Abstract
A novel deep learning (DL)-based attenuation correction (AC) framework was applied to clinical whole-body oncology studies using 18F-FDG, 68 Ga-DOTATATE, and 18F-Fluciclovine. The framework used activity (λ-MLAA) and attenuation (µ-MLAA) maps estimated by the maximum likelihood reconstruction of activity and attenuation (MLAA) algorithm as inputs to a modified U-net neural network with a novel imaging physics-based loss function to learn a CT-derived attenuation map (µ-CT).
Methods
Clinical whole-body PET/CT datasets of 18F-FDG (N = 113), 68 Ga-DOTATATE (N = 76), and 18F-Fluciclovine (N = 90) were used to train and test tracer-specific neural networks. For each tracer, forty subjects were used to train the neural network to predict attenuation maps (µ-DL). µ-DL and µ-MLAA were compared to the gold-standard µ-CT. PET images reconstructed using the OSEM algorithm with µ-DL (OSEMDL) and µ-MLAA (OSEMMLAA) were compared to the CT-based reconstruction (OSEMCT). Tumor regions of interest were segmented by two radiologists and tumor SUV and volume measures were reported, as well as evaluation using conventional image analysis metrics.
Results
µ-DL yielded high resolution and fine detail recovery of the attenuation map, which was superior in quality as compared to µ-MLAA in all metrics for all tracers. Using OSEMCT as the gold-standard, OSEMDL provided more accurate tumor quantification than OSEMMLAA for all three tracers, e.g., error in SUVmax for OSEMMLAA vs. OSEMDL: − 3.6 ± 4.4% vs. − 1.7 ± 4.5% for 18F-FDG (N = 152), − 4.3 ± 5.1% vs. 0.4 ± 2.8% for 68 Ga-DOTATATE (N = 70), and − 7.3 ± 2.9% vs. − 2.8 ± 2.3% for 18F-Fluciclovine (N = 44). OSEMDL also yielded more accurate tumor volume measures than OSEMMLAA, i.e., − 8.4 ± 14.5% (OSEMMLAA) vs. − 3.0 ± 15.0% for 18F-FDG, − 14.1 ± 19.7% vs. 1.8 ± 11.6% for 68 Ga-DOTATATE, and − 15.9 ± 9.1% vs. − 6.4 ± 6.4% for 18F-Fluciclovine.
Conclusions
The proposed framework provides accurate and robust attenuation correction for whole-body 18F-FDG, 68 Ga-DOTATATE and 18F-Fluciclovine in tumor SUV measures as well as tumor volume estimation. The proposed method provides clinically equivalent quality as compared to CT in attenuation correction for the three tracers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) is widely used for diagnosis, staging, and monitoring treatment effect in clinical oncology. Quantitative or semi-quantitative measures in PET, e.g., standard uptake value (SUV), are sensitive to different physics effect corrections, e.g., attenuation correction (AC). In PET/CT, CT scans provide high spatial resolution attenuation maps, but these can lead to artifacts in the PET images due to the CT itself, e.g., beam-hardening, metal artifacts, and count-starving [1], or from PET-CT mis-alignment. In addition, concerns of CT radiation exposure can be raised when multiple PET/CT scans are needed for the same patient during treatment evaluation, which is even more of a concern for pediatric patients. In PET/MR, attenuation map generation is more complex [2] since an MR image does not directly provide PET attenuation information, and the optimal AC solution for PET/MR has not been found [3]. Therefore, the development of an AC algorithm that does not depend on CT (or MR) is of strong clinical interest, since it may not only elimite the aforementioned AC artifacts but also substantially reduce the radiation dose.
Maximum likelihood reconstruction of activity and attenuation (MLAA) [4] was proposed to simultaneously reconstruct tracer activity (λ-MLAA) and attenuation maps (μ-MLAA) based on the time-of-flight (TOF) PET raw data only without CT or MR. However, μ-MLAA suffers from high noise and λ-MLAA suffers from quantitative error [5] as compared to the conventional CT-based (μ-CT) OSEM reconstruction. Recently, deep-learning (DL) frameworks were proposed to improve MLAA by predicting the CT attenuation map (μ-DL) from λ-MLAA and μ-MLAA [6, 7]. Specifically, Hwang et al. [6] used a convolutional neural network to predict μ-DL while Shi et al. [7] added an additional line-integral constraint into the loss function and further improved the μ-DL accuracy. However, the μ-DL in [6, 7] was only applied to datasets using 18F-FDG.
While 18F-FDG accounts for most of the PET scans in clinical oncology, many other oncological tracers have shown promising clinical efficacy in more specific cancer types, e.g., prostate-specific membrane antigen (PSMA) [8] or 18F-Fluciclovine [9] for prostate cancer and 68 Ga-DOTATATE [10] for neuroendocrine tumor imaging. Since the μ-DL prediction framework [6, 7] was data-driven, i.e., relying on the PET raw data itself, the neural network trained for 18F-FDG cannot be used for other tracers. In this work, we applied the μ-DL prediction framework, which was previously developed in [7], to clinical 18F-FDG, 18F-Fluciclovine, and 68 Ga-DOTATATE datasets. Evaluation of tumor uptake was performed by two radiologists. SUVmax, SUVmean and tumor volume are reported in addition to evaluation using conventional image analysis metrics.
Methods
Clinical data
In this study, 890 whole-body PET/CT datasets were used, which were previously acquired on a Siemens Biograph mCT 40 scanner at Yale New Haven Hospital, including 610 18F-FDG, 120 68 Ga-DOTATATE, and 160 18F-Fluciclovine studies. Scans with minimal body motion between PET and CT, based on visual examination, were selected for neural network training (N = 40 per tracer, M/F: 14/26, age: 59.7 ± 12.0 years old for 18F-FDG; M/F: 25/15, age: 62.1 ± 14.0 for 68 Ga-DOTATATE; M/F: 40/0, age: 70.1 ± 8.7 for 18F-Fluciclovine) and testing (N = 73, M/F: 25/48, age: 61.2 ± 17.9 for 18F-FDG; N = 36, M/F: 11/25, age: 62.3 ± 14.4 for 68 Ga-DOTATATE; N = 50, M/F: 50/0, age: 74.5 ± 7.6 for 18F-Fluciclovine). CT was acquired prior to the PET scan with regular radiation dose for diagnostic purpose. Both 18F-FDG and 68 Ga-DOTATATE scans started at ~ 60 min post-injection using a supine protocol. Approximately 10-mCi injection was used for 18F-FDG and ~ 0.054 mCi/kg (5.4 mCi max) was used for 68 Ga-DOTATATE. 18F-Fluciclovine scans started at 3–5 min post-injection of ~ 10 mCi using a pelvis-first supine protocol, which was used to minimize bladder fill-up. Continuous-bed-motion protocol was used for all the PET acquisitions. Two and three minutes per bed position-equivalent bed speed was used for 18F-FDG and 68 Ga-DOTATATE, respectively. For 18F-Fluciclovine, 4–5 min (2–3 min) per bed position-equivalent time was used over the pelvis (abdomen, chest, and head/neck) and the total imaging time was ~ 20 min.
MLAA and data preprocessing
Three iterations and 21 subsets were used for MLAA [11]. Both λ-MLAA and μ-MLAA images were reconstructed using 2 mm3 voxel size followed by 5-mm full-width-half-max Gaussian smoothing and were down-sampled to 4 mm3.
In deep learning applications, image pre-processing is key for effective network training [12]. Here, two-channel inputs, i.e., λ-MLAA and μ-MLAA, represent two different physical quantities with different numerical value ranges, i.e., λ-MLAA represents the radiotracer density measured in Bq/mL while μ-MLAA represents attenuation coefficient measured in cm−1. For λ-MLAA, converting Bq/mL to standardized uptake value (SUV) helps to normalize the tracer injection dose and the patient weight. This normalization, however, does not help with the broad range of biological tracer uptake in different organs. Here, λ-MLAA images were normalized using λnorm = tanh(λ/σ) before training and testing, where λ and λnorm are the λ-MLAA images (in SUV) before and after normalization. σ controls the active gradient range of the hyperbolic tangent (tanh) function. σ was set to 10 to ensure most organs of interests (except for bladder) which are in the active gradient range for tracers. Every voxel in μ-MLAA and μ-CT (training label) was divided by 0.15 cm−1, which corresponds to skull bone attenuation coefficient at 511 keV, to match the value range of λnorm. The normalized λ-MLAA and μ-MLAA were concatenated as a dual-channel input to the deep neural network for training and testing. All training input and label μ-maps were 4 mm3 isotropic voxel size.
Network structure, loss function, and training
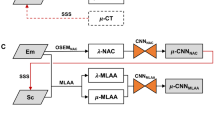
A modified fully convolutional 3D U-net architecture [13] was used for predicting the attenuation map (μ-DL) from λ-MLAA and μ-MLAA. Figure 1 shows the framework of the training process and Supplemental Fig. 1 shows the detailed U-net network structure. The network operates on 3D patches and uses 3 × 3 × 3 convolution kernels. Patch-based training, i.e., 64 × 64 × 32 voxels per patch, was used. Resolution reduction was performed by 2 × 2 × 2 convolution kernels with stride 2. Other network details can be found in Supplemental Fig. 1.
In this study, we used a novel loss function [14] that includes not only the conventional image intensity-based loss (IM-loss), but also a gradient-based loss (GDL-loss) as well as a line-integral projection (LIP-loss) to corporate with the PET attenuation physics. Formally, the loss function is defined as follows:
where \({\beta }_{1}\) and \({\beta }_{2}\) are the hyper-parameters for the GDL-loss (\({L}_{\text{GDL}}\)) and LIP-loss (\({L}_{\text{LIP}}\)) and were empirically set to 1.0 and 0.02, respectively. \({{\boldsymbol{\upmu}}}^{\text{DL}}\) and \({{\boldsymbol{\upmu}}}^{\text{CT}}\) represent patches of the μ-DL and the gold-standard μ-CT, respectively. The L1-norm IM-loss (\({L}_{\text{IM}}\)) is constructed as follows:
where \({\mu }_{j}^{\text{DL}}\) and \({\mu }_{j}^{\text{CT}}\) are the intensities of voxel j in \({{\boldsymbol{\upmu}}}^{\text{DL}}\) and \({{\boldsymbol{\upmu}}}^{\text{CT}}\), respectively. Nj is the number of voxels inside one patch.\({L}_{\text{GDL}}\) is an image gradient difference loss defined as follows:
where \({\nabla }_{x}\) is the gradient operator in the x direction (same for y and z). \({L}_{\text{GDL}}\) is shown to be effective in preventing μ-map blurring [15].
To enforce the additional similarity in the projection domain between μ-DL and μ-CT, the LIP-loss was used to measure the line-integral difference between the μ-map patch \({{\boldsymbol{\upmu}}}^{\text{DL}}\) and \({{\boldsymbol{\upmu}}}^{\text{CT}}\):
where \({\mathbb{k}}\) is the set of line-integral projection (LIP) angles, k indexes the projection angles, \({N}_{\mathbb{k}}\) represents the total number of angles in \({\mathbb{k}}\), \({P}_{k}\) is the LIP operator on the patch \({{\boldsymbol{\upmu}}}^{\text{DL}}\) and \({{\boldsymbol{\upmu}}}^{\text{CT}}\) at the k-th angle, i is the pixel index in the projection domain, and \({N}_{k}\) is the total number of pixels in \({P}_{k}{{\boldsymbol{\upmu}}}^{\text{DL}}\) and \({P}_{k}{{\boldsymbol{\upmu}}}^{\text{CT}}\). The set \({\mathbb{k}}\) was designed to uniformly sample \({N}_{\mathbb{k}}\) angles over 180°, e.g., \({\mathbb{k}}=[ 0^\circ ,45^\circ ,90^\circ ,135^\circ ]\) for \({N}_{\mathbb{k}}\) = 4 was used in this study.
For each tracer, an individual network was trained for 160 epochs at which point network training converged. In each epoch, 10,000 patches were randomly sampled from the training data and a mini-batch size of 16 patches was used to update the network. Adam optimization [16] was used with an initial learning rate of 10−3 with a decay factor of 0.975, which was applied after each epoch, was used. In the testing phase, instead of using the same patch size as in the training, we used a larger patch, i.e., 200 × 200 × 32 voxels and stride size of 200 × 200 × 16, to avoid striding in the first 2 dimensions. The fully convolutional architecture allows us to use different sizes of patches as inputs and this practice helps to reduce stitching artifacts caused by overlapping small image patches. Twenty-two FDG subjects were used for computing the validation loss during the training, which were independent from the cohorts of training and testing. The validation loss (Supplemental Fig. 2) results suggested that the network was sufficiently trained and there was no overfitting issue. The framework [17] was implemented in Python using TensorFlow (shared on the GitHub, https://github.com/j-onofrey/deep-image-pet). Network training took approximately 40 h on an NVIDIA RTX 8000 GPU with 48 GB memory.
Image reconstructions
PET image reconstructions were performed using the 3D-OSEM algorithm (3 iteration and 21 subsets) with the Siemens e7 toolkit. μ-DL, μ-MLAA, and μ-CT were used as the attenuation map to reconstruct PET images, i.e., OSEMDL, OSEMMLAA, and OSEMCT, respectively. Before reconstruction, μ-DL was up-sampled from 4 to 2 mm3 in voxel size while the original μ-MLAA and μ-CT with 2 mm3 were used. Note that OSEMMLAA is different from λ-MLAA since OSEMMLAA is reconstructed by the OSEM algorithm with μ-MLAA as the attenuation map whereas λ-MLAA is reconstructed by the MLAA algorithm.
Image analysis
Physics-based quantitative measurement
Using μ-CT as the reference, the quality of μ-DL was evaluated using normalized mean absolute error (NMAE), normalized root mean squared error (NRMSE), and mean normalized voxel error (MNVE), which are defined as follows:
where \({{\boldsymbol{\upmu}}}^{\text{CT}}\) represents the entire μ-map volume within a body-contour mask and M represents the number of voxels inside the mask. The mask was generated by setting all voxels with attenuation coefficient greater than 0.01 in μ-CT with bed-removed to 1 (rest as 0). The same metrics were used to evaluate μ-MLAA using the same μ-CT as reference.
The quality of the reconstructed PET images, i.e., OSEMMLAA and OSEMDL, was evaluated using NMAE, NRMSE, and normalized regional error (NRE) inside the same body-contour mask as for attenuation map evaluation. NRE is defined as \(\text{NRE}={~}^{{\sum }_{j=1}^{M}({\text{OSEM}}_{\text{DL},j} -{\text{OSEM}}_{\text{CT},j})}\!\left/ \!\!{~}_{{\sum }_{j=1}^{M}{\text{OSEM}}_{\text{CT},j}}\right.\). Note that the mean of OSEMCT, instead of the difference between maximum and minimum, was used as the denominator in the NMAE and NRMSE calculation for PET evaluation.
For both μ and PET evaluation, in addition to whole body, we evaluated within three sub-regions: neck-thorax, abdomen, and pelvis, which correspond to 0–35%, 35–65%, and 65–100% of the image volume.
To reflect the overall μ-map quality, we performed joint histogram analysis. Specifically, for each tracer, joint histograms were generated between μ-MLAA (or μ-DL) and μ-CT in both μ-map and projection domains. Projection μ-maps were generated by computing the line integral of a whole-body attenuation map, e.g., μ-MLAA, μ-DL, or μ-CT, at 0 and 90°. Image domain joint histogram analysis was also performed for PET images, i.e., between OSEMMLAA (or OSEMDL) and OSEMCT.
Tumor delineation and clinical quantitative measure
For the clinical evaluation, two experienced radiologists (TT and DS) identified 45/73 subjects for 18F-FDG, 20/36 for 68 Ga-DOTATATE, and 22/50 for 18F-Fluciclovine with tumor uptakes with high confidence by referring OSEMCT. Tumor region of interests (ROIs) were generated using in-house software, Metavol2 (modified from Metavol [18]), as follows: (1) voxels with SUV above a patient-specific threshold (see below) were extracted to form tumor clusters; (2) the radiologist dropped a digital tumor pin in each identified cluster; (3) additional non-tumor pins, adjacent to the tumor in step (2), were dropped in other high-uptake clusters to separate inflammatory/physiological uptake; and (4) automatic segmentation was performed in Metavol2 using information from all the pins, i.e., tumor and non-tumor. The above 4 steps were iteratively performed with manual adjustment of the threshold and the pin locations until satisfactory tumor segmentation was obtained. Supplemental Fig. 3 depicts the tumor delineation process. Note that the SUV thresholds as well as the tumor pins (not the same ROIs), which were defined on the OSEMCT to delineate the tumor, were used for OSEMMLAA and OSEMDL for the same subject. Metavol2 was used to generate ROIs for OSEMMLAA or OSEMDL based on the same pins and threshold as for OSEMCT. SUVmax, SUVmean, and tumor volume (Voltumor) were computed for each ROI.
Statistical analysis
All variables are presented as mean ± standard deviation. A paired t-test was applied to assess whether the means of each metric for DL and MLAA using μ-CT or OSEMCT as references were statistically different. p-values were adjusted by Bonferroni correction for multiple comparisons. Multiple comparison corrections were applied per tracer and u-map, OSEM images, and tumor ROIs were considered to be independent. Adjusted p-values of less than 0.05 were considered statistically significant.
Results
Attenuation map
Large spatial distribution differences were found across tracers, e.g., 68 Ga-DOTATATE exhibits high uptake in the spleen, liver, and kidney while 18F-Fluciclovine is broadly distributed with high muscle and bone marrow uptake (Fig. 2). Visually, μ-MLAA of 18F-FDG and 18F-Fluciclovine was found superior in quality, especially at bone areas, than 68 Ga-DOTATATE, which was likely due to the broader tracer distribution for 18F-FDG and 18F-Fluciclovine than 68 Ga-DOTATATE. Most anatomical details were successfully recovered in μ-DL for all the tracers. The ribs are distinguishable and the delineation between muscle and fat was mostly accurate for μ-DL. Notable differences between the μ-DL and μ-CT were observed in the abdominal regions, e.g., oral contrast agents in the stomach and intestine for 18F-FDG.
Numerically, μ-DL shows consistent superior performance as compared to μ-MLAA for all the tracers in NMAE, NRMSE, and MNVE with high statistical significance (Table 1). The whole-body NMAE of μ-MLAA was 7.3 ± 1.1% for 18F-FDG, 8.2 ± 1.3% for 68 Ga-DOTATATE, and 9.0 ± 0.8% for 18F-Fluciclovine, which were improved for μ-DL to 2.0 ± 0.4%, 1.4 ± 0.2%, and 2.5 ± 0.4%, respectively. Among the three sub-regions in μ-MLAA, the abdomen yielded higher NMAE for all three tracers (18F-FDG: μ-MLAA: 10.7 ± 1.5%, μ-DL: 2.9 ± 0.6%; 68 Ga-DOTATATE: 13.2 ± 2.9%, 1.8 ± 0.3%; 18F-Fluciclovine: 13.7 ± 1.4%, 3.4 ± 0.8%) than the thorax (7.2 ± 1.2%, 2.3 ± 0.6%; 7.6 ± 1.6%, 1.5 ± 0.3%; 8.2 ± 0.8%, 3.4 ± 0.8%) and the pelvis (8.2 ± 1.1%, 2.0 ± 0.6%; 9.1 ± 1.6%, 1.4 ± 0.3%; 11.6 ± 1.6%, 2.0 ± 0.5%). A scatter plot presentation of the NMAE results is shown in Fig. 3, where µ-MLAA shows larger variability than µ-DL. Sub-region NRMSE results were as follows: 18F-FDG at whole-body level: μ-MLAA: 9.8 ± 1.4%, μ-DL: 3.6 ± 0.8%; 68 Ga-DOTATATE: 10.6 ± 1.6%, 2.5 ± 0.6%; 18F-Fluciclovine: 11.7 ± 0.9%, 4.7 ± 0.9%. μ-MLAA showed larger negative MNVE comparing to μ-DL for all three tracers: 18F-FDG at whole-body level: μ-MLAA: − 10.6 ± 2.1%, μ-DL: − 1.5 ± 1.3%; 68 Ga-DOTATATE: − 5.7 ± 3.1%, − 0.5 ± 0.5%; 18F-Fluciclovine: − 9.2 ± 2.4%, − 2.0 ± 1.2%.
PET reconstruction
Overall, OSEMDL showed substantially smaller differences than OSEMMLAA for all tracers using OSEMCT as the reference (Fig. 4). OSEMMLAA showed larger differences in the abdomen than other regions for 68 Ga-DOTATATE and 18F-Fluciclovine, whereas the regional difference was not obvious for OSEMDL.
At the whole-body level, OSEMDL substantially outperformed OSEMMLAA in both NMAE (18F-FDG: OSEMMLAA: 7.1 ± 0.7%, OSEMDL: 4.4 ± 1.3%; 68 Ga-DOTATATE: 12.4 ± 4.5%, 3.1 ± 1.2%; 18F-Fluciclovine: 9.9 ± 6.4%, 4.9 ± 1.2%), and NRMSE (13.2 ± 4.9%, 10.9 ± 11.3%; 27.9 ± 11.6%, 11.7 ± 7.9%; 17.3 ± 13.1%, 9.1 ± 1.9%) (Table 2). Both OSEMDL and OSEMMLAA yielded low NRE at the whole-body level (− 1.9 ± 2.0% and − 1.7 ± 1.1% without statistical significance) for 18F-FDG while OSEMDL significantly outperformed OSEMMLAA for 68 Ga-DOTATATE (1.4 ± 1.7% and − 7.7 ± 4.1%, p < 0.0001) and 18F-Fluciclovine (− 2.8 ± 1.5% and − 5.0 ± 4.6%, p < 0.01). In the sub-regions, OSEMMLAA yielded larger NMAE, NRMSE, and NRE in the abdomen than the other two regions while OSEMDL yielded consistent performance at all regions with slightly higher error at thorax.
Joint histogram analysis
For all the tracers, μ-DL shows superior alliance with μ-CT than μ-MLAA in the image domain comparison (Fig. 5a). In the projection domain (b), for all the tracers, μ-MLAA shows larger error, i.e., deviating from the unity line, than the μ-DL, especially in the low value range. μ-MLAA also shows larger variability, i.e., more spread along the unity line, than the μ-DL. For PET images, OSEMDL also yielded smaller deviation from the OSEMCT than the OSEMMLAA (Supplemental Fig. 4). For OSEMMLAA, larger difference from OSEMCT was found for 18F-Fluciclovine as compared to the other two tracers, especially in the high SUV value range (> 7).
Tumor delieanation
In the 18F-FDG tumor delineation example (Supplemental Fig. 5), abnormal uptake was found in the inferior lobe of the left lung and the left hilar lymph nodes. The paramediastinal mass uptake was considered the primary tumor. In the 68 Ga-DOTATATE example, multiple high-uptake metastatic lesions of neuroendocrine tumor were found in the liver, the abdominal, and the pelvic lymph nodes. For 18F-Fluciclovine, abnormal uptake was found in the prostate and multiple retroperitoneal lymph nodes were identified. Among all the testing cases, 152 tumor ROIs (< 150 mL in volume) among 45/73 18F-FDG subjects, 70 ROIs (< 50 mL) among 20/36 68 Ga-DOTATATE subjects and 44 ROIs (< 20 mL) among 22/50 18F-Fluciclovine subjects were delineated for evaluation.
Clinical measure evaluation
Overall, OSEMMLAA yielded relatively small error (< 5%) for 18F-FDG and 68 Ga-DOTATATE but larger error (e.g., − 7.3 ± 2.9% in SUVmax) for 18F-Fluciclovine in all SUV measures whereas OSEMDL yielded excellent quantitation (< 2%, except for SUVmax of 18F-Fluciclovine) for all the three tracers (Table 3). Specifically, OSEMMLAA showed significantly lower % differences in SUVmax than OSEMDL (18F-FDG: OSEMMLAA: − 3.6 ± 4.4%, OSEMDL: − 1.7 ± 4.5%; 68 Ga-DOTATATE: − 4.3 ± 5.1%, 0.4 ± 2.8%; 18F-Fluciclovine: − 7.3 ± 2.9%, − 2.8 ± 2.3%, p < 0.001) (see Fig. 6 for the SUVmax scatter plot). Similar results were found for SUVmean.
OSEMDL yielded substantially smaller error in tumor volume measure (Table 3) than OSEMMLAA (18F-FDG: OSEMMLAA: − 8.4 ± 14.5%, OSEMDL: − 3.0 ± 15.0%; 68 Ga-DOTATATE: − 14.1 ± 19.7%, 1.8 ± 11.6%; 18F-Fluciclovine: − 15.9 ± 9.1%, − 6.4 ± 6.4%). Individual tumor volume results are shown in Fig. 7, where tumor volume measured in OSEMMLAA and OSEMDL was both significantly (p < 0.0001) correlated with it in OSEMCT. The slope for OSEMDL was closer to unity than it was for OSEMMLAA for all the tracers (18F-FDG: OSEMMLAA: 0.97, OSEMDL: 1.00; 68 Ga-DOTATATE: 0.96, 1.01; 18F-Fluciclovine: 0.86, 0.93), which suggested superior performance in volume measure accuracy from OSEMDL than OSEMMLAA.
a Correlation between OSEMDL (or OSEMMLAA) with OSEMCT in tumor volume measure. Both OSEMDL and OSEMMLAA showed significant correlation. Significance was evaluated using linear regression between OSEMDL (or OSEMMLAA) with OSEMCT b %differences between OSEMMLAA (or OSEMDL) with OSEMCT. Paired t-test was used to compare the %difference between OSEMDL and OSEMMLAA while OSEMCT was used as reference. Log scale is used in the x-axis
Discussion
In this study, we applied a deep learning–based attenuation map (μ-DL) prediction framework to three clinical oncology tracers with diverse imaging characteristics: 18F-FDG, 68 Ga-DOTATATE, and 18F-Fluciclovine. Using the CT-based attenuation map (μ-CT) as a gold standard, we evaluated both the μ-DL and the PET reconstructions using the μ-DL (OSEMDL) using both physics-based metrics and clinical-relevant measures, i.e., SUVmax, SUVmean, and tumor volume. μ-DL was compared to the MLAA attenuation map (μ-MLAA) and was found to be superior than the μ-MLAA for all the three tracers. OSEMDL yielded excellent tumor quantification for all tracers and was superior to OSEMMLAA.
In addition to the proposed framework, i.e., using the MLAA output as the neural network input [6, 7], many other machine-learning approaches have been proposed to improve the attenuation map in PET. Another well-explored framework is to use paired MRI images as the network input to synthesize pseudo-CT for PET AC [19, 20], but this approach only applies to PET/MR systems. Using non-attenuation corrected (NAC) PET as the network input is a more commonly used approach to synthesize pseudo-CT [21, 22] or PET with AC [23, 24], since it does not require MR or TOF capability. However, the quantification accuracy of this approach is still improving. As compared to our framework, the NAC-based framework [21, 22] makes use of less information from the PET data, i.e., abandoning all the attenuation information in the emission data. We refer readers to a comprehensive review article by Lee [25].
Here, although the OSEMDL was found more accurate in SUV measures than the OSEMMLAA, the radiologists (TT and DS) confirmed that it will be unlikely to make diagnositic differences whether OSEMMLAA or OSEMDL was used for a single PET scan. However, the excellent quantitation of OSEMDL will be invaluable in cancer treatment evaluations. For example, in FDG PET scans, any change in tumor uptake/volume between the baseline and the follow-up scans may alter treatment strategies where accurate tumor quantitation becomes crucial.
For a same patient, multiple PET/CT scans at multiple time points are typically performed. Therefore, radiation dose reduction is important (more important for pediatric patients). Many studies had been conducted to reduce PET injection dose without/with minimal compromise in the PET image quality [26, 27] while the radiation exposure for a patient from CT is also substantial. Even for a low-dose CT protocol [28], dose from CT is still comparable to a 10-mCi injection of 18F-FDG [28]. The proposed deep learning–based method provides a promising solution for CT-less PET. It is understood that in addition to providing AC for PET, CT also provides important anatomical guidance in a clinical PET read. Although our current solution cannot provide the same quality of anatomical guidance as compared to CT, for certain protocols in which CT may not be needed, e.g., follow-up scans in lymphoma treatment evaluation, the proposed method is sufficient for clinical use.
In this study, although we purposely excluded cases with mis-alignment between CT and PET due to patient motion, minor PET-CT mis-alignments were still found in many cases due to body motions (arms or legs) [29] and respiratory motion [30]. In the Supplemental Fig. 6, we show an example to demonstrate the application of the proposed method to eliminate the artifacts caused by patient motion between PET and CT.
Here, we point out other study limitations and discuss the future directions. (1) For each tracer, only 40 subjects were used for the neural network training. In order to train a more robust model for the patient population, especially for those with unusual anatomy or special conditions, e.g., subjects with limb amputation and pediatric patients or subjects with metal implants and pacemakers, datasets of more comprehensive patient cohort are needed. (2) The quality of μ-DL depends on the quality of μ-MLAA and λ-MLAA, which are PET dose-dependent. Here, we only studied the regular clinical dose used for the three tracers. In the future, we will explore the efficacy of applying the current method for low-dose PET studies. (3) The hyperparameters used in the loss functions and neural networks, e.g., \({\beta }_{1}\) and \({\beta }_{2}\), were not optimized. In the future, we will perform optimization studies. (4) In this study, the scatter estimate used in the MLAA reconstruction was generated based on the μ-CT. In a clinical application, detector background radiation [31, 32] can be used to provide the scatter estimate for MLAA reconstructions.
Conclusion
The proposed framework provides accurate and robust attenuation correction for clinical whole-body 18F-FDG, 68 Ga-DOTATATE, and 18F-Fluciclovine in tumor SUV measures as well as tumor volume estimation. The proposed framework provides clinically equivalent quality as compared to CT in attenuation correction for the three tracers.
References
Barrett JF, Keat N. Artifacts in CT: recognition and avoidance. Radiographics. 2004;24:1679–91. https://doi.org/10.1148/rg.246045065.
Ladefoged CN, Law I, Anazodo U, St Lawrence K, Izquierdo-Garcia D, Catana C, et al. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage. 2017;147:346–59. https://doi.org/10.1016/j.neuroimage.2016.12.010.
Chen Y, An H. Attenuation correction of PET/MR imaging. Magn Reson Imaging Clin N Am. 2017;25:245–55. https://doi.org/10.1016/j.mric.2016.12.001.
Rezaei A, Defrise M, Bal G, Michel C, Conti M, Watson C, et al. Simultaneous reconstruction of activity and attenuation in time-of-flight PET. IEEE Trans Med Imaging. 2012;31:2224–33. https://doi.org/10.1109/TMI.2012.2212719.
Rezaei A, Deroose CM, Vahle T, Boada F, Nuyts J. Joint reconstruction of activity and attenuation in time-of-flight pet: a quantitative analysis. J Nucl Med. 2018;59:1630. https://doi.org/10.2967/jnumed.117.204156.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight (18)F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60:1183–9. https://doi.org/10.2967/jnumed.118.219493.
Shi L, Onofrey J, Revilla EM, Toyonaga T, Menard D, Ankrah J, et al. A novel loss function incorporating imaging acquisition physics for PET attenuation map generation using deep learning. Med Image Comput Comput Assist Interv. 2019;11767:723–31. https://doi.org/10.1007/978-3-030-32251-9_79.
Maurer T, Eiber M, Schwaiger M, Gschwend JE. Current use of PSMA-PET in prostate cancer management. Nat Rev Urol. 2016;13:226–35. https://doi.org/10.1038/nrurol.2016.26.
Calais J, Ceci F, Eiber M, Hope TA, Hofman MS, Rischpler C, et al. (18)F-fluciclovine PET-CT and (68)Ga-PSMA-11 PET-CT in patients with early biochemical recurrence after prostatectomy: a prospective, single-centre, single-arm, comparative imaging trial. Lancet Oncol. 2019;20:1286–94. https://doi.org/10.1016/S1470-2045(19)30415-2.
Poeppel TD, Binse I, Petersenn S, Lahner H, Schott M, Antoch G, et al. 68Ga-DOTATOC versus 68Ga-DOTATATE PET/CT in functional imaging of neuroendocrine tumors. J Nucl Med. 2011;52:1864–70. https://doi.org/10.2967/jnumed.111.091165.
Panin VY, Aykac M, Casey ME. Simultaneous reconstruction of emission activity and attenuation coefficient distribution from TOF data, acquired with external transmission source. Phys Med Biol. 2013;58:3649–69. https://doi.org/10.1088/0031-9155/58/11/3649.
Onofrey JA, Casetti-Dinescu DI, Lauritzen AD, Sarkar S, Venkataraman R, Fan RE, et al. Generalizable multi-site training and testing of deep neural networks using image normalization. Biomedical Imaging (ISBI), 2019 IEEE 16th International Symposium on; 2019. p. pp. 1–4.
Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. Cham: Springer International Publishing; 2015. p. 234–41.
Shi L, Onofrey JA, Revilla EM, Toyonaga T, Menard D, Ankrah J, et al. A novel loss function incorporating imaging acquisition physics for PET attenuation map generation using deep learning. Cham: Springer International Publishing; 2019. p. 723–31.
Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, et al. Medical image synthesis with deep convolutional adversarial networks. Ieee T Bio-Med Eng. 2018;65:2720–30. https://doi.org/10.1109/Tbme.2018.2814538.
Kingma DP, Ba J. Adam: a method for stochastic optimization. 2014. p. arXiv:1412.6980.
Onofrey JA, Casetti-Dinescu DI, Lauritzen AD, Sarkar S, Venkataraman R, Fan RE, et al. Generalizable multi-site training and testing of deep neural networks using image normalization. Proc IEEE Int Symp Biomed Imaging. 2019;2019:348–51. https://doi.org/10.1109/isbi.2019.8759295.
Hirata K, Furuya S, Huang SC, Manabe O, Magota K, Kobayashi K, et al. A semi-automated method to separate tumor from physiological uptakes on FDG PET-CT for efficient generation of training data targeting deep learning. J Nucl Med. 2019;60:supplement 1213.
Bradshaw TJ, Zhao G, Jang H, Liu F, McMillan AB. Feasibility of deep learning-based PET/MR attenuation correction in the pelvis using only diagnostic MR images. Tomography. 2018;4:138–47. https://doi.org/10.18383/j.tom.2018.00016.
Arabi H, Zeng G, Zheng G, Zaidi H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur J Nucl Med Mol Imaging. 2019;46:2746–59. https://doi.org/10.1007/s00259-019-04380-x.
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for (18)F-FDG PET attenuation correction. EJNMMI Phys. 2018;5:24. https://doi.org/10.1186/s40658-018-0225-8.
Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. https://doi.org/10.1088/1361-6560/ab4eb7.
Shiri I, Ghafarian P, Geramifar P, Leung KH, Ghelichoghli M, Oveisi M, et al. Direct attenuation correction of brain PET images using only emission data via a deep convolutional encoder-decoder (Deep-DAC). Eur Radiol. 2019;29:6867–79. https://doi.org/10.1007/s00330-019-06229-1.
Hashimoto F, Ito M, Ote K, Isobe T, Okada H, Ouchi Y. Deep learning-based attenuation correction for brain PET with various radiotracers. Ann Nucl Med. 2021. https://doi.org/10.1007/s12149-021-01611-w.
Lee JS. A review of deep-learning-based approaches for attenuation correction in positron emission tomography. IEEE Transactions on Radiation and Plasma Medical Sciences. 2021;5:160–84. https://doi.org/10.1109/TRPMS.2020.3009269.
Gong K, Guan J, Kim K, Zhang X, Yang J, Seo Y, et al. Iterative PET Image reconstruction using convolutional neural network representation. Ieee T Med Imaging. 2019;38:675–85. https://doi.org/10.1109/TMI.2018.2869871.
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, et al. An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol. 2019;64:165019. https://doi.org/10.1088/1361-6560/ab3242.
Li Y, Jiang L, Wang H, Cai H, Xiang Y, Li L. Effective radiation dose of 18f-Fdg Pet/Ct: how much does diagnostic Ct contribute? Radiat Prot Dosimetry. 2019;187:183–90. https://doi.org/10.1093/rpd/ncz153.
Lu Y, Gallezot JD, Naganawa M, Ren S, Fontaine K, Wu J, et al. Data-driven voluntary body motion detection and non-rigid event-by-event correction for static and dynamic PET. Phys Med Biol. 2019;64:065002. https://doi.org/10.1088/1361-6560/ab02c2.
Lu Y, Fontaine K, Mulnix T, Onofrey JA, Ren S, Panin V, et al. Respiratory motion compensation for PET/CT with motion information derived from matched attenuation-corrected gated PET data. J Nucl Med. 2018;59:1480–6. https://doi.org/10.2967/jnumed.117.203000.
Teimoorisichani M, Sari H, Panin V, Bharkhada D, Rominger A, Conti M. Using LSO background radiation for CT-less attenuation correction of PET data in long axial FOV PET scanners. Journal of Nuclear Medicine. 2021;62:1530-.
Rothfuss H, Panin V, Moor A, Young J, Hong I, Michel C, et al. LSO background radiation as a transmission source using time of flight. Phys Med Biol. 2014;59:5483–500. https://doi.org/10.1088/0031-9155/59/18/5483.
Acknowledgements
We would like to thank Judson Jones and Vladimir Panin from the Siemens Healthcare for the reconstruction software support. We thank Zhongdong Sun for the IT support. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Funding
This work was supported by NIH grants R03EB027209 and R21EB028954.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Takuya Toyonaga and Dan Shao are the co-first authors.
This article is part of the Topical collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Toyonaga, T., Shao, D., Shi, L. et al. Deep learning–based attenuation correction for whole-body PET — a multi-tracer study with 18F-FDG, 68 Ga-DOTATATE, and 18F-Fluciclovine. Eur J Nucl Med Mol Imaging 49, 3086–3097 (2022). https://doi.org/10.1007/s00259-022-05748-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-022-05748-2