Abstract
Purpose
To assess the role of pretreatment multiparametric (mp)MRI-based radiomic features in predicting pathologic complete response (pCR) of locally advanced rectal cancer (LARC) to neoadjuvant chemoradiation therapy (nCRT).
Methods
This was a retrospective dual-center study including 98 patients (M/F 77/21, mean age 60 years) with LARC who underwent pretreatment mpMRI followed by nCRT and total mesorectal excision or watch and wait. Fifty-eight patients from institution 1 constituted the training set and 40 from institution 2 the validation set. Manual segmentation using volumes of interest was performed on T1WI pre-/post-contrast, T2WI and diffusion-weighted imaging (DWI) sequences. Demographic information and serum carcinoembryonic antigen (CEA) levels were collected. Shape, 1st and 2nd order radiomic features were extracted and entered in models based on principal component analysis used to predict pCR. The best model was obtained using a k-fold cross-validation method on the training set, and AUC, sensitivity and specificity for prediction of pCR were calculated on the validation set.
Results
Stage distribution was T3 (n = 79) or T4 (n = 19). Overall, 16 (16.3%) patients achieved pCR. Demographics, MRI TNM stage, and CEA were not predictive of pCR (p range 0.59–0.96), while several radiomic models achieved high diagnostic performance for prediction of pCR (in the validation set), with AUCs ranging from 0.7 to 0.9, with the best model based on high b-value DWI demonstrating AUC of 0.9 [95% confidence intervals: 0.67, 1], sensitivity of 100% [100%, 100%], and specificity of 81% [66%, 96%].
Conclusion
Radiomic models obtained from pre-treatment MRI show good to excellent performance for the prediction of pCR in patients with LARC, superior to clinical parameters and CEA. A larger study is needed for confirmation of these results.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the 3rd most common cancer worldwide and the 2nd most deadly cancer [1]. The incidence of rectal cancer has recently increased in patients younger than 50 years [2]. According to the National Comprehensive Cancer Network (NCCN) guidelines, the current standard treatment for patients with locally advanced rectal cancer (LARC) is neoadjuvant chemoradiation therapy (nCRT) followed by surgery with total mesorectal excision [3]. However, up to 25% of these patients will show complete pathological response (pCR) after nCRT, which may render surgery unnecessary [4]. As pCR can only be assessed with certainty at histopathology, imaging could represent a valuable noninvasive tool to predict which patients are more likely to respond to neoadjuvant therapy, sparing them surgery and its complications.
MRI is the imaging modality of choice for local rectal cancer staging and follow-up after neoadjuvant therapy, typically by combining T2-weighted imaging (T2WI) and diffusion-weighted imaging (DWI) [5]. Contrast-enhanced T1WI may have additional value, primarily in identifying tumor recurrence [6]. Moreover, radiomics, a method of algorithm-based computed extraction of a large number of quantitative image features from diagnostic medical images, has the potential for improving diagnostic and prognostic accuracy [7]. Several studies have reported the use of radiomic models based on pre-treatment MRI, with reported AUCs ranging between 0.69 and 0.94 for the prediction of pCR in LARC [8,9,10,11,12,13,14]. Most of these studies were single-center [8, 10, 12,13,14] and analyzed mostly T2WI sequences [8,9,10]. Three of these studies had external validation cohorts [9,10,11]. For example, a recent dual-center study that analyzed T2WI radiomic features on pre-treatment MRI (n = 132) achieved an AUC of 0.80 in the validation cohort [9]. Only two studies have analyzed radiomic features on DWI [11, 12], while one study assessed additional features from pre-contrast T1WI with T2WI [14] and developed a nomogram including clinical features, achieving an AUC of 0.912. Finally, another study analyzed contrast-enhanced T1WI and DWI (n = 48) with no validation cohort [12]. To our knowledge, there have not been significant multi-center efforts on evaluating the performance of radiomics using multiparametric MRI (mpMRI), across training and external validation cohorts.
The objective of our study was to assess the role of pretreatment mpMRI-based radiomic features in predicting pCR of LARC to neoadjuvant chemoradiation therapy (nCRT).
Materials and methods
Study population
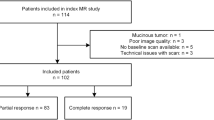
This was a retrospective dual-center study approved by the Institutional Review Board of both institutions (Institution 1, Ichan School of Medicine at Mount Sinai and Institution 2, Columbia University Medical Center) with a waiver for written informed consent. We examined the databases of both institutions for consecutive adult patients diagnosed with LARC (stage T3–4 or N+) who underwent pre-treatment MRI and who were subsequently treated by either neoadjuvant chemoradiation therapy followed by surgical excision or watch-and-wait between January 2013 and December 2019. Reasons for exclusion are listed in Fig. 1. Neoadjuvant therapy consisted of capecitabine (n = 69) or FOLFOX (combination of folinic acid, fluorouracil, and oxaliplatin, n = 11) combined with radiation therapy (dose ranging between 25 and 40 Gy divided into 5–28 fractions over a period of 35.6 days ± 10.3 days). The type of chemotherapy administered was missing in 18 patients. The final study population included 98 patients (77M/21F, average 58.3 years) including 58 from institution 1 and 40 from institution 2. The average time between the end of CRT and surgery was 80 days (range 5–315 days, Table 1).
MRI protocol
At institution 1, out of 58 rectal MRI examinations, 39/58 (67%) were performed in-house while the remaining 19/58 (33%) were done at outside facilities. A total of 26 MRIs were performed at 1.5 T and 32 at 3 T. At institution 2, all the MRIs were performed in-house (22 MRIs at 1.5 T and 18 MRIs at 3 T). The patients from institution 2 were included in a previous publication in which a radiomic model based on T2WI sequence in pre-treatment rectal MRI was used to predict pCR, neoadjuvant rectal score, and tumor regression score (blinded Ref). This previous publication did not assess T1WI pre- and post-contrast and DWI. MRI examinations were performed using different platforms including GE (n = 53), Siemens (n = 41), and Philips (n = 4). MRI sequence parameters are summarized in Supplementary Table 1.
Qualitative analysis
Tumor location and stage were retrieved by the study coordinator (observer 1, ME, a radiologist from institution 1, with 1 year experience in body MRI) from clinical reports of both institutions. Tumor location was identified according to the distance from the anal verge [15]. Tumor stage was assessed based on the American Joint Committee on Cancer Staging Manual of TNM staging (applied to MRI) and extracted from clinical MRI reports [16].
Quantitative analysis
Tumor segmentation
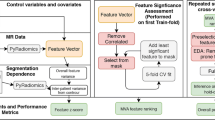
Tumors were manually segmented by observer 1, who prescribed volumes of interest (VOIs) in tumors on oblique axial T2WI, T1WI pre- and post-contrast (at 60 s), low (0–50) and high b-value (600–1500) DWI using software compliant with the Imaging Biomarker Standardization Initiative (IBSI) guidelines [17] (Olea Sphere 3.0.16, La Ciotat, France). ADC maps were not segmented. Tumor deposits outside the primary tumor and lymph nodes were not included in the segmentation. A second observer (observer 2, NV, from institution 1, with 1 year of experience in body MRI) segmented 24 patients following the same guidelines for the purpose of inter-observer reproducibility assessment. Both observers were blinded to outcome at time of image analysis. Tumor volume was measured on all segmented sequences.
Radiomic feature extraction
In order to homogenize image processing, spatial resampling was performed to create isotropic voxels (1.0 × 1.0 × 1.0 mm3) using the nearest neighbor as the interpolation algorithm. Signal intensity discretization was performed using a fixed bin number of 64, as recommended by the IBSI guidelines. Signal intensity normalization of the images was performed for all sequences, based on all pixel values in the image using z-score normalization:
where X and f(x) are the original and normalized intensity, respectively, and μx and σx are the mean and the standard deviation of the signal intensity values [18]. 108 Radiomic features for each image type (T2WI, low and high b-value, pre- and post-contrast T1WI) were extracted using the same software, for a total of 540 features per patient. These features were divided into the following categories: 16 shape features, 19 1st order features (histogram features), 23 original gray level co-occurrence (GLCM), 16 original gray level run length matrix (GLRLM), 15 original gray level size zone matrix (GLSZM), 14 original gray level dependence matrix (GLDM), and 5 original neighboring gray tone difference matrix (NGTDM).
Model building
Patients from institution 1 were used as a training set and those from institution 2 as validation set. A methodic feature selection with principal component analysis (PCA) was used to reduce the risk of overfitting and therefore a more generalizable model could be obtained. The top N radiomic features that best explained the variation between the target and non-target samples were used, with N = 20, 50, 100, and 108 (all). For generalizability, we conducted multiple experiments with different neural network architectures using a 20-k fold cross-validation method on the training set to identify the most suitable architectures for our predictive task. The cross-validation method was applied only on the training dataset; the hold-off validation set was only used for testing purposes. By evaluating the average performance across the 20 folds, this approach provided valuable insights into which network architectures were most suitable for our predictive task. Analyzing the average performance across the folds allowed us to identify the best-performing model architectures. These well-optimized architectures were subsequently tested on the validation dataset, ensuring a robust assessment of their predictive capabilities. We explored different combinations of layers and nodes, opting for a shallow network configuration (consisting in 1 input layer, 3 hidden layers and 1 output layer) due to the limited number of available samples [19, 20]. Once the optimal architecture was identified through the cross-validation approach, the selected models were trained using the entire training dataset and the performance was tested on the validation dataset. To overcome the limited number of samples, data augmentation was performed on the training set using Synthetic Minority Oversampling Technique (SMOTE) [21]. This is a popular technique in the field of machine learning to address class imbalance problems by generating synthetic samples to augment the minority class. To do so, the algorithm selects k-nearest neighbors of the minority class and creates new instances along the connecting multi-dimensional line between these samples. To apply this method, we used the Python imbalanced-learn module which provides a SMOTE function. In our application we kept the default parameters just keeping the same random state in order to have replicable results. This technique allowed us to balance our training dataset from 25% complete response and 75% non-complete response to a 50/50 distribution. The best results were obtained with a MLP (multilayer perceptron) model using the top 100 PCA (principal component analysis) features. This shallow MLP has 3 hidden layers of 8, 16, and 32 nodes, respectively, and an output layer predicting pCR vs. no pCR. The batch size corresponds to the number of samples for a single forward backward iteration for weights update. The output of the MLP is a value between 0 and 1 indicating a likelihood of a sample pertaining to the pCR group (1) or the non pCR group (0). This way, by thresholding the values at 0.5, the model made its predictions for every sample. Additional models were not evaluated because of the limited sample size. In order to obtain feature importance ranking, we used the permutation importance method [22]. The idea of this method is to compare the baseline score to the average score obtained from permutations of a specific feature to determine its importance in terms of predictive results.
Reference standard
The reference standard was the histopathologic clinical report of the surgical resection specimens (in operated patients). The modified tumor regression (TRG) score was used at both institutions as follows [16]: score 0: no viable cancer cells (pCR), score 1: single cells or rare small groups of cancer cells, score 2: residual cancer with evidence of tumor regression but more than a single cell and score 3: extensive residual cancer with no evidence of tumor regression. Patients with scores 0 are considered to have pCR. For 2 patients, a continued clinical CR with negative MRI examinations and endoscopy was considered a substitute for pCR for patients enrolled in watch-and-wait protocols [23].
Statistical analysis
Statistical analysis was performed using MATLAB R2018 (The MathWorks, Natick, MA) and Python. p-values < 0.05 were considered statistically significant. Inter-observer agreement was assessed by measuring the intraclass correlation coefficient (ICC); features with ICC > 0.7 were considered to have good inter-observer agreement. Feature redundancy, corresponding to features with high correlation (r > 0.95), was assessed using Spearman’s correlation coefficient between feature pairs. A radiomic score for the prediction of pCR using a combination of radiomic features was built using MLP modeling. The optimal sensitivity and specificity of the radiomic score was calculated by maximizing the Youden J statistic. Demographics and clinical characteristics (age, sex, baseline tumor stage, and serum CEA) were also analyzed using χ2-test for categorical variables and independent samples t-test for continuous variables.
Results
Patient characteristics
In total, 16/98 (16.3%) patients achieved pCR; 30/98 (30.6%) had a TRG score of 1, 38/98 (38.8%) a score of 2, and 14/98 (14.3%) a score of 3 (Table 1).
Prediction of pCR
Demographic data (age and sex), baseline TNM stage, and serum CEA were not significant predictors of pCR (p = 0.59–0.96).
Out of 540 extracted radiomic features (108 per sequence), 404 features had good to excellent inter-observer reliability (ICC > 0.7).
Table 2 summarizes the different models to predict pCR. For the prediction of pCR, the model based on high b-value DWI features achieved the best AUC of 0.9 [95% CI 0.67, 1] with a sensitivity of 100% [95% CI 100%, 100%] and specificity of 81% [95% CI 66%, 96%] in the validation set (Figs. 2, 3). Table 3 lists the top 10 features of the high b-value model in order of importance.
44-year-old man with locally advanced rectal cancer, stage T3. Axial oblique T2WI (A) and post-contrast T1WI (B) show near circumferential mid rectal tumor with enhancement (arrows). Histopathology on rectal specimen obtained after neoadjuvant chemoradiation therapy (C) shows no residual tumor with fibrotic and sclerotic vessels (arrow), compatible with pathologic complete response (TRG score 0). Radiomic maps of high b-value Gray Level Co-occurrence Matrix inverse difference normalized (D) and high b-value Gray Level Run Length Matrix (GLRLM) long run emphasis (E) show high value points. Radiomic map of high b-value Gray Level Run Length Matrix (GLRLM) LRHGL emphasis (F) shows sparse high value points
37-year-old man with locally advanced rectal cancer, stage T3. A Axial T2W1 and B post-contrast T1W1 show circumferentially enhancing low rectal tumor. C Histopathology on rectal specimen obtained after neoadjuvant chemoradiation therapy shows residual tumor cells in a background of fibrosis (arrow), TRG score 3. Radiomic maps of high b-value Gray Level Co-occurrence Matrix inverse difference normalized (D) and high b-value Gray Level Run Length Matrix (GLRLM) long run emphasis (E) show mostly intermediate values. Radiomic map of high b-value Gray Level Run Length Matrix (GLRLM) LRHGL emphasis (F) shows mostly low values with sparse high value points
Model performance across different MRI sequences and different numbers of input features are shown in Supplementary Table 2. For comparison purposes with the high b-value DWI model, two models built based on pre- and post-contrast T1W1 shape features, respectively, achieved AUCs of 0.83 [95% CI 0.53, 1] and 0.81 [95% CI 0.5, 1], specificities of 66% [95% CI 48%, 84%] and 63% [44%, 81%] and the same sensitivity of 100% [95% CI 100%, 100%]. The list of top shape features that were selected for inclusion in both these models is given in Table 4. Of note, there was no difference in performance between T1WI shape feature models and the high b-value DWI model as there was overlap in the 95% confidence intervals of their AUCs.
In addition, there was no difference in tumor volume on both low and high b-value DWI between patients with pCR vs. non pCR in both the training set (25.702 ± 22.853 mm3 vs. 37.713 ± 45.748 mm3, respectively, p = 0.35) and the validation set (19.578 ± 9.884 mm3 vs. 27.652 ± 20.325 mm3, respectively, p = 0.78).
Discussion
In our study, we developed and validated a radiomic model using mpMRI (including T2WI, pre- and post-contrast T1WI and DWI), obtaining excellent performance using high b-value DWI features, with an AUC of 0.9 (in the validation set), 100% sensitivity and 81% specificity for the prediction of pCR in LARC. Additionally, we developed two models using only shape features from T1WI pre- and post-contrast also to predict pCR, with observed good performance (AUCs of 0.81–0.83 in the validation set).
Our results show higher performance of radiomics compared with previous studies [8,9,10,11,12, 24]. For example, Petkovska et al. reported an AUC of 0.75 in 102 patients with LARC by combining radiomic features extracted from T2WI and staging criteria; however, their study lacked a validation cohort [8]. Antunes et al. reported an AUC of 0.712 in an external validation set by analyzing T2WI sequences in 104 patients [10]. Another study from Van Griethuysen et al. evaluating 133 patients from 2 centers showed AUCs between 0.73 and 0.77 in the validation set from a model obtained with DWI/ADC and T2WI [11]. Most of the published radiomics studies for the prediction of pCR using pre-treatment MRI were solely based on T2WI features [8,9,10, 25]. A few studies such as the one by Van Griethuysen et al. [11] built radiomic models combining T2WI and DWI with a few features selected from T2WI. Our study is similar to the single-center study of Nie et al. [12] in which features were extracted from mpMRI including post-contrast T1WI (n = 48), albeit Nie et al.’s models showed lower performance with AUCs of 0.54–0.73 for predicting pCR (without a validation cohort). Meanwhile, Cui et al. developed a radiomic model on 131 training datasets combining features from T2WI, post-contrast T1WI and ADC from a single center, with a reported excellent AUC of 0.944 for predicting pCR in their validation set comprising 55 patients, similar to our results [13]. The high performance of their model can be potentially explained by the homogeneity of their MRI exams, in which all MRIs were performed at 3 T using the same MRI parameters, as opposed to our study, where we used multiple field strengths, vendors, and protocols.
A portion of our cohort was included in a previously published radiomics study in patients with LARC using axial T2WI in 132 patients from 2 different centers, with a calculated AUC of 0.80 for prediction of pCR using repeated cross-validation [9]. This previous study built models for the prediction of pCR using 1,595 T2WI radiomic features in both tumor and mesorectal compartment, as well as clinical variables.
An important consideration when performing radiomics quantification is the inter-reader reproducibility of the measurement which we obtained in our study, as we only selected radiomic features with ICC > 0.7, as described before [11].
The identification of adverse imaging phenotypes based on radiomics at baseline (in addition to T staging) may trigger more aggressive neoadjuvant therapies (including immunotherapy) and closer follow-up of these patients [26]. Several studies showed that pCR after nCRT is associated with low recurrence and longer disease-free survival; thus, radiomic models can also potentially predict prognosis [27,28,29].
Radiomics is increasingly described for cancer diagnosis/characterization and for the prediction of cancer response to therapy. Radiomics quantification provides information on tumor heterogeneity and is associated with underlying gene expression and prognosis [30]. However, its technical complexity, inter-platform and inter-vendor variability and often limited sample size without validation cohorts limit its clinical impact [7]. Manual segmentation of tumors has been so far the mainstay in radiomics, yet it is time consuming, despite being reproducible (as confirmed by our study). There is an urgent need for automated tools using deep learning to help expedite these tasks [31], especially since it has been suggested that radiomic features are more robust when using automatic delineation of the tumor [32].
There are several limitations to our study. First, the retrospective nature of our study may have resulted in a selection bias of the patients. Second, the number of patients with pCR was relatively small even though we present one of the first approaches of multi-institutional validation of multi-parametric radiomics in LARC. Third, we did not attempt to correct for inter-platform and acquisition parameter variability.
Future directions include head-to-head comparison of radiomic biomarkers against blood [33] and tissue biomarkers [34] for predicting rectal cancer response.
In conclusion, radiomic models based on pre-treatment mpMRI are promising predictors of pCR in patients with LARC, which may help with personalized therapy and prognostication. Larger studies are needed to validate our results.
Data availability
Codes used to obtain radiomic models are available upon request.
Abbreviations
- DWI:
-
Diffusion-weighted imaging
- LARC:
-
Locally advanced rectal cancer
- MLP:
-
Multilayer perceptron
- mpMRI:
-
Multiparametric magnetic resonance imaging
- nCRT:
-
Neoadjuvant chemoradiation therapy
- PCA:
-
Principal component analysis
- pCR:
-
Pathologic complete response
- T1WI:
-
T1-weighted imaging
- T2WI:
-
T2-weighted imaging
- TRG:
-
Tumor regression grade
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68 (6):394–424. https://doi.org/10.3322/caac.21492
Kasi PM, Shahjehan F, Cochuyt JJ, Li Z, Colibaseanu DT, Merchea A (2019) Rising Proportion of Young Individuals With Rectal and Colon Cancer. Clin Colorectal Cancer 18 (1):e87–e95. https://doi.org/10.1016/j.clcc.2018.10.002
Benson AB, III, Venook AP, Al-Hawary MM, Cederquist L, Chen YJ, Ciombor KK, Cohen S, Cooper HS, Deming D, Engstrom PF, Grem JL, Grothey A, Hochster HS, Hoffe S, Hunt S, Kamel A, Kirilcuk N, Krishnamurthi S, Messersmith WA, Meyerhardt J, Mulcahy MF, Murphy JD, Nurkin S, Saltz L, Sharma S, Shibata D, Skibber JM, Sofocleous CT, Stoffel EM, Stotsky-Himelfarb E, Willett CG, Wuthrick E, Gregory KM, Gurski L, Freedman-Cass DA (2018) Rectal Cancer, Version 2.2018, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 16 (7):874–901. https://doi.org/10.6004/jnccn.2018.0061
Maas M, Nelemans PJ, Valentini V, Das P, Rödel C, Kuo L-J, Calvo FA, García-Aguilar J, Glynne-Jones R, Haustermans K, Mohiuddin M, Pucciarelli S, Small W, Jr., Suárez J, Theodoropoulos G, Biondo S, Beets-Tan RGH, Beets GL (2010) Long-term outcome in patients with a pathological complete response after chemoradiation for rectal cancer: a pooled analysis of individual patient data. The Lancet Oncology 11 (9):835–844. https://doi.org/10.1016/S1470-2045(10)70172-8
Maas M, Lambregts DMJ, Nelemans PJ, Heijnen LA, Martens MH, Leijtens JWA, Sosef M, Hulsewé KWE, Hoff C, Breukink SO, Stassen L, Beets-Tan RGH, Beets GL (2015) Assessment of Clinical Complete Response After Chemoradiation for Rectal Cancer with Digital Rectal Examination, Endoscopy, and MRI: Selection for Organ-Saving Treatment. Annals of Surgical Oncology 22 (12):3873–3880. https://doi.org/10.1245/s10434-015-4687-9
Vliegen RFA, Beets GL, Meyenfeldt MFv, Kessels AGH, Lemaire EEMT, Engelshoven JMAv, Beets-Tan RGH (2005) Rectal Cancer: MR Imaging in Local Staging—Is Gadolinium-based Contrast Material Helpful? 234 (1):179–188. https://doi.org/10.1148/radiol.2341031403
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278 (2):563–577. https://doi.org/10.1148/radiol.2015151169
Petkovska I, Tixier F, Ortiz EJ, Golia Pernicka JS, Paroder V, Bates DD, Horvat N, Fuqua J, Schilsky J, Gollub MJ, Garcia-Aguilar J, Veeraraghavan H (2020) Clinical utility of radiomics at baseline rectal MRI to predict complete response of rectal cancer after chemoradiation therapy. Abdominal Radiology. https://doi.org/10.1007/s00261-020-02502-w
Shaish H, Aukerman A, Vanguri R, Spinelli A, Armenta P, Jambawalikar S, Makkar J, Bentley-Hibbert S, Del Portillo A, Kiran R, Monti L, Bonifacio C, Kirienko M, Gardner KL, Schwartz L, Keller D (2020) Radiomics of MRI for pretreatment prediction of pathologic complete response, tumor regression grade, and neoadjuvant rectal score in patients with locally advanced rectal cancer undergoing neoadjuvant chemoradiation: an international multicenter study. European Radiology 30 (11):6263–6273. https://doi.org/10.1007/s00330-020-06968-6
Antunes JT, Ofshteyn A, Bera K, Wang EY, Brady JT, Willis JE, Friedman KA, Marderstein EL, Kalady MF, Stein SL, Purysko AS, Paspulati R, Gollamudi J, Madabhushi A, Viswanath SE (2020) Radiomic Features of Primary Rectal Cancers on Baseline T2-Weighted MRI Are Associated With Pathologic Complete Response to Neoadjuvant Chemoradiation: A Multisite Study. Journal of Magnetic Resonance Imaging n/a (n/a). https://doi.org/10.1002/jmri.27140
van Griethuysen JJM, Lambregts DMJ, Trebeschi S, Lahaye MJ, Bakers FCH, Vliegen RFA, Beets GL, Aerts HJWL, Beets-Tan RGH (2020) Radiomics performs comparable to morphologic assessment by expert radiologists for prediction of response to neoadjuvant chemoradiotherapy on baseline staging MRI in rectal cancer. Abdominal Radiology 45 (3):632–643. https://doi.org/10.1007/s00261-019-02321-8
Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, Niu T, Sun X (2016) Rectal Cancer: Assessment of Neoadjuvant Chemoradiation Outcome based on Radiomics of Multiparametric MRI. J Clinical Cancer Research 22 (21):5256–5264. https://doi.org/10.1158/1078-0432.CCR-15-2997%
Cui Y, Yang X, Shi Z, Yang Z, Du X, Zhao Z, Cheng X (2019) Radiomics analysis of multiparametric MRI for prediction of pathological complete response to neoadjuvant chemoradiotherapy in locally advanced rectal cancer. Eur Radiol 29 (3):1211–1220. https://doi.org/10.1007/s00330-018-5683-9
Cheng Y, Luo Y, Hu Y, Zhang Z, Wang X, Yu Q, Liu G, Cui E, Yu T, Jiang X (2021) Multiparametric MRI-based Radiomics approaches on predicting response to neoadjuvant chemoradiotherapy (nCRT) in patients with rectal cancer. Abdom Radiol (NY) 46 (11):5072–5085. https://doi.org/10.1007/s00261-021-03219-0
Nougaret S, Reinhold C, Mikhael HW, Rouanet P, Bibeau F, Brown G (2013) The use of MR imaging in treatment planning for patients with rectal carcinoma: have you checked the “DISTANCE”? Radiology 268 (2):330–344. https://doi.org/10.1148/radiol.13121361
Edge SB BD, Compton CC, Fritz AG, Greene FL, Trotti A. (2010) AJCC cancer staging manual. 7th edn. Springer-Verlag, New York
Zwanenburg A, Vallieres M, Abdalah MA, Aerts H, Andrearczyk V, Apte A, Ashrafinia S, Bakas S, Beukinga RJ, Boellaard R, Bogowicz M, Boldrini L, Buvat I, Cook GJR, Davatzikos C, Depeursinge A, Desseroit MC, Dinapoli N, Dinh CV, Echegaray S, El Naqa I, Fedorov AY, Gatta R, Gillies RJ, Goh V, Gotz M, Guckenberger M, Ha SM, Hatt M, Isensee F, Lambin P, Leger S, Leijenaar RTH, Lenkowicz J, Lippert F, Losnegard A, Maier-Hein KH, Morin O, Muller H, Napel S, Nioche C, Orlhac F, Pati S, Pfaehler EAG, Rahmim A, Rao AUK, Scherer J, Siddique MM, Sijtsema NM, Socarras Fernandez J, Spezi E, Steenbakkers R, Tanadini-Lang S, Thorwarth D, Troost EGC, Upadhaya T, Valentini V, van Dijk LV, van Griethuysen J, van Velden FHP, Whybra P, Richter C, Lock S (2020) The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 295 (2):328–338. https://doi.org/10.1148/radiol.2020191145
Bianchini L, Botta F, Origgi D, Rizzo S, Mariani M, Summers P, García-Polo P, Cremonesi M, Lascialfari A (2020) PETER PHAN: An MRI phantom for the optimisation of radiomic studies of the female pelvis. Phys Med 71:71–81. https://doi.org/10.1016/j.ejmp.2020.02.003
Agliari E, Alemanno F, Barra A, De Marzo G (2022) The emergence of a concept in shallow neural networks. Neural Netw 148:232–253. https://doi.org/10.1016/j.neunet.2022.01.017
Almeida LBJHoNCC (1997) C1. 2 Multilayer perceptrons. 1
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research 16:321–357
Breiman L (2001) Random Forests. Machine Learning 45 (1):5–32
Lambregts DM, Maas M, Bakers FC, Cappendijk VC, Lammering G, Beets GL, Beets-Tan RG (2011) Long-term follow-up features on rectal MRI during a wait-and-see approach after a clinical complete response in patients with rectal cancer treated with chemoradiotherapy. Dis Colon Rectum 54 (12):1521–1528. https://doi.org/10.1097/DCR.0b013e318232da89
Shin J, Seo N, Baek SE, Son NH, Lim JS, Kim NK, Koom WS, Kim S (2022) MRI Radiomics Model Predicts Pathologic Complete Response of Rectal Cancer Following Chemoradiotherapy. Radiology 303 (2):351–358. https://doi.org/10.1148/radiol.211986
Cusumano D, Dinapoli N, Boldrini L, Chiloiro G, Gatta R, Masciocchi C, Lenkowicz J, Casà C, Damiani A, Azario L, Van Soest J, Dekker A, Lambin P, De Spirito M, Valentini V (2018) Fractal-based radiomic approach to predict complete pathological response after chemo-radiotherapy in rectal cancer. Radiol Med 123 (4):286–295. https://doi.org/10.1007/s11547-017-0838-3
Kelly RJ, Bever K, Chao J, Ciombor KK, Eng C, Fakih M, Goyal L, Hubbard J, Iyer R, Kemberling HT, Krishnamurthi S, Ku G, Mordecai MM, Morris VK, II, Paulson AS, Peterson V, Shah MA, Le DT (2023) Society for Immunotherapy of Cancer (SITC) clinical practice guideline on immunotherapy for the treatment of gastrointestinal cancer. J Immunother Cancer https://doi.org/10.1136/jitc-2022-006658
Theodoropoulos G, Wise WE, Padmanabhan A, Kerner BA, Taylor CW, Aguilar PS, Khanduja KS (2002) T-level downstaging and complete pathologic response after preoperative chemoradiation for advanced rectal cancer result in decreased recurrence and improved disease-free survival. Dis Colon Rectum 45 (7):895–903. https://doi.org/10.1007/s10350-004-6325-7
Jalilian M, Davis S, Mohebbi M, Sugamaran B, Porter IW, Bell S, Warrier SK, Wale R (2016) Pathologic response to neoadjuvant treatment in locally advanced rectal cancer and impact on outcome. J Gastrointest Oncol 7 (4):603–608. https://doi.org/10.21037/jgo.2016.05.03
Park J-S, Baek J-H, Lee W-S, Yang J-Y, Lee W-K, Kim K-K, Park Y-H (2018) Long-term oncologic outcomes in pathologic tumor response after neoadjuvant chemoradiation for locally advanced rectal cancer. Korean J Clin Oncol 14 (1):37–42. https://doi.org/10.14216/kjco.18006
Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications 5 (1):4006. https://doi.org/10.1038/ncomms5006
Bobo MF, Bao S, Huo Y, Yao Y, Virostko J, Plassard AJ, Lyu I, Assad A, Abramson RG, Hilmes MA, Landman BA (2018) Fully Convolutional Neural Networks Improve Abdominal Organ Segmentation. Proceedings of SPIE—the International Society for Optical Engineering 10574:105742V. https://doi.org/10.1117/12.2293751
Traverso A, Kazmierski M, Shi Z, Kalendralis P, Welch M, Nissen HD, Jaffray D, Dekker A, Wee L (2019) Stability of radiomic features of apparent diffusion coefficient (ADC) maps for locally advanced rectal cancer in response to image pre-processing. Phys Med 61:44–51. https://doi.org/10.1016/j.ejmp.2019.04.009
Morais M, Pinto DM, Machado JC, Carneiro S (2022) ctDNA on liquid biopsy for predicting response and prognosis in locally advanced rectal cancer: A systematic review. Eur J Surg Oncol 48 (1):218–227. https://doi.org/10.1016/j.ejso.2021.08.034
Bedin C, Crotti S, D'Angelo E, D'Aronco S, Pucciarelli S, Agostini M (2020) Circulating Biomarkers for Response Prediction of Rectal Cancer to Neoadjuvant Chemoradiotherapy. Curr Med Chem 27 (25):4274–4294. https://doi.org/10.2174/0929867326666190507084839
Acknowledgments
The authors thank Joanne Chin, MFA, ELS, for editing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
El Homsi, M., Bane, O., Fauveau, V. et al. Prediction of locally advanced rectal cancer response to neoadjuvant chemoradiation therapy using volumetric multiparametric MRI-based radiomics. Abdom Radiol 49, 791–800 (2024). https://doi.org/10.1007/s00261-023-04128-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00261-023-04128-0