Abstract
Purpose
This study aims to compare two approaches using only emission PET data and a convolution neural network (CNN) to correct the attenuation (μ) of the annihilation photons in PET.
Methods
One of the approaches uses a CNN to generate μ-maps from the non-attenuation-corrected (NAC) PET images (μ-CNNNAC). In the other method, CNN is used to improve the accuracy of μ-maps generated using maximum likelihood estimation of activity and attenuation (MLAA) reconstruction (μ-CNNMLAA). We investigated the improvement in the CNN performance by combining the two methods (μ-CNNMLAA+NAC) and the suitability of μ-CNNNAC for providing the scatter distribution required for MLAA reconstruction. Image data from 18F-FDG (n = 100) or 68 Ga-DOTATOC (n = 50) PET/CT scans were used for neural network training and testing.
Results
The error of the attenuation correction factors estimated using μ-CT and μ-CNNNAC was over 7%, but that of scatter estimates was only 2.5%, indicating the validity of the scatter estimation from μ-CNNNAC. However, CNNNAC provided less accurate bone structures in the μ-maps, while the best results in recovering the fine bone structures were obtained by applying CNNMLAA+NAC. Additionally, the μ-values in the lungs were overestimated by CNNNAC. Activity images (λ) corrected for attenuation using μ-CNNMLAA and μ-CNNMLAA+NAC were superior to those corrected using μ-CNNNAC, in terms of their similarity to λ-CT. However, the improvement in the similarity with λ-CT by combining the CNNNAC and CNNMLAA approaches was insignificant (percent error for lung cancer lesions, λ-CNNNAC = 5.45% ± 7.88%; λ-CNNMLAA = 1.21% ± 5.74%; λ-CNNMLAA+NAC = 1.91% ± 4.78%; percent error for bone cancer lesions, λ-CNNNAC = 1.37% ± 5.16%; λ-CNNMLAA = 0.23% ± 3.81%; λ-CNNMLAA+NAC = 0.05% ± 3.49%).
Conclusion
The use of CNNNAC was feasible for scatter estimation to address the chicken-egg dilemma in MLAA reconstruction, but CNNMLAA outperformed CNNNAC.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Various physical and patient factors such as attenuation, scattering, and motion need to be corrected to accurately estimate the distribution of radioactive tracers in positron emission tomography (PET). As the scattered line-of-response (LOR) distribution is usually estimated based on the linear attenuation coefficient map (attenuation map: μ-map) for 511-keV annihilation photons [1,2,3], accurate μ-map generation is important for both attenuation correction (AC) and scatter correction (SC). In dual-modality hybrid PET systems, computed tomography (CT) or magnetic resonance (MR) images are converted into μ-maps with nearly no statistical noise [4,5,6,7,8,9]. However, CT artifacts often cause errors in attenuation-corrected PET images [10,11,12]. Additionally, the accuracy of MR-based PET AC has been proven only in adult brain PET images with normal anatomy [13, 14]. However, this has not yielded satisfactory results in whole-body scans [15, 16]. The spatial mismatch between the emission PET and μ-maps derived from CT or MR images is another source of error in anatomical image-based PET AC [17,18,19].
Deep learning (DL)-based PET AC methods using only PET emission data have several advantages over anatomical image-based AC methods [14]. DL-based emission-only approaches are free of errors due to the spatial mismatch between the emission and transmission data [14, 20, 21]. They can also be applied to standalone PET systems (e.g., brain-dedicated PET) without CT or MR images [22, 23]. One of the emission-only approaches is to use the deep neural network(s) to generate pseudo-CT or μ-maps from non-attenuation-corrected (NAC) PET images [20, 24, 25]. Although NAC PET images do not contain explicit information about photon attenuation, deep neural networks could predict μ-maps, including bone structures. However, the NAC PET-based method has shown relatively high error in the lungs, which generally has large inter-individual variability in μ-values [25]. Similarly, a method directly generating CT-based AC PET images from NAC PET without attenuation map generation has been proposed [26,27,28,29]. However, this approach is vulnerable to outliers and fails to recover quantitative accurate activity around the center of the head with complex anatomical structures [26]. There is an alternative approach to obtaining both AC PET and attenuation map from NAC PET [30]. However, this study was limited to 2D-based learning, which suffers from the problem of discontinuity across the slices [30].
Another DL-based emission-only approach is to improve the accuracy of μ-maps generated by simultaneous reconstruction of activity and attenuation only from emission PET data [21, 31,32,33,34]. Simultaneous activity and attenuation reconstruction has evolved by incorporating time-of-flight information into the sub-iterations estimating the activity distribution to apply spatial constraints on the activity origin [35,36,37,38]. The maximum likelihood estimation of activity and attenuation (MLAA) is an effective algorithm for simultaneous reconstruction [37]. However, the high noise level in the μ-map and the crosstalk between the activity and attenuation distribution are the main limitations of the MLAA algorithm, currently suffering from insufficient timing resolution of PET systems. To overcome the limitations of MLAA, we proposed a DL-based approach and improved the accuracy of the MLAA μ-map and the corresponding activity image (λ) [31,32,33]. Moreover, because the MLAA μ-maps are generated using monoenergetic 511-keV annihilation photons, metal artifacts caused by low-energy photon starving in X-ray CT are not observed in the DL-enhanced MLAA μ-maps [14].
Another limitation of the MLAA AC method is the chicken-egg dilemma of the scatter estimation [39]. Scatter event distribution needs to be known to conduct the MLAA. However, estimating scatter events requires μ-maps. Thus, scatter events were derived from CT μ-maps (μ-CT) and assumed to be known in our previous studies [31, 32]. This is a critical limitation.
This study’s purpose is three-fold. The first is to investigate whether the scatter distribution estimated from NAC PET activity images using DL is compatible with that estimated using the CT and the single-scatter simulation (SSS) algorithm [1, 2]. The second is to compare the two emission-only approaches (NAC and MLAA) proposed for the DL-based whole-body PET AC. Finally, the study addresses whether the accuracy of the DL-based whole-body PET AC improves by combining the NAC and MLAA approaches.
Materials and methods
Dataset
Image data from 150 oncology patients who underwent the 18F-FDG (n = 100; 38 men and 62 women; age, 57.3 ± 14.1 years) or 68 Ga-DOTATOC (n = 50; 29 men and 21 women; age, 53.5 ± 14.2 years) PET/CT scans were used for the training and testing of the neural network. The dataset was divided into training, validation, and test sets, as summarized in Table 1. The networks were trained separately for each tracer. Whole-body PET/CT scans were acquired using a Biograph mCT 40 scanner (Siemens Healthineers, Knoxville, TN; timing resolution = 580 ps) 60 min after the intravenous injection of the tracer (5.18 MBq/kg for 18F-FDG and 2.78 MBq/kg for 68 Ga-DOTATOC). Six-eight bed positions were used to cover the upper body in the PET scans with a scan time of 1 min per position. The institutional review board of our institute approved the retrospective use of the scan data and waiver of the need for informed consent.
The CT images, reconstructed in a 512 × 512 × 100 matrix and 1.52 × 1.52 × 2.03 mm voxel size, were converted into the μ-map for 511-keV photons (μ-CT; 200 × 200 × 109; 4.07 × 4.07 × 2.03 mm). Ground truth PET activity images (λ-CT) were reconstructed using an ordered-subset expectation maximization (OSEM) algorithm (3 iterations and 21 subsets, 5-mm Gaussian postfilter) with CT-based AC and SC. The CT-based scatter estimates were generated using the SSS algorithm. The correction factors were generated, and the OSEM reconstruction was performed using the vendor-supplied e7 toolkit. The size of the reconstructed PET images was 200 × 200 × 109 (4.07 × 4.07 × 2.03 mm voxel size) for each bed position.
The NAC PET activity images (λ-NAC) were reconstructed using the OSEM algorithm with the same reconstruction parameters, but AC and SC were not applied. The numbers of iteration and subset for MLAA reconstruction producing activity and attenuation maps (λ-MLAA and μ-MLAA) were 6 and 21, respectively. A boundary constraint was applied during the μ-MLAA estimation to resolve the problem of global scaling that is not unique in MLAA [37].
Network architectures
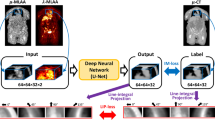
Convolutional neural networks (CNNs) were designed to predict the μ-CT, ground truth, from λ-NAC, λ-MLAA, and μ-MLAA (Fig. 1). The architecture of the CNNs was based on U-net [40], which is widely used in medical image processing analysis [41,42,43,44]. The CNNs’ architectures, except for the number of the input channel, were the same as those used in our previous study [31]. Network architecture detail is provided in Fig. 1 in [31]. The CNN’s inputs were the 32 × 32 × 32 matrix patches extracted from λ-NAC, λ-MLAA, and μ-MLAA. The training labels were equally sized patches from the μ-CT at that location. We stacked the inferences from the input patches on the image matrix to construct the output image with the trained network (μ-CNN). Patch-based min–max normalizations were performed on λ-NAC, λ-MLAA, and μ-MLAA before feeding the input patch to the network.
Strategies for attenuation (μ) map generation using CNNs. Non-attenuation-corrected PET activity image (λ-NAC) was used as input for the CNN in (A). Results (λ-MLAA and μ-MLAA) of the MLAA simultaneous reconstruction algorithm were used as CNN inputs in (B) and (C). All the λ-NAC, λ-MLAA, and μ-MLAA were used as CNN inputs in (D). Scatter distributions estimated using μ-CT were used in (B), but those estimated using the μ-map generated by CNNNAC (CNN output in A) were used in (C) and (D). Here, “Em” and “Sc” stand for the emission and scatter sinograms
Figure 1 compares the μ-map generation strategies used in this study. The CNNNAC takes λ-NAC as the input to produce a synthetic μ-map, μ-CNNNAC (Fig. 1A). The CNNMLAA* takes λ-MLAA* and μ-MLAA*, corrected for scatter using μ-CT, and produces μ-CNNMLAA* (Fig. 1B; a method used in our previous studies [31, 32]). The CNNMLAA takes the λ-MLAA and μ-MLAA corrected for scatter using μ-CNNNAC and produces the μ-CNNMLAA (Fig. 1C). Finally, the CNNMLAA+NAC takes λ-NAC along with λ-MLAA and μ-MLAA to produce μ-CNNMLAA+NAC (Fig. 1D). Note that the third and final methods do not require μ-CT, as scatter correction is performed using μ-CNNNAC.
Network training
The L1-norm between the output (μ-CNN) and ground truth (μ-CT) was chosen as the loss function for training the networks and was minimized using the adaptive moment estimation method. A learning rate of 0.001 was used as an initial value and decayed every two epochs with rate of 0.92. We adopted a batch size of 64 patches for all experiments. 3D patches for training the networks were selected randomly from the input images. To avoid meaningless computation with blank patches, the 3D patches whose centers were included in the body were only employed for the networks. Approximately 4000 patches for each bed were used for training. Each network was trained using a training set with a maximum of 200 epochs. When the training loss calculated using the validation set did not decrease for consecutive 10 epochs, training was stopped, and performance of the model was evaluated using the test set. The networks were implemented using the TensorFlow library and trained using NVIDIA RTX 3090 (24 GB VRAM).
Scatter estimate comparison
We compared the SSS scatter estimates derived from the μ-CNNNAC and μ-CT to determine whether the scatter estimates using μ-CNNNAC can solve the chicken-egg dilemma in the MLAA. The accuracy of the attenuation and scatter estimation was compared in the sinogram space in terms of absolute and percentage errors as AC and SC are performed in the sinogram space during the MLAA. The MLAA reconstruction and CNN-enhancement results obtained using scatter estimates from μ-CNNNAC and μ-CT were compared.
Comparison of attenuation and activity estimates
The attenuation and activity estimates obtained using the CNNs shown in Fig. 1 were compared to the ground truth using three different metrics: the structural similarity index measure (SSIM), peak signal-to-noise ratio (PSNR), and normalized root mean square error (NRMSE):
where \(\mu\), \(\sigma\), and \(c\) are average, standard deviation, and predefined constant. We used the default function for SSIM from MATLAB 2020b. \(MAX\) and \(MSE\) are the maximum intensity and mean square error. \({x}_{k}\) and \({\widehat{x}}_{k}\) are the \(k\)-th voxel of generated image and ground truth, respectively. Here, \(VOI\) is set to the patient body.
The voxel-wise correlation between the DL-based approaches and the ground truth was also estimated. The mean μ-values of the lungs and the standard uptake value (SUV) of lung and bone lesions were also compared to assess the accuracy of DL-based approaches further. The boundaries of the lungs were segmented from the μ-CT and eroded considering the mismatch between the PET and CT due to respiratory motion to calculate the mean μ-values of the lungs. Additionally, volumes of interest (VOIs) were semi-automatically drawn on 23 suspected lung cancer and 29 suspected bone cancer regions in the 18F-FDG PET scans of 20 patients by applying a threshold of 40% of the maximum SUV on the tumor, determined by averaging the SUV of the voxels with an SUV higher than 90% of the SUV peak. VOIs were drawn on λ-CT (reference images), and these VOIs were utilized in the other reconstructed images for evaluating SUV quantification.
Results
Scatter estimation using μ-CNNNAC
Figure 2 and Supplemental Fig. 1 compare the attenuation correction factors (ACFs) and scatter distributions estimated using μ-CT and μ-CNNNAC (μ-map inferred from NAC activity image using CNN as illustrated in Fig. 1A) and show the percent and absolute differences between them. While the mean squared percent error of the ACF was higher than 7% (7.2% ± 4.1%), that of the scatter estimates was only 2.5% ± 0.1%, indicating the validity of the scatter estimation using the μ-CNNNAC. Figure 3A compares the μ-MLAA* and μ-MLAA, which are the μ-maps estimated using the MLAA simultaneous reconstruction algorithm for which the scatter was estimated from μ-CT and μ-CNNNAC, as illustrated in Fig. 1B and C. As shown in Fig. 3B that compares μ-CNNMLAA* and μ-CNNMLAA, the difference between μ-MLAA* and μ-MLAA was further reduced by applying the CNN to the output images of the MLAA. The data shown in Figs. 2 and 3 have been obtained using 18F-FDG PET scans.
Attenuation maps
Figure 4 and Supplemental Figs. 2 show the sagittal and coronal slices of the CNN models’ input, output, and ground truth images for the 68 Ga-DOTATOC and 18F-FDG studies, respectively. Although the bone structures were not clearly resolved and the noise levels were high in the input images (λ-MLAA, μ-MLAA, and λ-NAC), the CNNs provided nearly noiseless μ-maps with improved bone delineation. As indicated by the orange arrows in Fig. 4, the CNN with only NAC input (CNNNAC) generated less accurate bone structures in the μ-maps for the 68 Ga-DOTATOC studies showing lower bone uptake and higher noise level than in the 18F-FDG studies. The best results for recovering the fine bone structures were obtained by providing all λ-MLAA, μ-MLAA, and λ-NAC to the CNN (CNNMLAA+NAC), as indicated by the white arrows in Fig. 4 and Supplemental Figs. 2.
Figure 5 shows the advantages of providing λ-MLAA and μ-MLAA as inputs to the CNN. The μ-map generation errors, indicated by orange arrows in the μ-CNNNAC, were observed less frequently in the μ-CNNMLAA and μ-CNNMLAA+NAC, which also resulted in better soft tissue and fat contrast than μ-CNNNAC, as shown in Fig. 5C. However, abdominal air was often misclassified as fat or soft tissue in all the CNN models.
The voxel-wise correction and quantitative measurements of the similarity between μ-CNNs and μ-CT confirmed the qualitative comparison results. As shown in Table 2 and Supplemental Figs. 3 and 4, the μ-CNNMLAA and μ-CNNMLAA+NAC achieved better voxel-wise correlation, higher PSNR and SSIM, and lower NRMSE than the μ-CNNNAC, which showed an especially poor correlation between the μ-values corresponding to the lung tissues. Figure 6 shows the percent errors of μ-CNNs relative to the μ-CT in whole lung tissues, indicating the overestimation and increased variability of μ-values using the CNNNAC in the lung. The difference in the performance of the CNN models was smaller in the 68 Ga-DOTATOC than in 18F-FDG studies. Figure 7 shows the μ- and λ-maps of a lung cancer patient who underwent 18F-FDG PET/CT study, demonstrating that the abnormal hot uptake in lung lesions prevented proper inference of the μ-map by CNNNAC.
Activity images
The activity images corrected for attenuation using μ-CNNMLAA and μ-CNNMLAA+NAC were also superior to those corrected using μ-CNNNAC in terms of their similarity to λ-CT, as shown in Supplemental Figs. 5, 6, and 7 (voxel-wise correlation plots, percent difference maps, and quantitative similarity measures (PSNR, SSIM, and NRMSE) between λ-CNNs and λ-CT) and Table 3. The improvement in the similarity with λ-CT achieved by employing λ-NAC in addition to λ-MLAA and μ-MLAA was not significant (CNNMLAA versus CNNMLAA+NAC).
Supplemental Fig. 8 shows the correlation between the SUV measurements in lung cancer lesions. λ-CT shows the highest correlation with λ-CNNMLAA+NAC. Although λ-CNNNAC was also correlated with λ-CT, it showed a higher positive bias and variability in the regional SUV than the other methods (percent error: λ-CNNNAC = 5.45% ± 7.88%; λ-CNNMLAA = 1.21% ± 5.74%; λ-CNNMLAA+NAC = 1.91% ± 4.78%). Supplemental Fig. 9 shows the correlation between SUV measurements in bone cancer lesions, showing no significant differences among results obtained using the different methods (percent error: λ-CNNNAC = 1.37% ± 5.16%; λ-CNNMLAA = 0.23% ± 3.81%; λ-CNNMLAA+NAC = 0.05% ± 3.49%).
Discussion
This study compared two approaches using only the emission PET data and a CNN to correct the attenuation of annihilation photons in PET: one used a CNN to generate μ-maps from NAC PET images (μ-CNNNAC), and in the other method, CNN was used to improve the accuracy of μ-maps generated through MLAA reconstruction (μ-CNNMLAA). It also investigated whether the CNN performance is improved by combining the two methods (μ-CNNMLAA+NAC) and whether μ-CNNNAC would be suitable for providing the scatter distribution required for MLAA reconstruction.
The use of CNN to generate μ-maps from NAC PET images is a relatively straightforward approach because it does not require special image reconstruction algorithms such as MLAA. Therefore, this method can be applied to any PET data, regardless of the PET scanner’s time-of-flight measurement capability. Additionally, this method allows for joint attenuation and scatter correction [26]. The feasibility of this method for brain PET studies using 18F-FDG and other tracers has been demonstrated by several groups using various DL models, including convolutional autoencoder and generative adversarial networks [20, 24, 25]. Recently, Dong et al. demonstrated the potential of this method in whole-body 18F-FDG PET studies. However, the errors in their study were large in the lung, mainly due to the heterogeneity and inter-individual variability of lung density [25]. The current study also highlights similar limitations of this method for whole-body PET scans, especially in the lung and metallic implants (Figs. 5, 6 and 7).
However, the μ-CNNNAC was useful in estimating the scatter distribution needed when applying the MLAA. Although the ACF error between the μ-CT and μ-CNNNAC was relatively high, the error of the scatter distribution in the sinogram space, estimated using the μ-maps, was only 2.5% on average. Therefore, the μ-CNNNAC appears to be a promising solution for addressing the chicken-egg dilemma [39] in MLAA reconstruction.
The results of this study show the superiority of the μ-CNNMLAA over the μ-CNNNAC in many ways. Bone and metallic implants were better delineated, and the error in tissue classification was smaller when applying μ-CNNMLAA (Figs. 4, 5, 6 and 7). This resulted in the improvement of the similarity between the reconstructed images and the ground truth and the accuracy in the quantitation of tumor SUVs (Supplemental Figs. 5-7). The difference in the accuracy between the μ-CNNMLAA and μ-CNNNAC was most pronounced in the lungs, as shown in Figs. 6 and 7. The difference in performance between the CNNMLAA and CNNNAC approaches was less significant in the 68 Ga-DOTATOC PET studies than in the 18F-FDG, potentially because of the erroneous estimation of the μ-map using the CNNNAC in the lung was mainly caused by the abnormal hot uptake in lung lesions (Fig. 7). This was less prevalent in the 68 Ga-DOTATOC PET studies than in 18F-FDG PET.
Interestingly, the CNNMLAA was able to predict metallic hip implants, despite there being no implants in any of the cases included in the training set (Fig. 5D). Moreover, streaking artifacts appearing around the metal on the μ-CT due to low-energy photon starvation were not observed in the μ-CNNMLAA. However, the μ-values of the metallic implants in the μ-CNNMLAA were slightly lower than those in the μ-CT, leading to an ~ 5% SUV difference in the lesions near the metallic implants. This is thought to be due to the lack of training data as high as those of the metal implants with the μ-values. Therefore, further investigation with the training and test sets, including many metallic implant cases, which are properly corrected for metal artifacts in CT, is necessary to improve the quantitative accuracy of the CNNMLAA approach.
No significant improvement in the similarity with the λ-CT by combining the CNNNAC and CNNMLAA approaches was observed in this study. Additional λ-NAC input to the CNN, along with the λ-MLAA and μ-MLAA, resulted in a better prediction of fine bone structures in the μ-maps (Fig. 4 and Supplemental Fig. 2). However, the improvement of the quantitative similarity measures on the μ- and λ-maps by the combined inputs was not as evident as the difference between the CNNNAC and CNNMLAA approaches (Fig. 6 and Supplemental Figs. 3-9).
An alternative approach for AC is the MRI-based methods, including the segmented-based and the atlas-based method [6, 45]. The use of CNN combined with MLAA in AC shows same or better performance compared to the MRI-based AC methods. Martinez-Moller et al. and Arabi et al. reported SUV quantification in the osseous lesions, which were an average decrease of 8.0% ± 3.3% using the segmentation-based method and an average increase of 1.5% ± 3.5% using the atlas-based method, respectively [6, 45]. The errors in CNNMLAA and CNNMLAA+NAC were only 0.23% ± 3.81% and 0.05% ± 3.49%. Martinez-Moller et al. reported that SUV in the lung lesions was underestimated, with an average decrease of 1.9% ± 2.3% [6], while errors in CNNMLAA and CNNMLAA+NAC were 1.21% ± 5.74% and 1.91% ± 3.49%. No evaluation for lung lesions was not performed in Arabi et al..
Different neural networks were trained and used individually in this study for two tracers to compare the emission-only approaches under the best conditions for each tracer. However, this resulted in the overfitting of the neural networks suitable only for a specific tracer and requiring network retraining for new tracers. We tried training the U-net model with two tracers to create a more general model, but the results were worse than the individual training results. Further investigation is required to develop a general model that provides the optimal performance for all tracers.
Summary and conclusion
We compared two DL-based approaches to PET AC that use only emission data. The use of CNNNAC for scatter estimation successfully addressed the chicken-egg dilemma in MLAA reconstruction. However, the CNNMLAA outperformed the CNNNAC. The benefits of combining these two approaches were not significant.
References
Ollinger JM. Model-based scatter correction for fully 3D PET. Phys Med Biol. 1996;41:153–76. https://doi.org/10.1088/0031-9155/41/1/012.
Watson CC. New, faster, image-based scatter correction for 3D PET. IEEE Trans Nucl Sci. 2000;47:1587–94.
Accorsi R, Adam LE, Werner ME, Karp JS. Optimization of a fully 3D single scatter simulation algorithm for 3D PET. Phys Med Biol. 2004;49:2577–98. https://doi.org/10.1088/0031-9155/49/12/008.
Kinahan PE, Townsend DW, Beyer T, Sashin D. Attenuation correction for a combined 3D PET/CT scanner. Med Phys. 1998;25:2046–53. https://doi.org/10.1118/1.598392.
Townsend DW. Dual-modality imaging: combining anatomy and function. J Nucl Med. 2008;49:938–55. https://doi.org/10.2967/jnumed.108.051276.
Martinez-Möller A, Souvatzoglou M, Delso G, Bundschuh RA, Chefd’hotel C, Ziegler SI, et al. Tissue classification as a potential approach for attenuation correction in whole-body PET/MRI: evaluation with PET/CT data. J Nucl Med. 2009;50:520–6.
Keereman V, Fierens Y, Broux T, De Deene Y, Lonneux M, Vandenberghe S. MRI-based attenuation correction for PET/MRI using ultrashort echo time sequences. J Nucl Med. 2010;51:812–8.
Catana C, van der Kouwe A, Benner T, Michel CJ, Hamm M, Fenchel M, et al. Toward implementing an MRI-based PET attenuation-correction method for neurologic studies on the MR-PET brain prototype. J Nucl Med. 2010;51:1431–8.
An HJ, Seo S, Kang H, Choi H, Cheon GJ, Kim H-J, et al. MRI-based attenuation correction for PET/MRI using multiphase level-set method. J Nucl Med. 2016;57:587–93.
Sureshbabu W, Mawlawi O. PET/CT imaging artifacts. J Nucl Med Technol. 2005;33:156–61.
Beyer T, Bockisch A, Kühl H, Martinez M-J. Whole-body 18F-FDG PET/CT in the presence of truncation artifacts. J Nucl Med. 2006;47:91–9.
Goerres GW, Ziegler SI, Burger C, Berthold T, Von Schulthess GK, Buck A. Artifacts at PET and PET/CT caused by metallic hip prosthetic material. Radiology. 2003;226:577–84. https://doi.org/10.1148/radiol.2262012141.
Ladefoged CN, Law I, Anazodo U, Lawrence KS, Izquierdo-Garcia D, Catana C, et al. A multi-centre evaluation of eleven clinically feasible brain PET/MRI attenuation correction techniques using a large cohort of patients. Neuroimage. 2017;147:346–59.
Lee JS. A review of deep-learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Radiat Plasma Med Sci. 2021;5:160–84. https://doi.org/10.1109/TRPMS.2020.3009269.
Kim JH, Lee JS, Song I-C, Lee DS. Comparison of segmentation-based attenuation correction methods for PET/MRI: evaluation of bone and liver standardized uptake value with oncologic PET/CT data. J Nucl Med. 2012;53:1878–82.
Samarin A, Burger C, Wollenweber SD, Crook DW, Burger IA, Schmid DT, et al. PET/MR imaging of bone lesions–implications for PET quantification from imperfect attenuation correction. Eur J Nucl Med Mol Imaging. 2012;39:1154–60.
Lodge MA, Mhlanga JC, Cho SY, Wahl RL. Effect of patient arm motion in whole-body PET/CT. J Nucl Med. 2011;52:1891–7.
Beyer T, Antoch G, Blodgett T, Freudenberg LF, Akhurst T, Mueller S. Dual-modality PET/CT imaging: the effect of respiratory motion on combined image quality in clinical oncology. Eur J Nucl Med Mol Imaging. 2003;30:588–96. https://doi.org/10.1007/s00259-002-1097-6.
Osman MM, Cohade C, Nakamoto Y, Wahl RL. Respiratory motion artifacts on PET emission images obtained using CT attenuation correction on PET-CT. Eur J Nucl Med Mol Imaging. 2003;30:603–6. https://doi.org/10.1007/s00259-002-1024-x.
Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for (18)F-FDG PET attenuation correction. Eur J Nucl Med Mol Imaging Phys. 2018;5:24. https://doi.org/10.1186/s40658-018-0225-8.
Shi L, Onofrey JA, Revilla EM, Toyonaga T, Menard D, Ankrah J, et al. A novel loss function incorporating imaging acquisition physics for PET attenuation map generation using deep learning. Proc Med Image Comput Comput Assist Interv. 2019:723–31.
Bauer CE, Brefczynski-Lewis J, Marano G, Mandich MB, Stolin A, Martone P, et al. Concept of an upright wearable positron emission tomography imager in humans. Brain Behav. 2016;6:e00530.
Tashima H, Yoshida E, Iwao Y, Wakizaka H, Maeda T, Seki C, et al. First prototyping of a dedicated PET system with the hemisphere detector arrangement. Phys Med Biol. 2019;64:065004. https://doi.org/10.1088/1361-6560/ab012c.
Armanious K, Kustner T, Reimold M, Nikolaou K, La Fougere C, Yang B, et al. Independent brain (18)F-FDG PET attenuation correction using a deep learning approach with Generative Adversarial Networks. Hell J Nucl Med. 2019;22:179–86. https://doi.org/10.1967/s002449911053.
Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. 2019;64:215016. https://doi.org/10.1088/1361-6560/ab4eb7.
Arabi H, Bortolin K, Ginovart N, Garibotto V, Zaidi H. Deep learning-guided joint attenuation and scatter correction in multitracer neuroimaging studies. Hum Brain Mapp. 2020;41:3667–79. https://doi.org/10.1002/hbm.25039.
Dong X, Lei Y, Wang T, Higgins K, Liu T, Curran WJ, et al. Deep learning-based attenuation correction in the absence of structural information for whole-body PET imaging. Phys Med Biol. 2019. https://doi.org/10.1088/1361-6560/ab652c.
Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, et al. Deep-JASC: joint attenuation and scatter correction in whole-body (18)F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging. 2020;47:2533–48. https://doi.org/10.1007/s00259-020-04852-5.
Yang J, Park D, Gullberg GT, Seo Y. Joint correction of attenuation and scatter in image space using deep convolutional neural networks for dedicated brain (18)F-FDG PET. Phys Med Biol. 2019;64:075019. https://doi.org/10.1088/1361-6560/ab0606.
Hu Z, Li Y, Zou S, Xue H, Sang Z, Liu X, et al. Obtaining PET/CT images from non-attenuation corrected PET images in a single PET system using Wasserstein generative adversarial networks. Phys Med Biol. 2020;65:215010. https://doi.org/10.1088/1361-6560/aba5e9.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60:1183–9.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9. https://doi.org/10.2967/jnumed.117.202317.
Hwang D, Kim KY, Kang SK, Choi H, Seo S, Paeng JC, et al. Accurate attenuation correction for whole-body Ga-68-DOTATOC PET studies using deep learning. J Nucl Med. 2019;60:568.
Hwang D, Kang SK, Kim KY, Choi H, Seo S, Lee JS. Data-driven respiratory phase-matched PET attenuation correction without CT. Phys Med Biol. 2021;66:115009.
Chun SY, Kim KY, Lee JS, Fessier JA. Joint estimation of activity distribution and attenuation map for TOF-PET using alternating direction method of multiplier. Proc IEEE Int Symp Biomed Imaging. 2016:86–9.
Defrise M, Rezaei A, Nuyts J. Time-of-flight PET data determine the attenuation sinogram up to a constant. Phys Med Biol. 2012;57:885–99. https://doi.org/10.1088/0031-9155/57/4/885.
Rezaei A, Defrise M, Bal G, Michel C, Conti M, Watson C, et al. Simultaneous reconstruction of activity and attenuation in time-of-flight PET. IEEE Trans Med Imaging. 2012;31:2224–33. https://doi.org/10.1109/tmi.2012.2212719.
Salomon A, Goedicke A, Schweizer B, Aach T, Schulz V. Simultaneous reconstruction of activity and attenuation for PET/MR. IEEE Trans Med Imaging. 2011;30:804–13. https://doi.org/10.1109/tmi.2010.2095464.
Rezaei A, Schramm G, Willekens SMA, Delso G, Van Laere K, Nuyts J. A Quantitative evaluation of joint activity and attenuation reconstruction in TOF PET/MR brain imaging. J Nucl Med. 2019;60:1649–55. https://doi.org/10.2967/jnumed.118.220871.
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Proc Med Image Comput Comput Assist Interv. 2015:234–41.
Hegazy MAA, Cho MH, Cho MH, Lee SY. U-net based metal segmentation on projection domain for metal artifact reduction in dental CT. Biomed Eng Lett. 2019;9:375–85. https://doi.org/10.1007/s13534-019-00110-2.
Lee MS, Hwang D, Kim JH, Lee JS. Deep-dose: a voxel dose estimation method using deep convolutional neural network for personalized internal dosimetry. Sci Rep. 2019;9:10308. https://doi.org/10.1038/s41598-019-46620-y.
Park J, Bae S, Seo S, Park S, Bang JI, Han JH, et al. Measurement of glomerular filtration rate using quantitative SPECT/CT and deep-learning-based kidney segmentation. Sci Rep. 2019;9:4223. https://doi.org/10.1038/s41598-019-40710-7.
Park J, Hwang D, Kim KY, Kang SK, Kim YK, Lee JS. Computed tomography super-resolution using deep convolutional neural network. Phys Med Biol. 2018;63:145011. https://doi.org/10.1088/1361-6560/aacdd4.
Arabi H, Zaidi H. Whole-body bone segmentation from MRI for PET/MRI attenuation correction using shape-based averaging. Med Phys. 2016;43:5848–61. https://doi.org/10.1118/1.4963809.
Funding
This study was supported by grants issued by the National Research Foundation of Korea (NRF), funded by the Korean Ministry of Science and ICT (Grant No. NRF-2016R1A2B3014645) and the Korea Medical Device Development Fund grant funded by the Korea government (the Ministry of Science and ICT, the Ministry of Trade, Industry and Energy, the Ministry of Health & Welfare, the Ministry of Food and Drug Safety) (Project Number: 1711137868, KMDF_PR_20200901_0006 ). The funding sources were not involved in the study design, collection, analysis, or interpretation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The institutional review board of Seoul National University Hospital approved the retrospective use of the scan data and waiver of the need for informed consent.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Advanced Image Analyses (Radiomics and Artificial Intelligence)
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hwang, D., Kang, S.K., Kim, K.Y. et al. Comparison of deep learning-based emission-only attenuation correction methods for positron emission tomography. Eur J Nucl Med Mol Imaging 49, 1833–1842 (2022). https://doi.org/10.1007/s00259-021-05637-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00259-021-05637-0