Sci USA 104(18):7426–7431. doi:10.1073/pnas.0609198104
Horn HF, Kim DI, Wright GD, Wong ES, Stewart CL, Burke B, Roux KJ (2013) A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J Cell Biol 202:1023–1039. doi:10.1083/jcb.201304004
Fridolfsson HN, Ly N, Meyerzon M, Starr DA (2010) UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol 338(2):237–250. doi:10.1016/j.ydbio.2009.12.004
Fridolfsson HN, Starr DA (2010) Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol 191(1):115–128. doi:10.1083/jcb.201004118
Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA (2009) UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development 136(16):2725–2733. doi:10.1242/dev.038596
Whited JL, Cassell A, Brouillette M, Garrity PA (2004) Dynactin is required to maintain nuclear position within postmitotic Drosophila photoreceptor neurons. Development 131(19):4677–4686. doi:10.1242/dev.01366
Wilson MH, Holzbaur EL (2012) Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci 125(Pt 17):4158–4169. doi:10.1242/jcs.108688
Cowan CR, Cande WZ (2002) Meiotic telomere clustering is inhibited by colchicine but does not require cytoplasmic microtubules. J Cell Sci 115(Pt 19):3747–3756
Trelles-Sticken E, Loidl J, Scherthan H (2003) Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J Cell Sci 116(Pt 12):2431–2442. doi:10.1242/jcs.00453
Drummond DR, Cross RA (2000) Dynamics of interphase microtubules in Schizosaccharomyces pombe. Curr Biol 10(13):766–775
Tran PT, Marsh L, Doye V, Inoue S, Chang F (2001) A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol 153(2):397–411
Sagolla MJ, Uzawa S, Cande WZ (2003) Individual microtubule dynamics contribute to the function of mitotic and cytoplasmic arrays in fission yeast. J Cell Sci 116(Pt 24):4891–4903. doi:10.1242/jcs.00796
Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M (1991) The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci 99(Pt 4):693–700
Funaya C, Samarasinghe S, Pruggnaller S, Ohta M, Connolly Y, Muller J, Murakami H, Grallert A, Yamamoto M, Smith D, Antony C, Tanaka K (2012) Transient structure associated with the spindle pole body directs meiotic microtubule reorganization in S. pombe. Curr Biol 22(7):562–574. doi:10.1016/j.cub.2012.02.042
Sawin KE, Tran PT (2006) Cytoplasmic microtubule organization in fission yeast. Yeast 23(13):1001–1014. doi:10.1002/yea.1404
Sawin KE, Lourenco PC, Snaith HA (2004) Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol 14(9):763–775. doi:10.1016/j.cub.2004.03.042
Venkatram S, Tasto JJ, Feoktistova A, Jennings JL, Link AJ, Gould KL (2004) Identification and characterization of two novel proteins affecting fission yeast gamma-tubulin complex function. Mol Biol Cell 15(5):2287–2301. doi:10.1091/mbc.E03-10-0728
Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M (2005) Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol 15(16):1479–1486. doi:10.1016/j.cub.2005.07.058
Samejima I, Miller VJ, Rincon SA, Sawin KE (2010) Fission yeast Mto1 regulates diversity of cytoplasmic microtubule organizing centers. Curr Biol 20(21):1959–1965. doi:10.1016/j.cub.2010.10.006
Manandhar G, Schatten H, Sutovsky P (2005) Centrosome reduction during gametogenesis and its significance. Biol Reprod 72(1):2–13. doi:10.1095/biolreprod.104.031245
Schatten H, Sun QY (2009) The functional significance of centrosomes in mammalian meiosis, fertilization, development, nuclear transfer, and stem cell differentiation. Environ Mol Mutagen 50(8):620–636. doi:10.1002/em.20493
Schuh M, Ellenberg J (2007) Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130(3):484–498. doi:10.1016/j.cell.2007.06.025
Khodjakov A, Cole RW, Oakley BR, Rieder CL (2000) Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10(2):59–67
Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E (1996) Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382(6590):420–425. doi:10.1038/382420a0
Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A (1997) Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol 138(3):615–628
Jaspersen SL, Giddings TH Jr, Winey M (2002) Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J Cell Biol 159(6):945–956. doi:10.1083/jcb.200208169
Tomita K, Cooper JP (2007) The telomere bouquet controls the meiotic spindle. Cell 130(1):113–126. doi:10.1016/j.cell.2007.05.024
Mitchison TJ, Kirschner MW (1985) Properties of the kinetochore in vitro. I. Microtubule nucleation and tubulin binding. J Cell Biol 101(3):755–765
Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor TM (2003) Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol 160(5):671–683. doi:10.1083/jcb.200208143
Maiato H, Rieder CL, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167(5):831–840. doi:10.1083/jcb.200407090
Kitamura E, Tanaka K, Komoto S, Kitamura Y, Antony C, Tanaka TU (2010) Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev Cell 18(2):248–259. doi:10.1016/j.devcel.2009.12.018
Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M (2010) The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol 12(2):164–169. doi:10.1038/ncb2016
Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F (2008) Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell 19(5):1873–1882. doi:10.1091/mbc.E07-10-1050
Salina D, Enarson P, Rattner JB, Burke B (2003) Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 162(6):991–1001. doi:10.1083/jcb.200304080
Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M (2004) The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 14(7):611–617. doi:10.1016/j.cub.2004.03.031
Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, Doye V (2007) The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J 26(7):1853–1864. doi:10.1038/sj.emboj.7601642
Feng J, Huang H, Yen TJ (2006) CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma 115(4):320–329. doi:10.1007/s00412-006-0049-5
Vergnolle MA, Taylor SS (2007) Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol 17(13):1173–1179. doi:10.1016/j.cub.2007.05.077
Moynihan KL, Pooley R, Miller PM, Kaverina I, Bader DM (2009) Murine CENP-F regulates centrosomal microtubule nucleation and interacts with Hook2 at the centrosome. Mol Biol Cell 20(22):4790–4803. doi:10.1091/mbc.E09-07-0560
Chen JS, Lu LX, Ohi MD, Creamer KM, English C, Partridge JF, Ohi R, Gould KL (2011) Cdk1 phosphorylation of the kinetochore protein Nsk1 prevents error-prone chromosome segregation. J Cell Biol 195(4):583–593. doi:10.1083/jcb.201105074
Buttrick GJ, Meadows JC, Lancaster TC, Vanoosthuyse V, Shepperd LA, Hoe KL, Kim DU, Park HO, Hardwick KG, Millar JB (2011) Nsk1 ensures accurate chromosome segregation by promoting association of kinetochores to spindle poles during anaphase B. Mol Biol Cell 22(23):4486–4502. doi:10.1091/mbc.E11-07-0608
Allan VJ, Thompson HM, McNiven MA (2002) Motoring around the Golgi. Nat Cell Biol 4(10):E236–E242. doi:10.1038/ncb1002-e236
Rogalski AA, Singer SJ (1984) Associations of elements of the Golgi apparatus with microtubules. J Cell Biol 99(3):1092–1100
Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB (1997) Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol 139(2):469–484
Corthesy-Theulaz I, Pauloin A, Pfeffer SR (1992) Cytoplasmic dynein participates in the centrosomal localization of the Golgi complex. J Cell Biol 118(6):1333–1345
Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N (1998) Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol 141(1):51–59
Fath KR, Trimbur GM, Burgess DR (1994) Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol 126(3):661–675
Efimov A, Kharitonov A, Efimova N, Loncarek J, Miller PM, Andreyeva N, Gleeson P, Galjart N, Maia AR, McLeod IX, Yates JR 3rd, Maiato H, Khodjakov A, Akhmanova A, Kaverina I (2007) Asymmetric CLASP-dependent nucleation of noncentrosomal microtubules at the trans-Golgi network. Dev Cell 12(6):917–930. doi:10.1016/j.devcel.2007.04.002
Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C (2001) The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell 12(7):2047–2060
Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M (2004) GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118(3):323–335. doi:10.1016/j.cell.2004.07.012
Verde I, Pahlke G, Salanova M, Zhang G, Wang S, Coletti D, Onuffer J, Jin SL, Conti M (2001) Myomegalin is a novel protein of the Golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J Biol Chem 276(14):11189–11198. doi:10.1074/jbc.M006546200
Ho WC, Allan VJ, van Meer G, Berger EG, Kreis TE (1989) Reclustering of scattered Golgi elements occurs along microtubules. Eur J Cell Biol 48(2):250–263
Reinsch S, Gonczy P (1998) Mechanisms of nuclear positioning. J Cell Sci 111(Pt 16):2283–2295
Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B (2002) Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108(1):97–107
Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS (2008) Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol 182(6):1063–1071. doi:10.1083/jcb.200803071
Burke B, Roux KJ (2009) Nuclei take a position: managing nuclear location. Dev Cell 17(5):587–597. doi:10.1016/j.devcel.2009.10.018
Mejat A, Misteli T (2010) LINC complexes in health and disease. Nucleus Austin 1(1):40–52. doi:10.4161/Nucl.1.1.10530
Rothballer A, Schwartz TU, Kutay U (2013) LINCing complex functions at the nuclear envelope: what the molecular architecture of the LINC complex can reveal about its function. Nucleus 4(1):29–36. doi:10.4161/nucl.23387
Tapley EC, Starr DA (2013) Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol 25(1):57–62. doi:10.1016/j.ceb.2012.10.014
Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115(7):825–836
Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA (2007) Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly 1(2):75–85
Anderson MA, Jodoin JN, Lee E, Hales KG, Hays TS, Lee LA (2009) Asunder is a critical regulator of dynein-dynactin localization during Drosophila spermatogenesis. Mol Biol Cell 20(11):2709–2721. doi:10.1091/mbc.E08-12-1165
Sitaram P, Anderson MA, Jodoin JN, Lee E, Lee LA (2012) Regulation of dynein localization and centrosome positioning by Lis-1 and asunder during Drosophila spermatogenesis. Development 139(16):2945–2954. doi:10.1242/dev.077511
Jodoin JN, Shboul M, Sitaram P, Zein-Sabatto H, Reversade B, Lee E, Lee LA (2012) Human Asunder promotes dynein recruitment and centrosomal tethering to the nucleus at mitotic entry. Mol Biol Cell 23(24):4713–4724. doi:10.1091/mbc.E12-07-0558
Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M (2009) SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron 64(2):173–187. doi:10.1016/j.neuron.2009.08.018
Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V (2011) A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol 192(5):855–871. doi:10.1083/jcb.201007118
Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, Akhmanova A (2010) Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8(4):e1000350. doi:10.1371/journal.pbio.1000350
Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG (2004) Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol 165(5):709–721. doi:10.1083/jcb.200309025
Erhardt M, Stoppin-Mellet V, Campagne S, Canaday J, Mutterer J, Fabian T, Sauter M, Muller T, Peter C, Lambert AM, Schmit AC (2002) The plant Spc98p homologue colocalizes with gamma-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J Cell Sci 115(Pt 11):2423–2431
Acknowledgments
I thank Akira Shinohara, Kayoko Tanaka, Takashi Ushimaru, and Masahiro Uritani for critical reading of the manuscript and helpful comments. This work was supported by Grants-in-aid for Scientific Research (C) to A. Y. and the Cooperative Research Program of the Institute for Protein Research, Osaka University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamamoto, A. Gathering up meiotic telomeres: a novel function of the microtubule-organizing center. Cell. Mol. Life Sci. 71, 2119–2134 (2014). https://doi.org/10.1007/s00018-013-1548-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1548-1