Abstract
Purpose
Most adjuvant breast cancer treatment regimens include the combination of an anthracycline (epirubicin or doxorubicin) and the alkylating agent cyclophosphamide. This study sought to investigate the influence of pharmacogenetics on the pharmacokinetics and metabolism of these agents.
Methods
Blood samples were taken from patients treated with cyclophosphamide (n = 51) and epirubicin (n = 35), with or without 5-fluorouracil (5-FU). The pharmacokinetics and metabolism of the three drugs were investigated, together with pharmacogenetic investigations for cyclophosphamide and epirubicin. Cyclophosphamide and its metabolites and also epirubicin and epirubicinol were measured in plasma. DNA was extracted from whole blood and genotyping performed using RT-PCR.
Results
Patients with at least one variant CYP2C19*17 allele had a longer CP half-life (p = 0.007), as did homozygous variants for the CYP2B6*6 allele. There was no significant effect of GSTP1, CYP2B6*2, CYP2B6*5 or CYP2C19*2 on any pharmacokinetic parameter of CP. An NQO2 exonic SNP was associated with a higher exposure to epirubicinol relative to epirubicin (p = 0.011). Other polymorphic variants of NQO1, carbonyl reductase, UGT enzymes and transporters had no influence on epirubicin or its metabolite.
Conclusion
Overall, pharmacogenetic factors had only a minor influence on cyclophosphamide or anthracycline-based adjuvant therapy of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adjuvant chemotherapy is commonly applied postsurgery with the aim of eradicating any residual disease and has been shown to improve outcomes in patients with early stage breast cancer. Although a variety of different regimens have been used, most include the combination of an anthracycline (epirubicin or doxorubicin) and the alkylating agent cyclophosphamide, often combined with 5-fluorouracil (5-FU).
Cyclophosphamide is a prodrug that requires metabolic activation to a 4-hydroxy metabolite, which subsequently breaks down to yield DNA-alkylating species [1]. The initial reaction, as well as inactivation of cyclophosphamide to keto (keto)- and dechloroethyl- (DCCP) metabolites, is catalyzed by a range of cytochrome P-450 enzymes. An additional inactivation reaction is catalyzed by aldehyde dehydrogenases to form carboxyethylphosphoramide mustard (CX), the major metabolite seen in urine. Previous studies in patients with breast cancer [2] or with non-Hodgkins lymphoma (NHL) [3] have shown an inverse relationship between plasma concentrations of the parent drug and anti-tumor effect. Conversely, high concentrations in plasma of the inactive metabolites have been associated with relapse in the NHL patients [3].
Epirubicin has often supplanted doxorubicin as the anthracycline component of adjuvant breast cancer regimens. The cardiotoxic effects of epirubicin are less than those associated with doxorubicin. Although not subject to the same degree of reduction by aldo-ketoreductase enzymes, epirubicin can be conjugated to form glucuronides, catalyzed by uridine glucuronosyl transferase enzymes (UGTs) [4, 5]. Epirubicin, and other anthracyclines, are also subject to transport by the ABC [6] and SLC [7, 8] families of membrane proteins, which contribute to the uptake of the drug in liver and to biliary excretion.
Genetic polymorphisms result in variations in expression and activity of CYP and UGT enzymes and of ABC and SLC transporters. Such variations can have multiple effects on the pharmacology of drugs used in cancer chemotherapy, including cyclophosphamide and epirubicin. For instance, a UGT2B7 polymorphism has been reported to influence the outcome of adjuvant breast cancer treatment [4]. Similarly, polymorphisms in GST enzymes, which inactivate DNA-alkylating metabolites, have been shown to influence outcome in breast cancer patients treated with cyclophosphamide [9] or with anthracyclines [10]. The impact of these and other polymorphisms on the pharmacokinetics and metabolism of cyclophosphamide and epirubicin is unknown.
In the study reported here, blood samples have been taken from patients receiving treatment with the combination of epirubicin and cyclophosphamide, combined with 5-FU in some patients. The pharmacokinetics and metabolism of all three drugs have been investigated, together with genotyping for the common polymorphisms in genes known to influence the pharmacology of cyclophosphamide and epirubicin.
Methods
A total of 51 patients were recruited at the Northern Centre for Cancer Care. The protocol for the study was approved by Newcastle and North Tyneside Ethics Committee 1. The women received between 3 and 6 cycles of either 5-FU, epirubicin and cyclophosphamide (FEC) (n = 32), epirubicin and cyclophosphamide (EC) (n = 3), doxorubicin and cyclophosphamide (AC) (n = 13) or 5-FU, doxorubicin and cyclophosphamide FAC (n = 3) adjuvant chemotherapy. Twelve FEC patients had 3 cycles of FEC followed by 3 cycles of docetaxel. Forty-six patients were treated with a planned CP dose of 600 mg/m2 and 5 (all FEC) with planned dose of 500 mg/m2. The dose of epirubicin was 75–100 mg/m2. Dose of 5-FU was 500–600 mg/m2. Each of the drugs was administered as a short infusion (3–20 min). Blood samples (10 ml) for analysis of drugs and metabolites were obtained in EDTA tubes pretreatment and at 0.25, 0.5, 1, 2, 4, 6 and 24 h postadministration. Plasma was separated by centrifugation. Cyclophosphamide and its stable inactivated metabolites; dechloroethylcyclophosphamide (DCCP), carboxyethylphosphoramide mustard (CX) and ketocyclophosphamide (keto) were measured by a validated LCMS method described previously [11]. Limits of quantitation were 0.5 μg/ml for cyclophosphamide and 0.05 μg/ml for the metabolites. Epirubicin and its major metabolite epirubicinol were quantified in plasma by a validated HPLC assay with fluorescence detection [12]. 5-FU was measured by a validated LCMS assay based on a previously published method [13]. Limits of quantitation for the epirubicin and epirubicinol were 2 and 5 ng/ml for 5-FU. Doxorubicin was not quantified in the 13 patients treated with this anthracycline.
DNA was extracted from whole blood samples using a Qiagen kit and working to the manufacturer’s instructions. Genotyping for the polymorphisms described in Table 1 was performed using RT-PCR by Taqman assays-on-demand (Applied Biosystems) and one custom Taqman assay (CYP2B6*4), again supplied by Applied Biosystems.
A non-compartmental analysis was performed using WinNonLin (version 5) for CP and its metabolites (CX, DCCP, Keto), using the linear/log trapezoidal method to calculate AUCs. A similar analysis was performed for epirubicin and epirubicinol and for 5-FU.
Data were analyzed using SPSS version 17.0. The Kolmogorov–Smirnoff test was used to test for the normal distribution of parameters. A statistically significant effect of genotype on CP or metabolite (CX, DCCP, Keto) pharmacokinetics (half-life, clearance, distribution and AUC) was tested using a univariate analysis (one-way ANOVA or independent t test depending on frequency of minor allele). Other factors that may have had an influence on the pharmacokinetics and toxicity were also tested, including differences between treatment program, age and concomitant medication using independent t test and Pearson’s correlation. A total of 594 statistical tests were carried out, and following Bonferonni correction for multiple testing, a threshold for statistical significance of <0.00008 was established for an Alpha of 0.05.
Results
Representative plasma concentration profiles for cyclophosphamide and metabolites, epirubicin and epirubicinol and for 5-FU are shown in Fig. 1a–c. Cmax values for cyclophosphamide varied from 18.6 to 44.9 μg/ml, with corresponding ranges of values of 57–148 and 2.9–26.4 μg/ml for epirubicin and 5-FU, respectively. Cyclophosphamide and epirubicin and metabolites could be quantitated throughout the 24-h sampling period. 5-FU was eliminated very rapidly and was not detectable beyond 4 h after administration. Pharmacokinetic parameters for each of the drugs were normally distributed and are given in Table 2, together with AUC values for the metabolites. With the exception of epirubicin clearance, which is higher than that reported previously (181 ± 46 l/h cf 61.7 (33.3–107.9) l/h [14]), all of the parameters are in keeping with previously published data [15, 16].
In relation to cyclophosphamide exposure, genotypes of CYP2B6*2, CYP2B6*4, CYP2B6*5, CYP2B6*9, CYP2C19, CYP2C19*2, CYP2C19*17 and GSTP1 were determined for all 51 patients. All SNPs were in Hardy–Weinberg equilibrium, and the frequencies were consistent with those of a European population. Linkage disequilibrium was investigated using a Fisher’s exact test and a 3 by 3 contingency table. Strong linkage disequilibrium was seen between the CYP2B6 SNPs 758A>G and 516G>T (p < 0.001), and this haplotype describes the CYP2B6*6 genotype. This linkage disequilibrium is consistent with previously reported data [17]. Since only one patient had the *4 allele (785A>G), patients who were homozygous for the CYP2B6*6 alleles were compared as a group to all other patients. None of the other SNPs were in linkage disequilibrium.
There was a significant effect of CYP2C19*17 on both CP and CX half-life. Having at least one variant allele was associated with a longer CP half-life (6.72 ± 1.14 h vs. 5.76 ± 1.12 h, p = 0.007, Fig. 2a) and a longer CX half-life (13.8 ± 6.0 h vs. 10.1 ± 2.4 h, p = 0.030, Fig. 2b). The magnitude of the effect was small, and there was no statistical effect of CYP2C19*17 on any of the other pharmacokinetic parameters. Although 4 patients were taking the CYP2C19 inhibitor omeprazole, the effect of CYP2C19*17 was retained when these patients were omitted from the analysis. There was similarly a significant effect of CYP2B6*6 on CP half-life, however, not on CX half-life. Homozygote CYP2B6*6 patients showed a longer half-life compared to all others (7.12 ± 1.43 h vs. 5.92 ± 1.11 h, p = 0.020, Fig. 2c). There were no significant effects of the CYP2B6*6 allele on any of the other pharmacokinetic parameters. There was also no significant effect of GSTP1, CYP2B6*2, CYP2B6*5, CYP2C19*2 or NQO1/2 on any pharmacokinetic parameter (Supplementary Fig. 1).
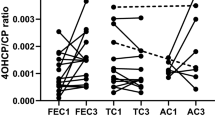
The NQO2 rs1143684 variant was associated with a higher ratio of epirubicinol/epirubicin AUC (Fig. 3). The mean and standard deviation of the ratio for carriers and minor allele homozygotes were 0.73 ± 0.26 compared with a value of 0.50 ± 0.11 for wild-type homozygotes (p = 0.011, t test). Although a wide range of genotypes linked to the pharmacology of epirubicin was explored, no other influence of genotype on pharmacokinetics was identified. Plots of epirubicin Cl for the genetic variants most frequently observed or reported to influence epirubicin pharmacokinetics are given in Supplementary Fig. 2.
None of the pharmacogenetic effects on cyclophosphamide or epirubicin pharmacokinetics retained statistical significance after Bonferroni correction for multiple testing.
There were statistically significant differences in epirubicin clearance (264 ± 47 vs. 173 ± 39 l/h, p < 0.0001) and volume of distribution (4,732 ± 1,361 vs. 3,159 ± 795 l, p = 0.004) between patients who had received EC (n = 3) compared with those who had received FEC (n = 32), resulting in a lower AUC in the EC group. However, this may have been confounded by age, with the EC patients tending to be older (64.3 ± 4.9 vs. 55.3 ± 9.4 years), although this difference was not significant (p = 0.194).
In four patients who received concomitant treatment with omeprazole, the half-life of cyclophosphamide was longer than in the other 47 patients (7.33 ± 0.64 h vs. 5.95 ± 1.18 h, p = 0.027). There was no impact on pharmacokinetics of any other concomitant medication.
Although the primary aim of this study was to analyze the effects of genotype on pharmacokinetics, toxicity was also considered. Due to the short follow-up time for this study, this was measured by investigating patients who experienced a dose delay or a dose reduction, inability to complete planned course, or who were suffering from a grade 3 infection. None of the SNPs had a significant effect on indicators of, or surrogates for, toxicity.
Discussion
The adjuvant treatment of early breast cancer now commonly includes an anthracycline, cyclophosphamide and possibly the addition of other agents such as 5-FU. In recent years, doxorubicin has been replaced by epirubicin as the preferred anthracycline, largely on the basis of a lower risk of cardiotoxicity. Although a regimen of EC or FEC is reasonably well tolerated, some patients experience toxicity that limits dose intensity, and some patients suffer recurrent disease. The hypothesis that pharmacogenetic variability, mediated by an effect on pharmacokinetics or metabolism of these adjuvant agents, influences response to treatment is explored in this study. Although the effect of dihydropyrimidine dehydrogenase on the pharmacology of 5-FU is profound, genetic variants in the DPYD gene are rare in a Caucasian population and the clinical impact of DYPD pharmacogenetics is unlikely to be seen in this small population [18, 19]. Although CPIC guidelines indicate that dose reduction may be appropriate for individuals heterozygous for DPYD variants, the frequency of these is cited to be only 3–5 %. Therefore, the primary focus of the study reported here is on cyclophosphamide (n = 51) and epirubicin (n = 35). These are relatively small numbers for a pharmacogenetic study, but the aim was to study the direct influence of pharmacogenetics on the metabolism of these two drugs rather than on outcome.
The only effect of pharmacogenetic variation on cyclophosphamide pharmacokinetics was the impact of CYP2C19 and CYP2B6 genotype on half-life. Individuals with a variant CYP2C19*17 allele or homozygous for CYP2B6*6 had a longer half-life than patients lacking these variants. These effects were fairly modest and were not linked to a significant consistent effect of genotype on clearance, as would be expected by the direct inverse relationship between these two parameters. Previous studies have reported a similarly modest effect of CYP2C19 genotype on cyclophosphamide pharmacokinetics [17, 20]. The CYP2C19*17 variant is usually associated with an ultrafast metabolizer phenotype [21], which seems inconsistent with the longer half-life of cyclophosphamide observed. However, the impact of genotype on metabolism is substrate dependent and may be confounded by differences between normal volunteers and cancer patients [22]. The modest interaction with omeprazole is also consistent with a significant role for CYP2C19 in the metabolism of cyclophosphamide. Interestingly, a similar impact of CYP2B6*9 variants (a constituent of the *6 genotype) on cyclophosphamide elimination rate constant, consistent with the effect seen on half-life here due to CYP2B6*6, was reported previously in patients with nephritis [23]. The clinical significance of these modest differences in pharmacokinetics is unclear; however, variants linked to CYP2B6*6 have previously been associated with worse overall survival [24], which is consistent with the slower rate of activation of cyclophosphamide.
A number of polymorphisms in genes believed to be associated with the metabolism (CBR1, CBR3, NQO1 and NQO2), conjugation (UGT2B7) and transport (ABCB1, ABCC1 and SLC22A16) of anthracyclines were investigated in this study. Individuals carrying at least one minor allele for the NQO2 SNP rs1143684 had a higher exposure to epirubicinol when corrected for epirubicin exposure. NQO2 is a homolog of the quinone reductase, NQO1, which has previously been shown to be associated with response to anthracycline-containing regimens [25, 26]. However, while NQO1 can use both NADH and NADPH to catalyze the obligate two-electron reduction in quinones to yield a hydroquinone product, NQO2 can use neither efficiently [27, 28], and it remains to be demonstrated that NQO2 retains a functional enzymatic activity. We have previously reported that the same NQO2 polymorphism is associated with a worse outcome following adjuvant AC therapy for hormone receptor negative breast cancer and that stable expression of the NQO2 rs1143684 mutant confers resistance to doxorubicin in MCF7 cells, while wild-type NQO2 sensitizes their isogenic partner cells [26]. The previous clinical and in vitro observations are consistent with the apparent increased accumulation of epiribicinol reported in this study. The mechanism behind this relationship remains to be elucidated, but given the lack of endogenous NQO2 activity and the lack of activity of the homologous NQO1 with anthracyclines as a substrate, it is unlikely to involve a direct reduction in the quinone group of epirubicin.
Although SNPS in SLC22A16 [29], CBR1 and CBR3 [7], or expression of ABCB1 [30] have been associated with the pharmacology of epirubicin and other anthracyclines, no significant effect of known, functionally significant SNPS was observed. A SNP in the UGT2B7 gene, associated with the glucuronidation of epirubicin [5], has previously been reported to have a profound influence on survival outcome [4]. However, this SNP was also found to have no impact on epirubicin pharmacokinetics or metabolism. The rs4148350 T allele in the ABCC1 gene has been associated with a greater risk of cardiotoxicity in children receiving anthracycline therapy [31] and febrile neutropenia in breast cancer patients treated with FEC [8]. However, only the SNP in NQO2 had any effect on epirubicin pharmacokinetics (Fig. 3).
This study sought to investigate the effect of variants in key genes thought to influence the pharmacology of cyclophosphamide and epirubicin. Detailed data on pharmacokinetics and metabolism, in relation to pharmacogenetic variants, were generated in a relatively small population. Although larger studies would be required to discern more subtle effects, the data presented here indicate only a modest impact of pharmacogenetics on FEC therapy for breast cancer.
References
Boddy AV, Yule SM (2000) Metabolism and pharmacokinetics of oxazaphosphorines. Clin Pharmacokinet 38:291–304
Ayash LJ, Wright JE, Tretyakov O, Gonin R, Elias A, Wheeler C et al (1992) Cyclophosphamide pharmacokinetics: correlation with cardiac toxicity and tumor response. J Clin Oncol 10:995–1000
Yule SM, Price L, McMahon AD, Pearson ADJ, Boddy AV (2004) Cyclophosphamide metabolism in children with non-Hodgkin’s lymphoma. Clin Cancer Res 10:455–460
Parmar S, Stingl J, Huber-Wechselberger A, Kainz A, Renner W, Langsenlehner U et al (2011) Impact of UGT2B7His268Tyr polymorphism on the outcome of adjuvant epirubicin treatment in breast cancer. Breast Cancer Res 13:R57
Innocenti F, Iyer L, Irez JRAM, Green MD, Ratain MJ, Pharmacology C (2001) Epirubicin glucuronidation is catalysed by human UDP-glucuronosyl transferase 2B7. Drug Metab Dispos 29:686–692
Kamiyama N, Takagi S, Yamamoto C, Kudo T, Nakagawa T, Takahashi M et al (2006) Expression of ABC transporters in human hepatocyte carcinoma cells with cross-resistance to epirubicin and mitoxantrone. Anticancer Res 26:885–888
Lal S, Wong Z, Sandanaraj E, Xiang X, Ang P, Lee E et al (2008) Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci 99:816–823
Vulsteke C, Lambrechts D, Dieudonné A, Hatse S, Brouwers B, van Brussel T et al (2013) Genetic variability in the multidrug resistance associated protein-1 (ABCC1/MRP1) predicts hematological toxicity in breast cancer patients receiving (neo-)adjuvant chemotherapy with 5-fluorouracil, epirubicin and cyclophosphamide (FEC). Ann Oncol 24:1513–1525
Petros WP, Hopkins PJ, Spruill S, Broadwater G, Vredenburgh JJ, Colvin OM, Peters WP, Jones RB, Hall J, Marks J (2005) Associations between drug metabolism genotype, chemotherapy pharmacokinetics, and overall survival in patients with breast cancer. J Clin Oncol 23:6117–6125
Oliveira AL, Rodrigues FFO, Santo RE, Aoki T, Rocha MN, Longui CAMM (2010) GSTT1, GSTM1 and GST1 polymorphisms and chemotherapy response in locally advanced breast cancer. Genet Mol Res 9:1045–1053
Chinnaswamy G, Errington J, Foot A, Boddy AV, Veal GJ, Cole M (2011) Pharmacokinetics of cyclophosphamide and its metabolites in paediatric patients receiving high-dose myeloablative therapy. Eur J Cancer 47:1556–1563
Dobbs NA, Twelves CJ (1991) Measurement of epidoxorubicin and its metabolites by high-performance liquid chromatography using an advanced automated sample processor. J Chromatogr 572:211–217
Remaud G, Morel A, Gamelin A (2005) Sensitive MS/MS-liquid chromatography assay for simultaneous determination of tegafur, 5-fluorouracil and 5-fluorodihydrouracil in plasma. J Chromatogr B Analyt Technol Biomed Life Sci 824:153–160
Prado CMM, Lima I, Baracos VE, Bies R, McCargar LJ, Reiman T et al (2011) An exploratory study of body composition as a determinant of epirubicin pharmacokinetics and toxicity. Cancer Chemother Pharmacol 67:93–101
Batey MA, Wright JG, Azzabi A, Newell DR, Lind MJ, Calvert AH et al (2002) Population pharmacokinetics of adjuvant cyclophosphamide, methotrexate and 5-fluorouracil (CMF). Eur J Cancer 38:1081–1089
Sandstrom BM, Freijs A, Larsson R, Nygren P, Fjillskog M, Bergh J et al (1996) Lack of relationship between systemic exposure for the component drugs of the fluorouracil, epirubicin, and 4-hydroxycyclophosphamide regimen in breast cancer patients. J Clin Oncol 14:1581–1588
Ekhart C, Doodeman VD, Rodenhuis S, Smits PHM, Beijnen JH, Huitema ADR (2008) Influence of polymorphisms of drug metabolizing enzymes of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genomics 18:515–523
Ciccolini J, Gross E, Dahan L, Lacarelle B, Mercier C (2011) Routine dihydropyrimidine dehydrogenase testing for anticipating 5-fluorouracil–related severe toxicities: hype or hope? Clin Colorectal Cancer 9:224–228
Caudle KE, Thorn CF, Klein TE, Swen JJ, McLeod HL, Diasio RB et al (2013) Clinical pharmacogenetics implementation consortium guidelines for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing. Clin Pharmacol Ther 94:640–645
Helsby N, Hui C, Goldthorpe M, Coller J, Soh M, Gow P et al (2010) The combined impact of CYP2C19 and CYP2B6 pharmacogenetics on cyclophosphamide bioactivation. Br J Clin Pharmacol 70:844–853
Baldwin RM, Ohlsson S, Pedersen RS, Mwinyi J, Ingelman-Sundberg M, Eliasson E et al (2008) Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol 65:767–774
Helsby N, Lo W, Sharples K, Riley G, Murray M, Spells K et al (2008) CYP2C19 pharmacogenetics in advanced cancer: compromised function independent of genotype. Br J Cancer 99:1251–1255
Joy M, La M, Wang J, Bridges A, Hu Y, Hogan S et al (2012) Cyclophosphamide and 4-hydroxycyclophosphamide pharmacokinetics in patients with glomerulonephritis secondary to lupus and small vessel vasculitis. Br J Clin Pharmacol 74:445–455
Bray J, Sludden J, Griffin MJ, Cole M, Verrill M, Jamieson D et al (2010) Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br J Cancer 102:1003–1009
Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjäkoski K et al (2008) NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat Genet 40:844–853
Jamieson D, Cresti N, Bray J, Sludden J, Griffin MJ, Hawsawi NM et al (2011) Two minor NQO1 and NQO2 alleles predict poor response of breast cancer patients to adjuvant doxorubicin and cyclophosphamide therapy. Pharmacogenet Genomics 21:808–819
Wu K, Knox R, Sun XZ, Joseph P, Jaiswal AK, Zhang D et al (1997) Catalytic properties of NAD(P)H:quinone oxidoreductase-2 (NQO2), a dihydronicotinamide riboside dependent oxidoreductase. Arch Biochem Biophys 347:221–228
Jamieson D, Tung ATY, Knox RJ, Boddy AV (2006) Reduction of mitomycin C is catalysed by human recombinant NRH : quinone oxidoreductase 2 using reduced nicotinamide adenine dinucleotide as an electron donating co-factor. Br J Cancer 95:1229–1233
Lal S, Wong ZW, Jada SR, Xiang X, Shu XC, Ang PCS, Figg WD, Lee EJD, Chowbay B (2007) Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics 8:567–575
Reed K, Hembruff SL, Sprowl JA, Parissenti AM (2010) The temporal relationship between ABCB1 promoter hypomethylation, ABCB1 expression and acquisition of drug resistance. Pharmacogenomics J 10:489–504
Visscher H, Ross C, Rassekh S, Barhdadi A, Dube M, Al-Saloos H et al (2012) Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol 30:1422–1428
Zimniak P, Nanduri B, Pikuła S, Bandorowicz-Pikuła J, Singhal SS, Srivastava SK et al (1994) Naturally occurring human glutathione S-transferase GSTP1-1 isoforms with isoleucine and valine in position 104 differ in enzymic properties. Eur J Biochem 224:893–899
Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML et al (2003) Hepatic CYP2B6 expression : gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther 307:906–922
Xie H, Yasar Ü, Lundgren S, Griskevicius L, Terelius Y, Hassan M et al (2003) Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J 3:53–61
Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL et al (2005) Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics 15:1–5
Satyanarayana C, Devendran A, Jayaraman M, Mannu J, Mathur PP, Gopal SD, Rajagopal K, Chandrasekaran A (2009) Influence of the genetic polymorphisms in the 5′ flanking and exonic regions of CYP2C19 on proguanil oxidation. Drug Metab Pharmacokinet 24:537–548
Sim SC, Risinger C, Dahl M-L, Aklillu E, Christensen M, Bertilsson L et al (2006) A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther 79:103–113
Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, Ross D (2001) Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:quinone oxidoreductase 1. Mol Pharmacol 59:263–268
Blanco J, Sun C, Landier W, Chen L, Esparza-Duran D, Leisenring W et al (2012) Anthracycline-related cardiomyopathy after childhood cancer: role of polymorphisms in carbonyl reductase genes—a report from the children’s oncology group. J Clin Oncol 30:1415–1421
Fan L, Goh B-C, Wong C-I, Sukri N, Lim S-E, Tan S-H et al (2008) Genotype of human carbonyl reductase CBR3 correlates with doxorubicin disposition and toxicity. Pharmacogenet Genomics 18:621–631
Bains O, Karkling M, Grigliatti T, Reid R, Riggs K (2009) Two nonsynonymous single nucleotide polymorphisms of human carbonyl reductase 1 demonstrate reduced in vitro metabolism of daunorubicin and doxorubicin. Drug Metab Dispos 37:1107–1114
Hoffmeyer S, Burk O, von Richter O, Arnold H, Brockmoller J, Johne A et al (2000) Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci 97:3473–3478
Acknowledgments
This study was supported by Cancer Research UK.
Conflict of interest
All authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jamieson, D., Lee, J., Cresti, N. et al. Pharmacogenetics of adjuvant breast cancer treatment with cyclophosphamide, epirubicin and 5-fluorouracil. Cancer Chemother Pharmacol 74, 667–674 (2014). https://doi.org/10.1007/s00280-014-2541-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-014-2541-6