Abstract
Purpose
In terms of drug resistance, cancer cells usually benefit from high clusterin (CLU) expression on chemotherapy. In contrast, CLU expression has been found to be a favorable prognostic factor in lung cancer patients. The aims of this study are to determine the association between CLU expression and chemotherapeutic sensitivity and the potential role of CLU in migration in human non-small-cell lung cancer (NSCLC) cell lines.
Methods
The levels of clusterin in NSCLC cell lines were altered by short hairpin RNA interference (shRNAi) and overexpression on chemosensitivity assay. Migratory ability of these cell lines was also investigated.
Results
H1355 cells with the highest level of CLU demonstrated the lowest sensitivities to Adriamycin (ADR), docetaxel (DOC), and gemcitabine (GEM) treatment. Inhibition of CLU expression in H1355 cells resulted in higher chemosensitivities. When CLU was stably overexpressed in A549 and H1299 cells, only the chemosensitivity to ADR was reduced. The migratory ability of CLU-overexpressing cells significantly decreased. Moreover, MMP2 transcription was inhibited in CLU-overexpressing H1299 cells. These results indicated lower metastatic potential for cancer cells with high CLU level.
Conclusion
Lung cancer cells with high level of CLU have reduced chemosensitivity. High level of CLU may result in migratory inhibition and thus favorable prognosis in lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the second most frequently diagnosed cancer in men and women and accounts for one-third of all cancer-related deaths in the United States every year [11]. Eighty percent of lung cancer cases are non-small-cell lung cancer (NSCLC), and the remaining 20% are small-cell lung cancer (SCLC).
In humans, clusterin (CLU) is a single-copy gene located at chromosome 8 [23]. CLU is a secreted glycoprotein that is translated from a single mRNA as a preprotein [2]. This preprotein, cytosolic clusterin (cCLU), is an intracellular 449 amino acid polypeptide chain, with an apparent MW of 60 kDa, in which the first 22 amino acids represent the classical hydrophobic secretory signal sequence [19]. Proteolytic cleaving of mature glycosylated cCLU results in α and β chains that are linked by five disulfide bridges [6] to produce the mature form of the secreted clusterin protein (sCLU). The processed clusterin protein appears as a smear of about 40 kDa on Western blot [13]. Nuclear clusterin (nCLU) is translated from the second ATG of the mRNA without leader peptide. The nCLU is unglycosylated and involved in apoptosis induction [13]. Overexpression of a truncated form of CLU lacking the hydrophobic secretion signal sequence that localizes to the nucleus of prostate epithelial cells causes the induction of apoptosis [3, 21].
The expression of CLU has been extensively investigated in prostate cancer [15, 24], cervical cancer [22], bladder cancer [10], and renal cell carcinoma [16, 20] for its prognostic significance, which includes chemotherapeutic sensitivity and metastatic potential. Inhibition of CLU expression in cancer cells results in higher sensitivity to stress and toxic conditions and, thus, higher susceptibility to cell death [24].
In trying to understand the association between CLU expression and lung cancer, investigators have faced some difficulties due to inconsistent in vitro and in vivo clinical data. Targeting of the CLU gene expression in lung cancer cell lines by antisense oligonucleotides (ASO) and siRNAs results in sensitization of cancer cells to radiotherapy [4] and chemotherapy [12] and reduces their migratory and invasive abilities [7]. From examinations of tumor specimens from patients with NSCLC, cytoplasmic CLU-positive staining is associated with better overall survival and lower recurrence rates than cytoplasmic CLU-negative staining [1]. These results suggest that CLU expression is a favorable prognostic factor in patients with NSCLC. Such contradictions between promising in vitro cell line data and clinical findings jeopardize the development of an effective therapy for lung cancer. Therefore, using human NSCLC cell lines, we altered the CLU expression and with various therapeutic drugs tested the association between CLU expression and chemotherapeutic sensitivity and investigated the potential role of CLU in cancer cell migration.
Materials and methods
Cell lines and cultures
Human NSCLC cell lines (A549, H1299, H1355, H460, and Calu-1) and lung cells (BEAS-2B) from the American Type Culture Collection were cultured on DMEM (GIBCO, Rockville, MD, USA). The human bronchial epithelial cell line BEAS-2B was cultured on LHC-9 medium (GIBCO). All lung cancer cell lines were maintained at 37°C in a 5% CO2 humidified atmosphere on medium containing 10% FBS and 100 ng/mL each of penicillin and streptomycin (GIBCO).
Drugs and chemicals
Docetaxel (DOC) was obtained from Aventis Pharmaceuticals Inc. (Bridgewater, NJ, USA). Adriamycin (ADR) was purchased from Sigma–Aldrich (St Louis, MO, USA). Gemcitabine (GEM) was provided by Eli Lilly (Indianapolis, IN, USA).
Chemosensitivity comparisons (MTS assay)
Cell viability was assessed on CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI, USA), according to the manufacturer’s instructions. In brief, 20 μL of MTS reagent was added to each well of a 96-well assay plate containing the samples in 100 μL of culture medium. Plates were incubated at 37°C in a humidified 5% CO2 atmosphere for 40 min. Absorbance at 490 nm was read by means of an ELISA plate reader. Each experiment was repeated at least three times. Final data were normalized and presented as percentage of controls.
Western blot analysis
The cells were washed with PBS and harvested for sonication in the presence of a protease inhibitor cocktail (Sigma–Aldrich). The concentration of protein was determined by Bradford assay, and the cell lysates (15 μg) were applied to SDS–PAGE gels. After transferring the proteins to a PVDF membrane (GE Healthcare Bioscience, Amersham Place, UK), they were reacted with anti-clusterin (Santa Cruz Biotechnology, CA, USA), anti-MMP2 (Cell Signaling, Boston, MA, USA), or anti-β-actin (Sigma–Aldrich), followed by anti-mouse or rabbit IgG conjugated with horseradish peroxidase (Calbiochem, San Diego, CA, USA). An ECL kit (GE Healthcare Bioscience) was used to determine the levels of protein expression.
Transfection of clusterin-shRNAi
V2LHS-150636-clusterin lentiviral shRNAmir (CLU-shRNA) and Non-silencing shRNAmir negative control (RHS4346, shRNA control) were purchased from Open Biosystems (Thermo Fisher Scientific, Rockford, IL, USA). TurboFectTM in vitro transfection reagent (Thermo Fisher Scientific) was used to transfect shRNA control and CLU-shRNA vectors into H1355 cells (4 × 105) on 6-cm dishes (3 μg of DNA/dish). This was followed by 48 h or 72 h incubation for Western blot analysis. For MTS assay, H1355 cells (7.5 × 104) seeded onto a 24-well plate were transfected (0.4 μg of DNA/well). After 60 h, the medium was collected for Western blot. Fresh medium with ADR, DOC, or GEM was added to the cells followed by incubation for 48 h (ADR, DOC) or 72 h (GEM) at the indicated drug concentrations.
Clusterin overexpression
Full-length open reading frame of human clusterin cDNA was generated by RT–PCR from a human Huh7 cell line using the forward primer 5-GTGACATATGATGAAGACTCTG and the reverse primer 5-AACGCGTCGACATCTCACTCCTCCCT. To express the gene, the obtained DNA fragment was subcloned into p3XFLAG-CMV-10 (Sigma–Aldrich). This was followed by DNA sequencing. The CLU expression vector was transfected with TransFastTM Transfection Reagent (Promega) into A549 and H1299 cells. This was followed by G418 selection (1.5 mg/mL, Sigma–Aldrich). We isolated the well-separated clone of cells on a 96-well plate and reamplified until the cells grew on 10-cm dishes for immunoblot analysis.
Cell migration assay with Boyden chamber
To each well of a 48-well bottom chamber, 32 μL of DMEM with 10% FBS was added. We used forceps to handle the 8-μm-porosity polycarbonate membrane, over which a silicone gasket was placed. Then, the top chamber was positioned over the gasket, and the Boyden chamber was assembled. A total of 52 μL of DMEM solution containing 2% FBS and cancer cells was loaded into each well of the top chamber, and the whole chamber was incubated at 37°C and 5% CO2 for 6 h for A549 cells or 12 h for H1299 cells. Thereafter, the chamber was disassembled, and the membrane, through which cells had passed and adhered, was immersed in cooled 95% methanol for 10 min to fix migrated cells. The membrane was then stained in 20% Giemsa stain overnight. The stained cells on the underside of the membrane were counted under a microscope.
Gelatin zymography
Production of MMPs by cancer cells was analyzed by gelatin zymography. To avoid MMP contamination, cells were cultured in serum-free medium for 24 h, and conditioned media were collected. Equal amounts of conditioned medium samples were mixed with SDS sample buffer containing 2% SDS without β-mercaptoethanol and applied to 10% SDS–polyacrylamide gels copolymerized with 0.1% gelatin (Sigma–Aldrich) without boiling. After electrophoresis, gels were washed for 1 h at room temperature with gentle agitation in a renaturing buffer (2.7% Triton X-100 in H2O) to remove SDS. The gels were then equilibrated in developing buffer (50 mM Tris–HCl, pH 7.4, 0.2 M NaCl, 5 mM CaCl2, and 0.2% Brji 35) at 37°C overnight. This was followed by staining with 0.5% Coomassie Brilliant Blue and destaining. The MMP activities were visualized as clear bands against the blue background of the stained gels.
Semi-quantitative RT–PCR of MMP2 and MMP9
To analyze the mRNA levels of MMP2 and MMP9, total RNA was extracted from H1299 cells that stably expressed clusterin using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The purified RNA (2 μg) was reverse-transcribed to cDNA by oligo-dT primers. For MMP2, a forward primer (5′-TTTTCTCGAATCCATGATGG) and a reverse primer (5′-CTGGTGCAGCTCTCATATTT) were used. For MMP9, a forward primer (5′-AAGATGCTGCTGTTCAGCGGG) and a reverse primer (5′-GTCCTCAGGGCACTGCAGGAT) were also used. For GAPDH, a forward primer (5′-GCCAAGGTCATCCATGACAAC) and a reverse primer (5′-CAGTAGAGGCAGGGATGATGTTC) served as the calibration control. The amplified PCR products were analyzed on 1% agarose gel.
Statistical analysis
All values are expressed as mean ± SD from triplicate experiments. Independent t test was performed for comparison of data from independent samples. A probability (P) value < 0.05 was considered significant.
Results
Expression levels of CLU in human NSCLC and lung cells
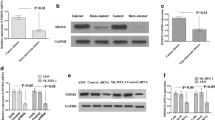
Levels of cytosolic and secreted CLU protein expressions in adenocarcinoma cell lines A549 and H1355, large cell carcinoma cell lines H1299 and H460, and squamous cell carcinoma cell line Calu-1 were determined on immunoblot analysis. The highest level of precursor form of the secreted CLU (pre-sCLU) was present in H1355 cells followed by A549, H460, and Calu-1 cells (Fig. 1a). sCLU was present in cell lysate and culture medium (Fig. 1b). The human bronchial epithelial cell line BEAS-2B and H1299 cells showed only insignificant levels of cytosolic CLU. According to the CLU levels, we chose H1355, A549, and H1299 cells to investigate the correlation of CLU expression with chemosensitivity in cancer cells.
Expression levels of clusterin were examined on Western blot analysis. H1299, H460, A549, H1355, and Calu-1 are human NSCLC cell lines. BEAS-2B cells were isolated from normal human bronchial epithelium obtained during autopsies of non-cancerous individuals. a Total cellular lysates (15 μg) from the cultured cells were analyzed for cytosolic clusterin expression. b The media collected from individual cultures (15 μL) were also analyzed
Chemosensitivities of NSCLC cells to ADR, GEM, and DOC are reversely correlated with CLU levels
The sensitivities of H1355, A549, and H1299 cells to ADR (Fig. 2a), GEM (Fig. 2b), and DOC (Fig. 2c) were evaluated on MTS assay. Although ADR is not used in lung cancer chemotherapy, it has been shown that development of resistance to ADR depends on CLU upregulation in human osteosarcoma cell lines [14]. Our data showed that the H1355 cells were the least sensitive to ADR, GEM, and DOC, whereas the A549 cells were the most sensitive to ADR, GEM, and DOC. The absence of p53 in H1299 cells may partly contribute to their insensitivity to drug treatment. H1299 cells with the lowest expression level of CLU have been shown to be less sensitive than A549 cells with preserved wild-type p53 [5]. Our results suggested that high level of CLU expression is associated with drug resistance in H1355 cells.
Inhibition of CLU expression results in sensitization of cells to drug treatment
To determine the effect of CLU on drug sensitivity, H1355 cells were transiently transfected with small hairpin interference RNA construct (CLU-shRNA) that inhibits CLU expression. The expression of CLU was significantly reduced after 72 h of transfection on immunoblot (Fig. 3a). When CLU expression was reduced, the chemosensitivities of H1355 to ADR (Fig. 3b), GEM (Fig. 3c), and DOC (Fig. 3d) increased on MTS assay. The levels of sCLU proteins in culture medium were determined on Western blot analysis. By calculating the resistance ratios (IC50 in vector-transfected cells)/(IC50 in siCLU-transfected cells) of H1355 cells, vector-transfected cells were found to be 1.47-fold, 3.85-fold, and 2.56-fold more resistant to ADR, GEM, and DOC, respectively. Therefore, CLU inhibition sensitizes H1355 cells to chemotherapy.
Inhibition of clusterin expression by shRNAi increases chemosensitivity. a To measure the effect of CLU inhibited by shRNAi, H1355 cells were transfected with CLU-shRNA and Non-silencing shRNAmir negative control (control). After 48 and 72 h, cells were harvested for Western blot analysis. b For chemosensitivity determinations, cells were treated with b 0.344–2.58 μM ADR c 0.5–2 μM GEM or d 10–80 nM DOC as described in “Materials and methods”. *P < 0.05
Secretory CLU collected from H1355 medium reduces drug sensitivity of H1299 cells
To examine whether chemosensitivity is regulated by sCLU, the medium of H1355 cells was harvested after 24 h of incubation. The conditioned medium used for cell treatment was a mixture of one part fresh to one part collected medium to avoid metabolic toxicity from the collected medium. As CLU level is lower in H1299 cells, chemosensitivities of H1299 cells were compared in the presence of conditioned medium and fresh medium. A control with 50% fresh medium and 50% conditioned medium from cultured H1299 cells was also included and exhibited similar effect to the fresh medium. The chemosensitivities of H1299 cells to ADR (Fig. 4a), GEM (Fig. 4b), and DOC (Fig. 4c) decreased with the addition of conditioned medium on MTS assay. Apparently, sCLU protein in the conditioned medium protects H1299 cells against these drug toxicities.
Secretory CLU collected from H1355 medium reduces drug sensitivity of H1299 cells. H1299 cells (2 × 103 cells) were seeded onto 96-well plate and conditioned medium was added together with a 86, 172, 344, 688, or 1,032 nM ADR for 36 h; b 20, 40, 80, 160, or 200 nM GEM for 48 h; or c 10, 20, 40, 80, or 100 nM DOC for 48 h. This was followed by MTS assay. *P < 0.05
Overexpression of CLU reduces ADR but not GEM and DOC chemosensitivities in A549 and H1299 cells
Since sCLU is produced from pre-sCLU with modification, we constructed a full-length gene of CLU that contains the leader signaling sequence for expression. Individual clones of A549 and H1299 cells that stably express pre-sCLU were selected by G418 and examined on Western blot (Fig. 5a). Two clones of A549 (A-1, A-2) and H1299 (H-1, H-2) cells that overexpress CLU were examined on chemosensitivity assay. The chemosensitivity to ADR was reduced in all A-1, A-2 (Fig. 5b) and H-1, H-2 (Fig. 5c) CLU-overexpressing cells. Interestingly, the chemosensitivities to GEM (Fig. 5d, e) and DOC did not significantly decrease (Fig. 5f, g).
Overexpression of CLU reduces ADR but not GEM and DOC chemosensitivities in A549 and H1299 cells. a Two CLU-overexpressing H1299 sublines (H1, H2) and two CLU-overexpressing A549 sublines (A-1, A-2) were selected by G418 and verified on Western blot analysis. Cells (2 × 103 cells) were seeded onto 96-well plate and treated with b, c 86–1,032 nM ADR for 36 h; d, e10–80 nM GEM for 48 h; or f, g 5–60 nM DOC for 48 h. This was followed by MTS assay. *P < 0.05
Migration is reduced in CLU-overexpressing H1299 and A549 cells
The alteration in cell migration by CLU was further investigated in vitro with modified Boyden chamber. When compared with parental H1299 cells, both clones of H1299 cells that overexpress CLU (H-1 and H-2) showed significantly reduced migratory ability (Fig. 6a). One clone of A549 cells that overexpresses CLU (A-2) showed reduced migratory ability (Fig. 6b), while the migratory ability of the other subline (A-1) was only slightly inhibited. Similar migratory inhibition was observed in H-1 and H-2 sublines on wound-healing assay (data not shown).
Migratory ability is reduced in CLU-overexpressing H1299 and A549 cells. Modified Boyden chamber was used for assay and the migratory a H1299, H-1 and H2 cells and b A549, A-1 and A-2 cells were counted. *P < 0.05. The transcription of MMP2 but not MMP9 was inhibited in H1299 cells that overexpress clusterin. c The activities of MMP2 and 9 from H1299, H-1 and H-2 were characterized by gelatin zymography. The conditioned medium (45 μL) was analyzed. d Semi-quantitative RT–PCR was conducted for MMP2, MMP9, and GAPDH. e Western blot analysis of MMP2 in H1299, H-1 and H-2 cells with whole-cell lysate (100 μg)
The transcription of MMP2 but not MMP9 is inhibited in H1299 cells that overexpress clusterin
To further explore the factors that may be inhibited by clusterin overexpression, we performed gelatin zymography to detect changes in MMP2 and MMP9 activities. Interestingly, MMP2 activity was lost in H-1 and H-2 CLU-overexpressing cells (Fig. 6c). The loss of MMP2 activity was confirmed on RT–PCR analysis of MMP2 and MMP9 mRNA transcripts. Only MMP2 transcription was inhibited in H-1 and H-2 CLU-overexpressing cells (Fig. 6d). To detect the proteins of MMP2, the lysates of H1299, H-1 and H-2 were collected and analyzed on Western blot. Reduced levels of MMP2 were observed (Fig. 6e). The H-1 subline exhibited the least migration potential and also showed the lowest MMP2 protein level.
Discussion
Clusterin (CLU), a protein with many identities, is not only present in cells, but also in circulating proteins. The extracellular form of CLU is a highly glycosylated α-β-heterodimer linked by five disulphide bonds [8]. Although clusterin expression levels have been extensively investigated and reviewed for their potential associations with chemo- and radiosensitivity [9, 17], results have been contradictory. For example, CLU-positive expression in human lung cancer patients is associated with better overall and disease-free survival than CLU-negative expression [1]. However, in a lung cancer cell line, CLU silenced by siRNA resulted in reduced migration and invasion [7]. Thus, it has not been possible to confirm a link between the effects of CLU inhibition and CLU overexpression. As the clinical data does not correlate well with cell line data, CLU’s potential as a favorable prognostic marker remains inconclusive. Therefore, CLU expression altered by inhibition and CLU overexpression in human NSCLC cell lines deserve further investigation.
We examined CLU levels in several human lung cancer cell lines and identified H1355 as a CLU-rich cell line. Inhibition of CLU expression by transient shRNA transfection reduced the chemosensitivity of H1355 cells to ADR, GEM, and DOC. Addition of CLU-rich conditioned medium to H1299 cells also resulted in reduced chemosensitivity. Our data are similar to the findings of July et al. [12] for targeted A549 CLU by siRNA, ASO sensitized paclitaxel, and GEM sensitivity in vitro and in vivo. To investigate whether a high level of CLU results in reduced chemosensitivity, we expressed exogenous CLU in A549 and H1299 cells. Interestingly, the chemosensitivity of CLU-overexpressing cells was not markedly altered, especially for GEM and DOC which are used in first-line lung cancer chemotherapy. The possible explanation is that nCLU in the CLU-overexpressing cells regulates pro-apoptotic activities simultaneously in contrast to sCLU. Although we did not test H460 cells, Cao et al. [4] reported that H460 cells treated with ASO against CLU (OGX-011) followed by radiotherapy induce tumor regression in xenograft model. Apparently, inhibition of CLU expression results in high chemosensitivity of NSCLC cell lines H1355, A549, and H460 cells, which is independent of the expression level of CLU.
Interestingly, when the extracellular CLU concentration was high, chemosensitivity was reduced in cancer cells with less endogenous CLU. This result suggests that as long as cancer cells exist in CLU-rich microenvironment, not all require expression of a high level of CLU to protect them from drug treatment. The sCLU from neighboring cells provides a shielding effect for cells expressing low-level CLU.
If a high level of CLU benefits cancer cells, why did the observed lung cancer cells express different levels of CLU? Is there any possible disadvantage to a high level of CLU for cancer cells? When the cells overexpressed CLU, they grew in bunches, clustering together on petri dishes (data not shown). Therefore, we examined the migratory ability of the CLU-overexpressing cells. According to our data, migratory ability was reduced and MMP2 transcription was inhibited in the cells overexpressing CLU. We have not yet identified whether nCLU or sCLU is responsible for migratory inhibition. It is postulated that CLU overexpression reduces the ability of cancer cells to spread out and results in better prognosis in lung cancer patients with positive CLU expression.
Human MMPs are a family of over 20 different endopeptidases that are able to degrade various components of the extracellular matrix (ECM). MMP2 is an enzyme with an important role in invasion to the basement membrane. The role of MMP2 in the survival of patients with NSCLC has been studied by Qian et al. [18], who supported the inclusion of MMP2 in further prospective trials on prognostic factors of NSCLC.
Our data pointed to a possible explanation for better overall and disease-free survival in human lung cancer patients with CLU-positive expression. In these patients, cancer cells may be less active in metastasis than in patients with CLU-negative expression. The application of ASO against CLU expression in lung cancer therapy requires further investigation. When CLU expression is inhibited by ASO, targeted cancer cells not killed by medicine or radiation treatment may turn into migrating active cells which can move to other tissues to rebuild a tumor.
References
Albert JM, Gonzalez A, Massion PP, Chen H, Olson SJ, Shyr Y, Diaz R, Lambright ES, Sandler A, Carbone DP, Putnam JB Jr, Johnson DH, Lu B (2007) Cytoplasmic clusterin expression is associated with longer survival in patients with resected non small cell lung cancer. Cancer Epidemiol Biomarkers Prev 16:1845–1851
Blaschuk O, Burdzy K, Fritz IB (1983) Purification and characterization of a cell-aggregating factor (clusterin), the major glycoprotein in ram rete testis fluid. J Biol Chem 258:7714–7720
Caccamo AE, Scaltriti M, Caporali A, D’Arca D, Corti A, Corvetta D, Sala A, Bettuzzi S (2005) Ca2 + depletion induces nuclear clusterin, a novel effector of apoptosis in immortalized human prostate cells. Cell Death Differ 12:101–104
Cao C, Shinohara ET, Li H, Niermann KJ, Kim KW, Sekhar KR, Gleave M, Freeman M, Lu B (2005) Clusterin as a therapeutic target for radiation sensitization in a lung cancer model. Int J Radiat Oncol Biol Phys 63:1228–1236
Chang JT, Chang GC, Ko JL, Liao HY, Liu HJ, Chen CC, Su JM, Lee H, Sheu GT (2006) Induction of tubulin by docetaxel is associated with p53 status in human non small cell lung cancer cell lines. Int J Cancer 118:317–325
Choi-Miura NH, Takahashi Y, Nakano Y, Tobe T, Tomita M (1992) Identification of the disulfide bonds in human plasma protein SP-40, 40 (apolipoprotein-J). J Biochem 112:557–561
Chou TY, Chen WC, Lee AC, Hung SM, Shih NY, Chen MY (2009) Clusterin silencing in human lung adenocarcinoma cells induces a mesenchymal-to-epithelial transition through modulating the ERK/Slug pathway. Cell Signal 21:704–711
de Silva HV, Harmony JA, Stuart WD, Gil CM, Robbins J (1990) Apolipoprotein J: structure and tissue distribution. Biochemistry 29:5380–5389
Djeu JY, Wei S (2009) Clusterin and chemoresistance. Adv Cancer Res 105:77–92
Hazzaa SM, Elashry OM, Afifi IK (2010) Clusterin as a diagnostic and prognostic marker for transitional cell carcinoma of the bladder. Pathol Oncol Res 16:101–109
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics, 2009. CA Cancer J Clin 59:225–249
July LV, Beraldi E, So A, Fazli L, Evans K, English JC, Gleave ME (2004) Nucleotide-based therapies targeting clusterin chemosensitize human lung adenocarcinoma cells both in vitro and in vivo. Mol Cancer Ther 3:223–232
Leskov KS, Klokov DY, Li J, Kinsella TJ, Boothman DA (2003) Synthesis and functional analyses of nuclear clusterin, a cell death protein. J Biol Chem 278:11590–11600
Lourda M, Trougakos IP, Gonos ES (2007) Development of resistance to chemotherapeutic drugs in human osteosarcoma cell lines largely depends on up-regulation of Clusterin/Apolipoprotein J. Int J Cancer 120:611–622
Miyake H, Hara I, Fujisawa M, Gleave ME (2006) The potential of clusterin inhibiting antisense oligodeoxynucleotide therapy for prostate cancer. Expert Opin Investig Drugs 15:507–517
Miyake H, Hara S, Arakawa S, Kamidono S, Hara I (2002) Over expression of clusterin is an independent prognostic factor for nonpapillary renal cell carcinoma. J Urol 167:703–706
Panico F, Rizzi F, Fabbri LM, Bettuzzi S, Luppi F (2009) Clusterin (CLU) and lung cancer. Adv Cancer Res 105:63–76
Qian Q, Wang Q, Zhan P, Peng L, Wei SZ, Shi Y, Song Y (2010) The role of matrix metalloproteinase 2 on the survival of patients with non-small cell lung cancer: a systematic review with meta-analysis. Cancer Invest 28:661–669
Reddy KB, Jin G, Karode MC, Harmony JA, Howe PH (1996) Transforming growth factor beta (TGF beta)-induced nuclear localization of apolipoprotein J/clusterin in epithelial cells. Biochemistry 35:6157–6163
Sakai I, Miyake H, Takenaka A, Fujisawa M (2009) Expression of potential molecular markers in renal cell carcinoma: impact on clinicopathological outcomes in patients undergoing radical nephrectomy. BJU Int 104:942–946
Scaltriti M, Bettuzzi S, Sharrard RM, Caporali A, Caccamo AE, Maitland NJ (2004) Clusterin overexpression in both malignant and nonmalignant prostate epithelial cells induces cell cycle arrest and apoptosis. Br J Cancer 91:1842–1850
Watari H, Kanuma T, Ohta Y, Hassan MK, Mitamura T, Hosaka M, Minegishi T, Sakuragi N (2010) Clusterin expression inversely correlates with chemosensitivity and predicts poor survival in patients with locally advanced cervical cancer treated with cisplatin-based neoadjuvant chemotherapy and radical hysterectomy. Pathol Oncol Res 16:345–352
Wong P, Taillefer D, Lakins J, Pineault J, Chader G, Tenniswood M (1994) Molecular characterization of human TRPM-2/clusterin, a gene associated with sperm maturation, apoptosis and neurodegeneration. Eur J Biochem 221:917–925
Zoubeidi A, Chi K, Gleave M (2010) Targeting the cytoprotective chaperone, clusterin, for treatment of advanced cancer. Clin Cancer Res 16:1088–1093
Acknowledgments
This work was supported by grants (NSC-96-2314-B-040-017-MY3 and 98-CCH-CSMU-02) to G-T Sheu. We would also like to thank the staff of the Instrument Center of Chung Shan Medical University for their technical support. This center is partly supported by the National Science Council, Ministry of Education and Chung Shan Medical University.
Conflict of interest
The authors declare that there are no potential conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cheng, CY., Cherng, SH., Wu, WJ. et al. Regulation of chemosensitivity and migration by clusterin in non-small cell lung cancer cells. Cancer Chemother Pharmacol 69, 145–154 (2012). https://doi.org/10.1007/s00280-011-1682-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00280-011-1682-0