Abstract
Purpose
Positron emission tomography (PET) image quality can be improved by higher injected activity and/or longer acquisition time, but both may often not be practical in preclinical imaging. Common preclinical radioactive doses (10 MBq) have been shown to cause deterministic changes in biological pathways. Reducing the injected tracer activity and/or shortening the scan time inevitably results in low-count acquisitions which poses a challenge because of the inherent noise introduction. We present an image-based deep learning (DL) framework for denoising lower count micro-PET images.
Procedures
For 36 mice, a 15-min [18F]FDG (8.15 ± 1.34 MBq) PET scan was acquired at 40 min post-injection on the Molecubes β-CUBE (in list mode). The 15-min acquisition (high-count) was parsed into smaller time fractions of 7.50, 3.75, 1.50, and 0.75 min to emulate images reconstructed at 50, 25, 10, and 5% of the full counts, respectively. A 2D U-Net was trained with mean-squared-error loss on 28 high-low count image pairs.
Results
The DL algorithms were visually and quantitatively compared to spatial and edge-preserving denoising filters; the DL-based methods effectively removed image noise and recovered image details much better while keeping quantitative (SUV) accuracy. The largest improvement in image quality was seen in the images reconstructed with 10 and 5% of the counts (equivalent to sub-1 MBq or sub-1 min mouse imaging). The DL-based denoising framework was also successfully applied on the NEMA-NU4 phantom and different tracer studies ([18F]PSMA, [18F]FAPI, and [68 Ga]FAPI).
Conclusion
Visual and quantitative results support the superior performance and robustness in image denoising of the implemented DL models for low statistics micro-PET. This offers much more flexibility in optimizing preclinical, longitudinal imaging protocols with reduced tracer doses or shorter durations.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Positron emission tomography (PET) is a functional imaging modality that enables changes in physiological and metabolic processes to be visualized and quantified. In preclinical practice, micro-PET refers to imaging mice and rats using dedicated small-animal, high-resolution PET scanners [1, 2]. A radiotracer is injected into a living rodent and the 3D distribution of that labeled compound can be measured non-invasively. Imaging of specific molecular targets with micro-PET allows to study in vivo (patho)physiologic processes and molecular abnormalities with the added value of enabling longitudinal scans. The most widely used PET tracer is [18F]FDG to image glucose metabolism [2]. Other emerging radiopharmaceuticals are F-18 or Ga-68 radiolabeled prostate-specific membrane antigen (PSMA) [3] or fibroblast activation protein inhibitor (FAPI) [4] ligands. Preclinical PET imaging serves as a translational tool in tracer development. New compounds are first scanned in rodents to assess pharmacokinetics and monitor toxicity to specific organs before doing human studies [2, 5, 6].
PET imaging involves ionizing radiation, and there are higher dose requirements in preclinical set-ups compared to clinical imaging because the subject size and the spatial resolution needed to detect anatomical or functional changes are profoundly different [7]. Common preclinical radioactive doses (10 MBq) that are used to obtain good image quality have been shown to cause deterministic changes in biological pathways [8], especially in longitudinal studies. The maximum dose that can be administered to a rodent is influenced by several factors, including animal welfare considerations and the specific activity of injected radiotracer. However, the relatively higher blood volume-to-body weight ratio for small animals as well as equipment saturation can also be determining factors in establishing the upper limit for the injected radioactivity. Apart from radiation concerns, there are other advantages to lowering the injected dose in rodents. For example, in hormone and neuroreceptor imaging with micro-PET, limiting the radiotracer activity aids in alleviating concerns about toxicity and pharmacological effects which could violate the tracer principle and lead to inaccurate quantification [6]. Radio- and chemo-toxicity is generally not an issue when high affinity/high molar activity radiopharmaceuticals are employed. However, imaging with lower molar activity radiopharmaceuticals can pose challenges for ligands that target specific receptors with limited binding capacity. Micro-PET is often combined with micro-computed tomography (CT) and such dual-modality protocols deliver additional radiation dose to the rodents [9, 10].
In this context, low-dose micro-PET acquisitions have attracted much attention and any preclinical imaging setup should aim for dose reduction. Besides ensuring animal care and maintaining integrity in longitudinal experiments, the radiation exposure to the technicians using the micro-PET imaging devices can be close to clinical radiation doses [8]. However, reducing the injected dose and/or scan time inevitably results in lower count statistics which inherently introduces noise. This trade-off between image quality (noise), radiation dose and scan time remains of particular concern in small animal studies because of the cumulative impact of dose and anesthesia on animal physiology.
To address the challenges associated with count deficiency in low statistics PET, image denoising techniques can be applied. State-of-the-art methods explore the potential of deep learning (DL) to predict high-count like images from low-count PET data. Such DL-based denoising frameworks have shown superior performance with important benefits for dose reduction capabilities [11,12,13,14,15]. However, while there are clear driving factors for low-dose imaging in preclinical setups, the incentive for DL-based denoising of low-count micro-PET has been lacking compared to the extensive research efforts spent on clinical low-dose PET. To our knowledge, only two previous studies have investigated the use of DL for denoising low-dose micro-PET data [16, 17], and both studies showed that the DL approach achieved lower bias in [18F]FDG quantification than non-DL denoising techniques, but they did not consider cross-tracer applicability.
A critical concern with DL-based methods is their reproducibility and robustness to new studies. The growing range of radiopharmaceuticals used in practice consistently challenges the adaptability and dependability of any trained DL algorithm when applied to cross-tracer and cross-scanner datasets [12, 13]. This presents a particular hurdle in preclinical research environments because a larger diversity of (new) radiotracers is investigated with micro-PET given its crucial role in the early stages of tracer development [2, 5, 6]. This work presents an image-based DL framework for denoising low-count micro-PET images while aiming for improved quantitative accuracy. This work puts the emphasis for preclinical imaging on achieving low bias and better precision with lower count datasets. We consider four count level reductions which go as low as to emulate sub-1 MBq and/or sub-1 min acquisitions. We further investigate the applicability of the DL denoising algorithm (trained only with [18F]FDG data) on non-FDG and non-F-18 labeled radiotracer studies.
Materials and Methods
Image Data Acquisitions
All image datasets used for training were collected from the Molecubes β-CUBE [18]. The dataset consisted of 36 [18F]FDG PET scans from retrospective studies conducted in our lab (UGent’s Animal Experiments Ethics Committee: ECD 20/01) that investigated murine inflammation models. For each study, a 15-min whole-body micro-PET scan was acquired at 40 min post-injection after an intravenous injection of 8.15 ± 1.34 MBq [18F]FDG. The data were acquired in list mode and reconstructed using an ordered subsets expectation maximization (OSEM) algorithm at 400 µm voxel size (with 30 iterations and a number of subsets varying depending on the number of coincidences present in the time frame to be reconstructed—which is defined by the Molecubes system). For reconstruction, attenuation correction was not considered since most of the retrospective studies did not have a co-registered CT scan. Micro-PET voxel values were expressed in kilo-Becquerel per milliliter (kBq/mL) which represent the activity concentration. The standardized uptake value (SUV) normalizes the activity in the images by the injected dose and body weight of the mouse.
To train the DL image denoising model, low and high (standard) count image pairs were required, and the advantage of list-mode was that the low- and high-count datasets were perfectly aligned. The 15-min scan (standard-count) served as target image during training. This full time frame was then parsed into 2 sequential time frames of 7.5 min, 4 sequential time frames of 3.75 min, 10 sequential time frames of 1.5 min, and 20 sequential time frames of 0.75 min to emulate four low-count datasets with 50, 25, 10, and 5% of the standard-count events, respectively. Resultant sub-sampled list mode data were also reconstructed using OSEM at a voxel size of 400 µm.
Network Architecture
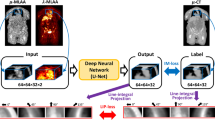
The DL model comprises a 2D convolutional network based on a four-layer U-Net [19] topology (Suppl. Figure 1). The feature maps in the encoding path are increased from 64 to 512 with three max pooling layers. The decoding path uses transposed convolutions. Skip connections copy and concatenate the encoder layer output in the channel dimension with the decoder layer input. A ReLU activation function [20] is added in the final layer to enforce non-negativity at the output (essential for PET voxel values). The network reads three adjacent image slices at the input layer to leverage spatial context from the neighboring slices.
Training Strategy
From the 36 [18F]FDG mice datasets, training, validation, and testing were split with a ratio 28:4:4, respectively. For training, image slices in transverse, coronal and sagittal orientations were used as inputs. During pre-processing, the slices that lacked enough relevant image content were not included in the training set (see Suppl. Information 1.2. for more details) which resulted in a total selection of 10,897 slices (transverse: 6026, coronal: 2413, sagittal: 2458). During inference, the transverse slices were denoised and then stacked to generate the CNN-denoised micro-PET image in DICOM format. Three orientations were considered during training to increase the training dataset and to provide a form of regularization, preventing overfit to a specific orientation and promoting generalization to unseen data.
Separately accounting for each low-count level, we trained a 2D CNN with input data originating from that count level only. This resulted in the development of four DL models which are denoted as X2Std CNN (where X refers to the respective low count level: half, quarter, tenth, or twentieth). Another 2D CNN model was trained with mixed input data from all four low-count levels which implied that a total of 43,588 slices (4 × 10,897) were used to train the M2Std CNN (“Mixed to Standard”). All five CNN models were implemented to randomly select one of the replicate time frames.
The voxel values in the input slices were normalized by the mean intensity of each individual image slice. This normalization factor was also applied to normalize the target (standard-count) image. The mean squared error (loss function) and the Adam optimizer [21] were used for compilation during back-propagation with a learning rate of 1E-4 and a batch size of 16. For data augmentation, random vertical and horizontal flipping were included. Suppl. Information 1.3. details software packages and versions used for training the CNNs.
Image Analyses and Performance Evaluations
Comparator Denoising Techniques
To compare the CNN-denoised images against other more conventional denoising methods, the low statistics micro-PET images reconstructed at different lower count levels were also denoised by the following spatial filters: a Gaussian filter (with the standard deviation set to 1.0 voxel which matches the spatial resolution of the β-CUBE scanner), a median filter (with a kernel size of 3), and a bilateral filter (i.e., edge-preserving filtering method).
Quantitative Analysis for Image Denoising of [18F]FDG Micro-PET
Accuracy and Reproducibility of SUV Measurements
By placing cuboidal volumes-of-interest (VOI) in the mouse liver and high tracer uptake regions (such as muscle inflammation sites), the performance of the denoising methods in improving the accuracy of SUV measurements in a noisy image was assessed. SUVmean and SUVmax measures obtained from the low-count and denoised images were compared to the standard-count image. These measurements were done over the number of replicate images available at the particular count level to conduct statistical analyses (see the “Statistical Analysis” section).
Quantitative Image Quality Analysis
Image enhancement performance was compared among different denoising methods in terms of peak signal-to-noise ratio (PSNR), structural similarity index metric (SSIM), and mean absolute error (MAE) computed with respect to the standard-count (15 min) image slice. In some cases, the signal-to-noise ratio (SNR) was calculated as the ratio of the mean to the standard deviation in SUV measured in the VOI.
Physical Image Quality Metrics Evaluated with Respect to an Estimated Ground-Truth Image Obtained from a 5-h (Ex Vivo) [18F]FDG Mouse Scan
Root-mean-squared error (RMSE), PSNR, and SSIM were computed between the low-count and denoised images with respect to an estimated ground-truth image obtained from a 5-h scan. A mouse (18.7 g) was injected with 11.19 MBq [18F]FDG and sacrificed 68 min after injection to perform a 5-h scan on the β-CUBE. The rationale for such a long acquisition was to reduce the statistical noise as much as possible and obtain a very high-count image. To match the reconstructed frames used in training, the 5-h data (in list mode) was sub-sampled into one time fraction of 7.5, 3.75, 1.5, and 0.75 min selected around the mid time point.
Additional intermediate frame reconstructions were performed to reconstruct images over a range of count levels: 20 separate images with frame durations from 90 s to 30 min (in incremental steps of 90 s) were selected from the mid time point. The RMSE was then calculated as a function of different low-count levels. This analysis aims to estimate the scan time (frame duration) needed to obtain the same image quality (in RMSE) as measured on differently denoised images reconstructed from a 45 s scan.
NEMA-NU4 Image Quality Phantom for Small-Animal PET
The NEMA-NU4 phantom [22] was filled with 3.7 MBq [18F]FDG and scanned for 20 min on the β-CUBE. The 20-min data was sub-sampled into time fractions of 10, 5, 2, and 1 min to emulate low statistics images at 50, 25, 10, and 5% of the counts. The NEMA-NU4 phantom was re-scanned in the same position for 20 min at four later time points, when the tracer activity decayed to a half, quarter, tenth and twentieth of the original injected dose. The rationale was to test the DL denoising model on “real” low-count datasets. The data was reconstructed with all available corrections including CT-based attenuation using OSEM into 400 µm voxels. Image uniformity and contrast recovery coefficients (CRCs) were compared between Gaussian-filtering and M2Std CNN denoising (see Suppl. Information 2.1.).
Applicability to Other Tracer Studies
Assessing neural network performance requires to consider the characteristics of the training data, specifically in this case, the DL method was trained only with [18F]FDG images that show a distinct distribution. It is necessary to test the network’s ability to denoise images derived from other tracers that have different distributions and uptake patterns. So, in this regard, low-count studies with [18F]PSMA, [18F]FAPI, and [68 Ga]FAPI were qualitatively and quantitatively evaluated. The rationale for using these particular tracers is to consider (i) two F-18 labeled tracers with different uptake profiles than FDG and (ii) one non-F-18 labeled radiotracer with different physical properties. Ga-68 has a higher positron energy and so a larger positron range compared to F-18 which consequently degrades image quality as a result of blurring and reduced resolution [23], especially in small animal imaging systems with a resolution of about 1 mm. The cross-tracer applicability is studied with the M2Std CNN denoising model and compared to Gaussian filtering. Further details on the acquired datasets and quantitative analyses used for each tracer study are specified in Suppl. Information. 2.
Statistical Analysis
Image quality metrics and SUV measures were quantified over the number of independent replicate images available at the particular count level (namely: 4 at 25% of the counts, 10 at 10% and 20 at 5%). We omitted the analysis for reconstructions at 50% of the counts, as no meaningful statistics can be deduced from two replicates. The X2Std CNN and M2Std CNN algorithms were compared to other denoising methods using two-tailed paired-sample t-tests with Bonferroni corrected p values lower than 0.05 considered as statistically significant.
Results
Qualitative and Quantitative Analysis for Denoising of [18F]FDG Micro-PET
Figure 1 shows an example of a [18F]FDG mouse with a visual comparison of the standard-count (here: 15 min, 9.9 MBq), low-count and denoised images at 50, 25, 10, and 5% of the counts, where two CNN-based denoising approaches are compared to other techniques for noise filtering. All denoising methods removed the image noise to different extents, with no clear visual differences between the denoised images at 50 and 25%, except for a slight tendency of the non-CNN denoised images to present more spatial blurriness and reduced contrast resolution recovery. This effect was also quantified with the NEMA-NU4 phantom (Fig. 5). For the image shown in Fig. 1, PSNR, SSIM, and MAE values were calculated between the low-count and denoised images with respect to the standard-count image (see Suppl. Information 3 for detailed summary statistics: Suppl. Tables 1–3). The quantitative differences in these physical image quality metrics observed between the CNN-based methods and conventional denoising filters (Gaussian, median, bilateral) were consistently found to be statistically significant (p < 0.05) for the reconstructions at 10% and 5% of the counts, with significantly higher PSNR, higher SSIM, and lower MAE for the CNN-based methods.
Visual comparison between the standard-count (15-min), low-count, and denoised images showing a coronal image of a mouse (18 g) which was administered a tracer activity of 9.9 MBq [.18F]FDG. There is FDG uptake in the fore- and hindlimb which indicates a muscular inflammation. Each row illustrates one of the low-count levels. Note that these images are not attenuation corrected. For this mouse study, SUV measurements (accuracy and reproducibility) were performed in the liver and inflammation site, see Fig. 2
Figure 2 summarizes the [18F]FDG SUV quantification measurements conducted for the mouse shown in Fig. 1. In Fig. 2a, the variability within the liver was measured, assuming that the liver SUVs are uniform. SUVmean in the liver showed a bias below 10% between the standard-count and all denoised low-count images. Compared to the filtering methods, the CNN-denoised images presented slightly smaller error bars on SUVmean (especially for the tenth and twentieth), suggesting that the CNN-based denoising models resulted in a larger noise reduction. Statistical analysis applied on a number of independent replicate images indicated that both CNN-denoising algorithms were able to significantly recover the SUVmax from the low-count image to an acceptable margin within the standard-count SUVmax (Fig. 2b and c). The improvement in SUVmax measurements (reduced bias) for the CNN-denoising methods relative to conventional techniques showed to be more significant with increased count reductions.
Micro-PET [18F]FDG SUV quantification results (for the mouse shown in Fig. 1) are compared across the low-count levels (on the X-axis) and for different denoising methods. a Mean SUV in a liver VOI is reported with standard deviation as a measure of noise and measured in one image replicate to assess quantitative accuracy. Maximum SUV in the liver (b) and inflammation hot spot (c) are reported as the average (± standard deviation) measured across the number of replicate images available at the particular count level. Statistical significance levels are provided for the pair-wise comparison between both CNN-denoising models and each of the three conventional filtering methods
Figure 3 presents another mouse study for which qualitative and quantitative analyses were conducted to compare the CNN-based denoising methods to conventional filtering techniques with an emphasis on SNR. For the lower count levels (10% and 5% of the counts) SNR could be significantly improved using the CNN-based methods compared to the conventional filtering methods. Additional results presented in Suppl. Information 4 (Suppl. Tables 4–6) further confirm that CNN-based methods also improved PSNR, SSIM, and MAE measures compared to conventional denoising filters.
Visual comparison between the standard-count (15-min), low-count, and denoised images showing a sagittal image of a mouse (21 g) which was administered a tracer activity of 11.5 MBq [18F]FDG. The FDG uptake in the heart, as well as some muscle inflammation spots can be seen. Each row illustrates one of the low-count levels. The bottom panel of this figure presents the summary statistics for the SNR quantified in a brain VOI (delineated in the top left image). SNR results are reported as the average (± standard deviations) measured across the number of replicate images available at the particular count level. Statistical significance levels (p value ranges) are given for the pair-wise comparisons between both CNN-denoising models and the three conventional filters
Suppl. Figure 2 compares the estimated “ground-truth” image (5-h ex vivo mouse scan), standard-count (15-min), low-count, and denoised images. Suppl. Table 7 reports the RMSE, PSNR and SSIM computed between the low-count and denoised images with respect to the estimated ground-truth. The CNN approaches resulted in the lowest RMSE, highest PSNR and highest SSIM at all count levels. Figure 4 a compares the conventional denoising techniques (blue) to the CNN methods (green); it can clearly be appreciated that the CNN-denoised images decreased the RMSE with the most significant improvement for the images reconstructed at 10 and 5% of the counts. The red curve in Fig. 4b shows the decrease in RMSE with longer frame durations. The outcome of the analysis in Fig. 4b implies that the X2Std and M2Std CNN methods (in green) can enhance the quality of an image (in terms of RMSE) obtained from a 45 s scan to a level as if acquired during a 7.6 and 6.4 min scan, respectively.
RMSE is determined with respect to the 5-h ex vivo mouse scan that was acquired to generate an estimated ground-truth image. a The 3D bar graph compares the RMSE at all four low-count levels for the different denoising methods. The red bars show the increase in RMSE for images of reduced counts but the application of denoising methods (in blue and green) decreases the RMSE. b Further quantitative analyses looked at the change in RMSE as a function of the frame time length selected to reconstruct lower count datasets, with the aim to estimate the level of scan time reduction that could be achieved
NEMA-NU4 Image Quality Phantom for Small-Animal PET
Image noise was increased when the scan time (i.e., count level) was reduced. Through application of the M2Std CNN-denoising algorithm, this noise could be restored to a level comparable of the standard scan (20 min with 3.7 MBq) based on visual inspection from Fig. 5b. The uniformity measurements reported in Fig. 5a confirm these observations; both Gaussian filtering and M2Std CNN denoising reduced the variability measured in the uniform phantom section (decreased COV%). Figure 5b compares the low-count reconstructions of the hot rod section to the Gaussian-filtered and M2Std CNN-denoised images. For CNN denoising, the intensity of the two smallest contrast rods visually matched closer to the 20-min standard scan compared to the Gaussian filtering which induced some spatial blurriness. This visual interpretation was also confirmed by the CRC values of the Gaussian-filter which were lower for each rod size than those of CNN-denoising (Fig. 5c and Suppl. Information 6). The difference in CRCs between the low-count and CNN-denoised images with respect to the standard-count image fall within 10% except for 1-mm rod.
a Uniformity measurements: coefficients of variation and maximum-to-mean ratios compared between 20-min, low-count, Gaussian-filtered and M2Std CNN denoised images of the uniform NEMA-NU4 phantom section. b Visual comparison of a transverse slice from the hot rod region in the phantom. c Contrast recovery curves compared between the 20-min (red dashed line), low-count (dashed), Gaussian-filtered (dot-dashed), and M2Std CNN denoised (dotted). See Suppl. Table 8 for the CRC values.
Applicability to Other Tracer Studies
For our cross-tracer applicability investigation, Fig. 6 presents visual comparisons and quantitative analyses for the [18F]FAPI and [68 Ga]FAPI mouse study. Suppl. Information 7 provides more examples (see Suppl. Tables 9–11 and Suppl. Figures 3–5).
Visual and quantitative results assess the cross-tracer applicability of the M2Std CNN denoising algorithm. Left (a, d): visual comparison between standard-count, low-count, Gaussian-filtered and M2Std CNN- denoised images. Top part [18F]FAPI: tumor-to-muscle ratios determined from mean (b) and maximum (c) uptake values measured across the number of replicate images available at the particular count level. Bottom part [68 Ga]FAPI: (e) mean kBq/mL value (± standard deviation) in a liver VOI measured for one image replicate. f SNR in the liver reported as average (± standard deviation) measured across the number of replicate images available at the particular count level. For b, c, and f: statistical significance levels (p value ranges) are given for the pair-wise comparison between Gaussian filtering and M2Std CNN denoising.
Discussion
This paper presents an image-based DL framework for denoising low statistics micro-PET images which could be leveraged for reducing the injected radiotracer dose and/or performing faster scans. Qualitative and quantitative results validated that the presented DL denoising frameworks enable to perform total-body mouse PET studies with doses below 1 MBq (or equivalently with sub-1 min acquisitions). The motivation behind this study was to explore the potential and feasibility of DL denoising solutions for preclinical (small animal) imaging, given that almost all research efforts for DL-enabled low-dose imaging have been spent on clinical PET, while there is a strong advocacy for low-dose micro-PET imaging.
Low-dose capabilities have already been offered through system design adaptations. The β-CUBE (Molecubes) [18] and the Si78 (Bruker Biospin) [8] scanner with an axial length of 13 cm and 15 cm, respectively, provide full animal coverage and the increase in solid angle leads to a higher sensitivity. This allows to reduce dose by at least a factor of 4 [8] while offering comparable image quality as previous generations of micro-PET/CTs. In the clinic, DL denoising techniques have increasingly gained in popularity thanks to their promising potential to improve image quality with clear benefits for dose reduction. However, there has been very little effort to translate such frameworks to the preclinical imaging field [16, 17].
In this study, we successfully implemented a DL model for denoising low-count micro-PET images in seconds (see Suppl. Table 12 for training and testing computation times). The presented DL frameworks did not only improve image quality by reducing noise but also maintained quantitative accuracy and reproducibility for SUV measurements. Quantification results also demonstrated statistically significant enhancements in SUVmax recovery when CNN-based denoising was applied (compared to Gaussian, Median or Bilateral filtering), indicating the effectiveness of these algorithms in mitigating noise-induced variability, especially for reconstructions at 10% and 5% of the counts. Both X2Std and M2Std CNN algorithms showed similar performance in terms of image quality metrics, resulting in the lowest RMSE, highest PSNR, highest SSIM, and lowest MAE at most low-count levels compared to non-CNN denoising. The largest improvement relative to the filtering methods was found for the images reconstructed with 10% and 5% of the counts. The results from the NEMA-NU4 phantom showed that the M2Std CNN denoising model was able to maintain image uniformity and reliably reduce the noise present in the low-count image to a level equivalent to the standard-count image. The CRC measures indicated that the M2Std CNN model could hamper detectability for smaller structures (< 3 mm) but compared to Gaussian-filtering the M2Std CNN-denoised images achieved a higher contrast recovery at all rod diameter sizes. So, it is worth to highlight that while Gaussian filtering and CNN denoising showed similar noise reduction performance at modest count reductions (i.e., 50% and 25%), CNN-based approaches demonstrated better contrast recovery performance.
Visually compared to the standard-count image, the CNN denoising introduced a “smoother” noise (image) texture but quantitative analyses showed no significant impact on accuracy. Previous studies using U-Net models for low-dose clinical PET [14, 15] reported similar effects on their denoised images. The explanation might be that the standard (high) count images used as training targets are still plagued by some noise (not ground-truth), and when minimizing the voxel-wise error between CNN-predicted and target values, the CNN solution converges towards smoother images than the standard-count (target) image.
To train our DL networks, high-low matching image pairs were required and the methodology used in this work to obtain low-count datasets was through sequential sub-sampling of the list-mode data without considering the differences in random and scatter fraction estimates at low and standard doses. The approach is valid for emulating low-count true coincidence data which linearly scales with the in-field activity. However, the “real” contribution of randoms (which follow a quadratic relationship) and scatter (which becomes challenging for ultra-low-dose cases when noisy scatter tails can lead to overestimations of scatter) is neglected. To address this point, image quality evaluated with the NEMA-NU4 phantom was compared between low-count images obtained by reconstructions of shorter time frames and low-count images from “real” lower dose scans. In terms of the CNN-denoising performance, no differences were noted between low-count acquisitions from scan time and dose reduction (see Suppl. Information 9: Suppl. Table 13 and Suppl. Figures 6–9). Another point of discussion is that training image pairs, and all test mice data presented in the result section of this paper, were not attenuation corrected. In order to demonstrate that the inclusion of attenuation correction did not affect the denoising performance of the M2Std model, the NEMA-NU4 phantom was attenuation-corrected. Suppl. Figures 9 and 10 provides further results to show that the CNN denoising approach performed equally well on images with or without attenuation correction.
In some cases, the micro-PET study is combined with a micro-CT scan to provide an anatomical reference image or for attenuation correction. Typical micro-CT doses range from 50 to 1000 mGy [9] while [18F]FDG doses in mice are within 100 mGy [10]. Dose reduction strategies with DL denoising are also applicable in micro-CT imaging. In our previous work [24] a neural network was trained and evaluated for denoising low-dose micro-CT acquisitions to realize higher quality micro-CT imaging at reduced doses (by a factor of 3). In preclinical research, low-dose multi-modality studies offer promising future prospects for managing the cumulative effects of radiation in longitudinal imaging studies. Moreover, low-dose imaging provides an opportunity to image more rodents given a certain amount of radioactivity which is particularly advantageous for situations when only limited amounts of tracer are available for daily preclinical practice.
A critical concern with DL-based methods is its reproducibility and robustness to new (unknown) studies. The diversity of tracers, scanners, imaging protocols, and reconstruction settings used in practice continues to question the reliability of any trained DL model when applied to external datasets [12, 13]. In preclinical settings, we are faced with the extra challenge of increased diversity in radiotracers investigated among the variety of rodent models. To our knowledge, this paper is the first to study the extensible applicability of an [18F]FDG-only trained DL model to other radiopharmaceuticals including [18F]PSMA, [18F]FAPI and [68 Ga]FAPI studies. While image noise does not depend on the tracer distribution for different F-18 based tracers, the performance of the neural network to denoise images with different uptake patterns than those seen in the [18F]FDG training images was tested with [18F]PSMA and [18F]FAPI. Besides the notable improvement in image quality, quantitative assessments showed very good agreement in mean and maximum uptake values between the standard-count and M2Std CNN-denoised images with smaller bias and higher reproducibility compared to the low-count images. The results with [68 Ga]FAPI were also encouraging to further investigate how a DL-based denoising algorithm handles images produced using tracers of different resolutions. Ga-68 exhibits different physical properties (positron range, decay fractions, etc.) to F-18, so this will lead to differences in noise pattern and image resolution. Our findings suggest that the methodology is able to handle different types of studies, focusing on the broader impact, while not yet fully capturing or considering subtle differences that exist across different study scenarios.
This work only focused on static low-count micro-PET imaging. A future consideration is to investigate cases of ultra-low statistics. For early, very short (2–10 s) frames during fast dynamic studies, or for imaging studies with short-lived isotopes (e.g., O-15, C-11, Rb-82) which decay very fast during scanning, or with long-lived isotopes (e.g., Zr-89) which exhibit low positron fraction, even higher noise levels of varying textures will corrupt the data. There is another biologic argument to be made about delayed-time-point imaging studies where denoising can improve the visualization and quantification for low density targets (e.g., bacterial and antibody imaging applications). In this regard, an extensive phantom study evaluating the effect of DL-denoising with positron emitters other than F-18 [25] should be performed in the future to study the noise contribution from differences in positron ranges.
In summary, this paper demonstrates the feasibility of applying DL-based denoising techniques for low-count preclinical acquisitions of mice. While the image quality obtained with DL methods is notably improved especially at very low count levels, this work also focuses on the importance of quantitative results; low bias and high precision in SUV was measured with the CNN-denoised images. The adoption of DL-based denoising frameworks allows for low-dose and/or faster imaging which might be the goal for some preclinical set-ups. However, with other scenarios such as imaging with very short-lived radionuclides or imaging long after injection (with Zr-89 for antibody imaging), DL denoising can also prove valuable to extract more (quantitative) information under extreme conditions of ultra-low statistics.
Conclusions
Results support the merits of CNN-based image enhancement in low statistics micro-PET, especially for ultra-low-count images (reconstructed at 5% and 10% of the counts). The presented 2D CNN denoising approach is able to enhance micro-PET images of improved quality while keeping quantitative accuracy from noisy input images. Sub-1 MBq and sub-1 min [18F]FDG PET mouse imaging becomes possible with DL-based denoising. The adoption of these denoising frameworks holds potential for various scenarios where it is necessary to limit the amount of injected dose (in addition to the concerns related to toxicity and safety). This offers much more flexibility to the investigators in optimizing preclinical, longitudinal imaging setups.
References
Yao R, Lecomte R, Crawford ES (2012) Small-animal PET: what is it, and why do we need it? J Nucl Med Technol 40(3):157–165
Miyaoka RS, Lehnert AL (2020) Small animal PET: a review of what we have done and where we are going. Phys Med Biol 65(24):24TR04
Piron S, Verhoeven J, Courtyn J, Kersemans K, Descamps B, Pieters L, Vral A, Vanhove C, De Vos F (2022) Preclinical comparative study of [18F]AlF-PSMA-11 and [18F]PSMA-1007 in varying PSMA expressing tumors. Sci Rep 12(1):15744
Lee IK, Noguera-Ortega E, Xiao Z, Todd L, Scholler J, Song D, Liousia M, Lohith K, Xu K, Edwards KJ, Farwell MD, June CH, Albelda SM, Puré E, Sellmyer MA (2022) Monitoring therapeutic response to anti-FAP CAR T cells using [18F]AlF-FAPI-74. Clin Cancer Res 28(24):5330–5342
Lau J, Rousseau E, Kwon D, Lin KS, Bénard F, Chen X (2020) Insight into the development of PET radiopharmaceuticals for oncology. Cancers (Basel) 12(5):1312
Hume SP, Jones T (1998) Positron emission tomography (PET) methodology for small animals and its application in radiopharmaceutical preclinical investigation. Nucl Med Biol 25(8):729–732
Vanhove C, Bankstahl JP, Krämer SD, Visser E, Belcari N, Vandenberghe S (2015) Accurate molecular imaging of small animals taking into account animal models, handling, anaesthesia, quality control and imaging system performance. EJNMMI Phys 2(1):31. https://doi.org/10.1186/s40658-015-0135-y
Molinos C, Sasser T, Salmon P, Gsell W, Viertl D, Massey JC, Mińczuk K, Li J, Kundu BK, Berr S, Correcher C, Bahadur A, Attarwala AA, Stark S, Junge S, Himmelreich U, Prior JO, Laperre K, Van Wyk S, Heidenreich M (2019) Low-dose imaging in a new preclinical total-body PET/CT scanner. Front Med (Lausanne) 6:88
McDougald WA, Collins R, Green M, Tavares AAS (2017) High dose microCT does not contribute toward improved microPET/CT image quantitative accuracy and can limit longitudinal scanning of small animals. Front Phys 5:50. https://doi.org/10.3389/fphy.2017.00050
Taschereau R, Chatziioannou AF (2007) Monte Carlo simulations of absorbed dose in a mouse phantom from 18-fluorine compounds. Med Phys 34(3):1026–1036
Decuyper M, Maebe J, Van Holen R, Vandenberghe S (2021) Artificial intelligence with deep learning in nuclear medicine and radiology. EJNMMI Phys 8(1):81
Liu H, Wu J, Lu W, Onofrey JA, Liu YH, Liu C (2020) Noise reduction with cross-tracer and cross-protocol deep transfer learning for low-dose PET. Phys Med Biol 65(18):185006
Xue S, Guo R, Bohn KP, Matzke J, Viscione M, Alberts I, Meng H, Sun C, Zhang M, Zhang M, Sznitman R, El Fakhri G, Rominger A, Li B, Shi K (2022) A cross-scanner and cross-tracer deep learning method for the recovery of standard-dose imaging quality from low-dose PET. EJNMMI 49(6):1843–1856
Kaplan S, Zhu YM (2019) Full-dose PET image estimation from low-dose PET image using deep learning: a pilot study. J Digit Imaging 32(5):773–778
Lu W, Onofrey JA, Lu Y, Shi L, Ma T, Liu Y, Liu C (2019) An investigation of quantitative accuracy for deep learning based denoising in oncological PET. Phys Med Biol 64(16):165019
Dutta K, Liu Z, Laforest R, Jha A, Shoghi KI (2022) Deep learning framework to synthesize high-count preclinical PET images from low-count preclinical PET images. Medical Imaging 2022: Physics of Medical Imaging, vol 12031. SPIE, pp 351–360
Amirrashedi M, Sarkar S, Mamizadeh H, Ghadiri H, Ghafarian P, Zaidi H, Ay MR (2021) Leveraging deep neural networks to improve numerical and perceptual image quality in low-dose preclinical PET imaging. Comput Med Imaging Graph 94:102010
Krishnamoorthy S, Blankemeyer E, Mollet P, Surti S, Van Holen R, Karp JS (2018) Performance evaluation of the MOLECUBES β-CUBE-a high spatial resolution and high sensitivity small animal PET scanner utilizing monolithic LYSO scintillation detectors. Phys Med Biol 63(15):155013
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5-9, 2015, Proceedings, Part III 18. Springer International Publishing, pp 234–241
Nair V, Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines. Proceedings of the 27th International Conference on Machine Learning (ICML-10). pp 807–814
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.698
National Electrical Manufacturers Association (2008) Performance measurements of small animal positron emission tomographs (PETs). NEMA Standards Publication, NU4-2008. 1-23
Gavriilidis P, Koole M, Annunziata S, Mottaghy FM, Wierts R (2022) Positron range corrections and denoising techniques for gallium-68 PET imaging: a literature review. Diagnostics 12(10):2335
Muller FM, Maebe J, Vanhove C, Vandenberghe S (2023) Dose reduction and image enhancement in micro-CT using deep learning. Med Phys 50:5643
Teuho J, Riehakainen L, Honkaniemi A, Moisio O, Han C, Tirri M, Liu S, Grönroos TJ, Liu J, Wan L, Liang X, Ling Y, Hua Y, Roivainen A, Knuuti J, Xie Q, Teräs M, D’Ascenzo N, Klén R (2020) Evaluation of image quality with four positron emitters and three preclinical PET/CT systems. EJNMMI Res 10(1):155
Acknowledgements
The authors would like to thank Dr. Pieter Mollet and Dr. Bert Vandeghinste (Molecubes) for their technical support with the β-CUBE and Dr. Elizabeth Li and Dr. Srilalan Krishnamoorthy for their suggestions on data visualization. The authors would like to acknowledge support from Small Animal Imaging Facility at UPenn.
Funding
This work was supported by FM’s UGent BOF doctoral grant. FM is financially supported by the Belgian American Educational Foundation and Fulbright Foreign Student Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Florence M. Muller and Boris Vervenne are co-first authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Muller, F.M., Vervenne, B., Maebe, J. et al. Image Denoising of Low-Dose PET Mouse Scans with Deep Learning: Validation Study for Preclinical Imaging Applicability. Mol Imaging Biol 26, 101–113 (2024). https://doi.org/10.1007/s11307-023-01866-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11307-023-01866-x