Abstract
The microbial composition of polyurethane degrading communities has been barely addressed, and it is unknown if microenvironmental conditions modify its composition, affecting its biodegradative capacity. The polyurethanolytic activity and taxonomic composition of five microbial communities, selected by enrichment in the polyether-polyurethane-acrylic (PE-PU-A) coating PolyLack®, from deteriorated PU foams collected at different microenvironments in a municipal landfill (El Bordo Poniente, BP) were explored. All BP communities grew similarly in PolyLack® as the sole carbon source, although BP1, BP4, and BP5 showed better performance than BP2 and BP7. FTIR spectroscopy showed that ester, urethane, ether, aromatic and aliphatic groups, and the acrylate component were targets of the biodegradative activity. Extracellular esterase activity was higher at 5 days of cultivation and decreased at 21 days, while urease activity showed the opposite. Microbial composition analysis, assessed by 16S rDNA V3 region PCR-DGGE, revealed a preponderance of Rhizobiales and Micrococcales. The reported PU-degrading genera Paracoccus, Acinetobacter, and Pseudomonas were identified. In contrast, Advenella, Bordetella, Microbacterium, Castellaniella, and Populibacterium, some of them xenobiotics degraders, can be considered potentially PU-degrading genera. Correspondence analysis identified independent groups for all communities, except the BP4 and BP5. Although partial taxonomic redundancy was detected, unique OTUs were identified, e.g., three members of the Weeksellaceae family were present only in the BP4/BP5 group. These results suggest that the microenvironmental conditions where the landfill microbial communities were collected shaped their taxonomical composition, impacting their PE-PU biodegradative capacities. These BP communities represent valuable biological material for the treatment of PU waste and other xenobiotics.
Key points
• Landfill microbial communities display slightly different capacities for growing in polyether-polyurethane-acrylic.
• Ester, urethane, ether, aromatic, aliphatic, and acrylate groups were attacked.
• Esterase activity was more significant at early culture times while urease activity at latter.
• Landfill microenvironments shape partial taxonomical redundancy in the communities.
• Best communities’ performance seems to be related to unique members’ composition.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plastics are among the most used xenobiotic materials in our daily lives. They are synthetic polymers with high resilience and a long lifespan, and one of the most widespread pollutants on Earth (Ali et al. 2021). Polyurethanes (PU) are plastics synthesized by condensation between polyols and diisocyanates, and, depending on the polyol, they can be classified as polyester (PS) or polyether (PE) type. PU are widely used as shoe soles, foams, sponges, building and refrigerator insulators, coatings, adhesives, wettings, elastomers, synthetic leathers, tires, gaskets, bumpers, and rubber goods (Akindoyo et al. 2016). PU have been ranked as the sixth most used polymer, with a global production of 30 million tons in 2019 (PlasticsEurope 2020). The massive PU amounts produced and their high persistence, because of their non-degradable nature, have led to an excessive increase in waste that threatens diverse ecosystems’ integrity (Ali et al. 2021). Even though several physical or chemical treatments for various PU wastes exist, biodegradation can be considered an incipient green alternative. Many isolated bacterial strains and fungal species can attack PS-PU, but few microorganisms attack PE-PU, which occurs at a slow rate because of its high recalcitrance (reviewed by Magnin et al. 2020; Roy et al. 2021).
In nature, microbial communities are of great importance due to their multifunctional degradation capabilities (Leadbeater et al. 2021; Raimundo et al. 2021). Moreover, various microbial communities can degrade different organic pollutants and xenobiotics, such as aryloxyphenoxypropionate herbicides, polycyclic aromatic hydrocarbons, and polyethylene or polystyrene films (Aziz et al. 2018; Dong et al. 2017; Obafemi et al. 2018; Syranidou et al. 2019). However, few studies on microbial communities capable of degrading PU have been published, and most of them have focused on the degradation of PS-PU. The microbial composition of PS-PU-degrading communities has been addressed microscopically, biochemically, or by 16S rDNA gene sequencing analyses, in microorganisms acting on films in liquid mineral medium (Shah et al. 2008), on films in compost (Das et al. 2017), and growing in acrylic and PS-PU coatings (Vargas-Suárez et al. 2019). Also, by using denaturing gradient gel electrophoresis (DGGE), terminal restriction fragment length polymorphisms, and pyrosequencing, the microbial composition of fungal communities growing on the surface of PS-PU coupons, either buried in soil or compost, has been deciphered (Cosgrove et al. 2007; Zafar et al. 2013). Moreover, by using bacterial species isolated from soil, consortia capable of degrading PS-PU films (Shah et al. 2016) or lignin-modified PS-based thermoplastic PU (Fernandes et al. 2016) have been assembled.
In contrast, microbial communities capable of biodegrading PE-PU have been less studied. Limited biodegradation of PE-PU foams by microbial communities from garbage landfill leakage water was observed based on changes in weight and FTIR patterns (Filip 1978), and similarly, soil microorganisms caused a discrete weight loss in soil buried PE-PU foams (Ge et al. 2000). In contrast, an aerobic thermophilic consortium from the sludge of a wastewater treatment plant showed a limited capability to degrade PE-PU elastomeric films, as evaluated by weight loss and microscopic analysis (Obruca et al. 2011). A microbial consortium isolated from the soil where PU foam was buried could degrade pulverized PE-PU foams at a low rate, as measured by O2 consumption, CO2 release, and Raman spectroscopy (Cregut et al. 2014). Unfortunately, none of the previously described studies approached the microbial communities’ composition. The landfill community BP8 was recently shown to degrade an aromatic polyether-polyurethane-acrylic (PE-PU-A) copolymer and the xenobiotic additives present in a commercial coating (PolyLack®), as demonstrated by different chemical and physical analyses. The microbial composition of the BP8 community was explored by Hi-C proximity ligation technology, revealing that landfills are a valuable source for finding novel microorganisms potentially involved in PU biodegradation (Gaytán et al. 2020). Therefore, exploring landfill ecosystems for discovering other potential PU-degrading microorganisms will increase the reservoir of biological specimens with this capability.
The microbial diversity of communities and their functions are influenced by environmental conditions (Sekhohola-Dlamini and Tekere 2020). However, in landfill microbial communities, the influence of microenvironmental conditions on PU biodegradation performance and their microbial composition have not been addressed yet. This work investigated the polyurethanolytic capability of five microbial communities (BP1, BP2, BP4, BP5, and BP7), isolated from different pieces of deteriorated PU foams collected from different microenvironments in a municipal landfill, to biodegrade the PE-PU-A coating PolyLack®. We measured the esterase and urease activities associated with the PU biodegradative capacity and investigated the communities’ microbial composition to assess their polyurethane biodegradation performance and discover novel potential PU-degrading microorganisms.
Materials and methods
Selection of microbial communities and growth quantification
Microbial communities were selected from five deteriorated PU foam samples collected at El Bordo Poniente (BP) landfill, near Mexico City, and named correspondingly, BP1, BP2, BP4, BP5, and BP7 (Fig. 1). The PU foams were collected at different sites representing microhabitats with distinct microenvironmental conditions. Collection of foam samples, enrichment procedure, and establishment of experimental cultures were previously described (Gaytán et al. 2020). Enrichment and culture were performed in a mineral medium (MM) (components in mmol L−1 were KH2PO4, 14.7; K2HPO4, 40.2; NH4NO3, 12.5; MgSO4·7H2O, 0.4; ZnSO4·7H2O, 0.0035; CuSO4·7H2O, 0.0004; FeSO4·7H2O, 0.036; MnSO4·6H2O, 0.0077; pH 7.2) (Nakajima-Kambe et al. 1995) supplemented with the PE-PU-A coating PolyLack® Aqua Brillante (Sayer Lack, Prod. Num. UB-0800, México) (0.3% w/v) as the only carbon source (MM-PolyLack). The BP communities’ growth in MM-PolyLack was measured over a 120-h period by quantifying dry biomass weight. For that, cultures were pelleted, the cells were washed three times with phosphate buffer (50 mM, pH 7) and dried in a vacuum oven at 65 °C to constant weight (Gaytán et al. 2020). Each experiment was done in triplicate.
Foam samples collect at El Bordo Poniente landfill (near Mexico City). a Size comparison between a truck and the garbage piles at El Bordo Poniente landfill. b A general landscape. c Landfill leachate drained from garbage piles as a result of rotting and decomposition of organic matter. d Manual collection of samples. e–i Foam samples collected at different microenvironments and used as inocula for enrichment selection in MM-PolyLack. e BP1, f BP2, g BP4, h BP5, and i BP7
Fourier-transform infrared spectroscopy analysis
The action of the BP communities on MM-PolyLack was analyzed at 21 days in cell-free supernatants. Briefly, cells were pelleted at 6723 × g at 4 °C for 15 min, and 1 mL aliquots of supernatants were evaporated to complete dryness at 37 °C. The remaining powder was analyzed in a 1605 spectrometer (Perkin Elmer), from 800 to 4000 cm−1. Supernatants from non-inoculated media were processed similarly and used as negative controls. For each condition, experiments were done in triplicate. Identification of functional groups in the FTIR spectra was performed according to McCarthy et al. (1997).
Enzymatic assays
Selected communities growing in MM-PolyLack were harvested at 5 and 21 days by centrifugation at 11,953 × g at 4 °C for 15 min. The remaining PolyLack in the supernatants was pelleted at 208,400 × g at 4 °C for 45 min. For better sample volumes’ handling, supernatants were first concentrated in an ultrafiltration cell (Model 8400, Amicon) with a 10-kDa ultrafiltration membrane and then in an Amicon Ultra-4 10-kDa filter, where they were washed with 50 mL of 50 mM potassium phosphate buffer (pH 7). Esterase and urease activities were measured spectrophotometrically in triplicate as previously described (Vargas-Suárez et al. 2019). Esterase activity was determined at 405 nm by hydrolysis of p-nitrophenyl acetate (p-NPA), and urease activity was determined using a phenol hypochlorite assay, quantifying ammonia release at 636 nm. Lipase from Aspergillus oryzae (0.1 U) (Sigma Aldrich, Cat. Num. 62285) and Canavalia ensiformis urease (0.25 U) (Sigma Aldrich, Cat. Num. U-1500) were used as positive controls in esterase and urease assays, respectively. According to the Bradford method (Bradford 1976), protein concentration was measured using bovine serum albumin as standard. Non-inoculated MM-PolyLack cultures were processed similarly and used as negative controls.
Statistical analysis
The data obtained from the community growth experiments and the enzymatic assays were submitted to statistical analysis (ANOVA One Way) using the software Statistica version 7 (StatSoft, Inc. 2004). The normality of the data and homogeneity of variance was verified using the Kolmogorov–Smirnov and Brown-Forsythe tests, respectively (Massey 1951; Brown and Forsythe 1974). Multiple comparisons were carried out based on Duncan’s and Tukey’s tests. The level of significance was set as α < 0.05.
DNA extraction and PCR amplification of the 16S rDNA V3 region
Genomic DNA extraction was carried out from 1.5 mL aliquots of MM-PolyLack cultures incubated for 48 h, according to Ausubel et al. (1994), with minor changes. Cells were pelleted, resuspended in 450 μL TE buffer, and lysed with 50 μL lysozyme (10 μg μL−1) (Sigma) and 8 μL RNAse A (25 μg μL−1 Sigma) for 1 h at 37 °C, and later incubated with 50 μL SDS (10% w/v) for 35 min at 37 °C. Subsequent additions of 6 μL proteinase K (20 μg μL−1 Thermo Scientific™) incubating for 30 min at 37 °C; 100 μL 5 M NaCl incubating at 65 °C for 10 min and 100 μL 10% (w/v, in 0.7 M NaCl) hexadecyltrimethylammonium bromide (Sigma) incubating at 65 °C for 15 min were done. Subsequent steps were as described in the original protocol. The V3 region of the 16S rDNA genes was PCR amplified with buffer HF (Thermo Scientific™) 1 × , 0.2 mM dNTPs, 0.02 U μL−1 Phusion high-fidelity DNA polymerase (Thermo Scientific™), 0.4 μM of each primer, 341F-GC (5’ CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG 3’) and 518R (5’ ATTACCGCGGCTGCTGG 3’), and 200 ng of genomic DNA from each community in a 50 μL final volume (Muyzer et al. 1993). PCR conditions were as follows: initial denaturation at 98 °C for 3 min; 30 cycles of denaturation at 98 °C for 10 s, annealing at 67.7 °C for 30 s, and extension at 72 °C for 30 s; and a final extension at 72 °C for 10 min, run in a Veriti thermocycler (Applied Biosystems). The expected amplicons were 220 bp long.
Denaturing gradient gel electrophoresis (DGGE), V3 region PCR reamplification, and bacterial identification
DGGE was run in polyacrylamide gels (8%, acrylamide:bisacrylamide 37.5:1) in a 35–70% gradient (urea and formamide) at 60 V, 60 °C for 16 h, in the DCode™ Universal Mutation Detection System (BioRad), according to the manufacturer instructions. The total PCR reactions from each community were concentrated to a final volume of 20 μL by evaporation (Centrivap, Labconco) and loaded in the gel wells. After gel running, the resolved bands were silver stained, and the more conspicuous ones were cut off with sterile razor blades. From selected bands, DNA was extracted by elution with 50 μL sterile deionized water for 1 h at 37 °C. Eluates were used as templates for PCR reamplification of the 16S rDNA V3 region using the conditions described above. Each reamplified amplicon was cloned into the EcoRV site of pBS-KS plasmid using T4 DNA ligase (Thermo Scientific™) in 30-μL reactions, and E. coli DH5α competent cells were consequently transformed with each construction. White/blue screening identified colonies containing the cloned insert. Plasmids were verified for expected cloned inserts by using EcoRI and HindIII restriction enzymes (Thermo Scientific™). At least two transformants were selected for plasmids purification with the PCR GeneJet purification kit (Thermo Scientific™) from each reamplified band. Plasmid DNA aliquots (100 ng μL−1) were sequenced at Macrogen, Inc. (Seoul, South Korea) with the primer M13F supplied by the service provider. Sequences were analyzed with ApE software (A plasmid Editor, version 2.0.47, M. Wayne Davis), and the regions between primers 341F and 518R were examined (150–190-bp length). Megablast compared the V3 regions’ sequences to the non-redundant nucleotide collection (nr/nt) database (NCBI) to explore neighbors’ taxa against the 500 best matches as initial seed. The search was restricted to the subset from type material (intended for cross-species comparisons), excluding uncultured/environmental sample sequences; other algorithm parameters were set as default. We used a correspondence analysis between communities and the minimum taxa level assigned to all the analyzed sequences to explore the influence of the microenvironment on communities’ structure. For this, a presence/absence matrix of standardized frequencies was constructed (multi-way Community*Order cross-tabulation table). The standardization was based on row and column coordinates profiles, and the number of dimensions was set to 2 (StatSoft, Inc. 2004). For establishing taxonomic associations, a cladogram with significant aligned sequences was constructed based on the distances retrieved by the Jukes-Cantor model. Intermediary trees were then built using the Neighbor-Joining method (Saitou and Nei 1987) to identify regular groupings and close phylogenetic neighborhoods.
BP communities’ deposit and 16S rDNA V3 region accession numbers
The BP communities were deposited in the Culture Collection at Cepario Facultad de Química, UNAM, World Data Centre for Microorganisms CFQ100, under the accession numbers: BP1 community CFQ-B-291; BP2 community, CFQ-B-292; BP4 community, CFQ-B-293; BP5 community, CFQ-B-294; BP7 community, CFQ-B-295. Nucleotide sequences of the 16S rDNA V3 region obtained from DGGE were deposited in the GenBank database under the accession numbers MT123979 to MT124065.
Results
Some BP communities exhibit different growths and PE-PU-A biodegradation
The capability of the BP communities to use PolyLack as the sole carbon source was estimated by measuring cell growth. The five BP communities showed similar growth tendencies in MM-PolyLack at early cultivation times, and they reached the stationary phase around 48 h. However, since that time, statistically, significantly better performances (α < 0.05) were observed for the BP4 community (1.03 mg dry weight mL−1) at 48 h and BP5 community at 96 h (1.21 mg dry weight mL−1). At 120 h, the highest growth of BP5, BP4, and BP1 were statistically significant with 1.31, 1.24, and 1.21 mg dry weight mL−1, respectively, over BP2 and BP7 that showed the lowest growth, with 0.88 and 0.94 mg dry weight mL−1, respectively (Fig. 2).
The capacity of the BP communities for degrading the PE-PU-A polymer was demonstrated by analyzing its structural changes at 21 days of culture by FTIR spectroscopy. All BP communities generated changes in the PE-PU-A functional groups compared to non-inoculated control, although at different intensities (Fig. 3). At 3260 cm−1, associated with the N–H stretch, BP1 decreased the signals, whereas BP2, BP4, BP5, and BP7 increased them, suggesting the generation of amines due to urethane hydrolysis. At 2919 and 2851 cm−1, associated with the aliphatic CH2 stretch, BP2 and BP4 almost disappeared the signals whereas BP1, BP5 (only at 2919 cm−1), and BP7 decreased them, indicating cleavage of the carbon backbone. At 1726 cm−1, associated with the C = O stretch from urethane and acrylate carbonyl groups, all communities generated significant decrements, pointing out the hydrolysis of urethanes and acrylates. At 1600 cm−1, associated with the stretching of C = O and C = C from aromatic groups, BP2 and BP4 seemed to disappear the signal, indicating cleavage of aromatic rings. At 1532 (N–H bending plus C-N stretch) and 1223 cm−1 (C-N stretching from the urethane group), BP1 generated a significant decrease in the signals, BP5 and BP7 small decreases, BP2 and BP4 disappeared them, indicating urethane attack. All the communities disappeared the 1452 cm−1 signal, associated with the CH2 bending of the aliphatic carbons, indicating C–C backbone cleavage. Signals at 1104 cm−1 associated with the urethane C–O–C stretch and 977 cm−1 associated with the C–O–C symmetric stretch, respectively, were also affected. The 1104 cm−1 signal was diminished by BP7, whereas the 977 cm−1 signal slightly increased in all the communities. These changes suggest cleavage of the ether bond. The 850 cm−1 signal, associated with the vinyl group C = CH2 out of the plane from acrylate, was increased by all communities, implying the acrylate component’s cleavage.
FTIR spectra of PolyLack attacked by microbial BP communities. Spectra were obtained from MM-PolyLack cultures’ supernatants inoculated with BP1, BP2, BP4, BP5, or BP7 communities. The non-inoculated medium was used as the negative control. Samples were taken after 21 days of incubation at 37 °C and 220 rpm. n = 3
Extracellular esterase and urease activities are displayed during BP communities’ growth in PolyLack
PU biodegradation involves attack to C = O and C-N bonds performed by esterases and ureases, respectively (reviewed by Magnin et al. 2020). To assess the involvement of these enzymatic activities in PolyLack’s PE-PU-A polymer biodegradation, we measured esterase and urease extracellular activities in cell-free supernatants of BP1, BP2, and BP7 grown in MM-PolyLack at 5 and 21 days. These communities were chosen based on the contrasting intensities of the urethane group’s FTIR signals at 1532/1223 cm−1. The specific differences between the communities were short signals in BP1, absent signals in BP2, and extended signals in BP7. Each enzymatic activity exhibited a particular pattern that was qualitatively similar in the three communities. Esterase activity was high at 5 days and decreased at 21 days of incubation, 56% for BP1 (from 11.9 to 5.2 mmol h−1 mg protein−1), and 64% for BP2 (from 18.1 to 6.5 mmol h−1 mg protein−1), but remained almost similar for BP7 (13.1 and 12.8 mmol h−1 mg protein−1 at 5 and 21 days, respectively). Esterase activity was statistically significantly higher for BP2 at 5 days (α < 0.05), followed by BP1 and BP7 at 5 days and BP7 at 21 days (Fig. 4a). On the contrary, urease activity increased during the same period: in BP1, 1.3-fold (from 14 to 19 μmol h−1 mg protein−1); in BP2, 2.5-fold (from 8 to 20 μmol h−1 mg protein−1); and in BP7, 2.2-fold (from 9 to 20 μmol h−1 mg protein−1). Statistical analysis showed that urease activity for BP1 at 5 days and the three communities at 21 days was significantly higher (α < 0.05) (Fig. 4b).
Extracellular enzymatic activities of BP communities growing in PolyLack. a Esterase and b urease activities from cell-free supernatants of BP1 (white columns), BP2 (gray columns), and BP7 (black columns) communities, cultured in MM-PolyLack (0.3%), were measured at 0 (no activity detected), 5, and 21 days. n = 3. Bars represent standard deviation. The same letters indicate no statistical differences between the conditions
BP communities share some OTUs, but some others are unique and reveal novel, potentially PU-degrading taxa
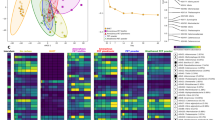
We explored the microbial composition of the polyurethanolytic BP communities by the culture-independent PCR-DGGE fingerprinting analysis. This technique resolved DNA bands from microorganisms present in the BP communities, showing differences and similarities among them (Fig. 5a). In our experience, bands located at a similar position in the different DGGE gel lanes not always corresponded to the same microorganisms (Fig. 5a). Therefore, no co-migration analysis based on bands’ position was carried out. The analysis was based on identifying sequences at the closer taxonomic category possible by sequencing the DNA bands eluted from the gel and PCR amplified. From a total of 87 sequenced clones, 51 were assigned. Identical sequences in one community were considered redundant, whereas if found in different communities, they were considered unique because of the different environmental contexts. From them, 21 sequences were assigned to 8 genera (Acinetobacter, Advenella, Bordetella, Castellaniella, Microbacterium, Paracoccus, Populibacterium, and Pseudomonas), 8 sequences to 6 families (Microbacteriaceae, Micrococcaceae, Phyllobacteriaceae, Propionibacteriaceae, Rhodobacteraceae, and Weeksellaceae), and 22 sequences to 2 orders (Micrococcales and Rhizobiales) (Table 1).
Since the deteriorated foams harboring the BP communities were collected in landfill areas with different microenvironmental conditions (Fig. 1), we explored whether these differences influenced the microbial composition. Therefore, we examined the degree of taxonomic relatedness by correspondence analysis at the order level since it was the taxonomic category at which all the identified 16S rDNA V3 sequences were assigned. This analysis detected four groups, each corresponding to one community, except that BP4 and BP5 were clustered. The four groups shared Micrococcales and Rhizobiales; Burkholderiales, which were present in all except BP1; BP1 and BP2 shared Pseudomonadales; BP2 and BP7, Rhodobacterales; Flavobacteriales was exclusive of BP4/BP5 group; and Propionibacteriales was exclusive of BP2 group. The BP2 community was the one that showed the broader microbial landscape (Fig. 5b).
Identical sequences identified in the BP communities were grouped into operational taxonomic units (OTUs), and their phylogenetic relationships are shown as a cladogram where taxa assigned to each of the resulting 20 OTUs are indicated (Fig. 6). A variable number of sequences corresponding to distinct BP communities, depicted in the cladogram by different colors, are shown for each OTU. Taxa associated with seven of these OTUs are shared by several BP communities as follows: four genera, Advenella (BP4, BP5, and BP7), Microbacterium (BP1 and BP2), Paracoccus (BP2 and BP7), Populibacterium (BP2 and BP5); one family, Weeksellaceae (BP4 and BP5); two orders, Rhizobiales (BP1, BP2, BP4, BP5, and BP7), and Micrococcales (BP4 and BP7). The other nine taxa associated with the OTUs were exclusively found in one community, i.e., Acinetobacter (MT 123990) in BP1; Bordetella (MT124048), Castellaniella (MT124040), Pseudomonas (MT124027), Propionibacteriaceae (MT124029), Micrococcaceae (MT124038), Microbacteriaceae (MT124031) and Rhodobacteraceae (MT124033) in BP2; and Phyllobacteriaceae (MT124056) in BP4. Three taxa Populibacterium, Micrococcales, and Rhizobiales, were associated with more than one OTU representing paraphyletic groups. Most of the sequences within each OTU (66.66%) corresponded to the phylum Proteobacteria: 23 to Alphaproteobacteria, 9 to Betaproteobacteria, and 2 to Gammaproteobacteria; 14 sequences (27.45%) to the phylum Actinobacteria; and 3 sequences (5.88%) to the phylum Bacteroidetes.
Cladogram based on 16S rDNA V3 region sequences. Keys at the branches’ tips are the accession number for each sequence, clustered as OTUs (different bracketed sequences groups with 100% identity). Names at the far right are the taxa assigned to each OTU. Colors on the accession numbers correspond to the different BP communities. Taxa redundancy is distinguished by the association of a specific taxon with two or more communities (multicolored OTUs). Sequences assigned to Micrococcales, Populibacterium, and Rhizobiales that were clustered in different OTUs represent paraphyletic groups
Discussion
The polyurethanolytic activity and the microbial composition of five landfill microbial communities, BP1, BP2, BP4, BP5, and BP7, isolated from deteriorated foams by culture enrichment in a medium with a polyether-polyurethane-acrylic coating as the only carbon source, were analyzed. Since the foam samples were collected at different microenvironments from the landfill (Fig. 1), we hypothesized we would find some differences in the biodegradative capacity and the microbial composition in the communities, which will allow us to identify some taxa with better PU biodegradative capacity. The BP communities grew similarly in PolyLack, although BP4, BP5, and, to some extent, BP1 showed significantly better performance at different times during the analysis (Fig. 2). Because cell growth reflects the abilities to degrade substrates and assimilate their products (Lucas et al. 2008), the BP communities’ capacity to thrive in PolyLack indicates they can be taxonomically redundant, sharing biochemical pathways. However, some communities’ significantly better performance suggests that specific taxa with more effective PU-degrading biochemical capacities could exist in them. The capability of the BP communities to degrade the PE-PU-A from PolyLack was evidenced by attacking not only the more susceptible ester bonds but also aliphatic and aromatic carbons and the acrylate component, and the more recalcitrant urethane and ether groups (Fig. 3). Previously, we reported the effects of the BP1h and BP7h communities, selected from the same deteriorated foams as BP1 and BP7, generated on polyacrylic and PS-PU coatings (Vargas-Suárez et al. 2019). Subsequently, we reported the biodegradative activity of the BP8 community, selected from another foam collected at the same landfill on PolyLack (Gaytán et al. 2020). These works have shown that the selected BP communities share the capability to attack PU functional groups and grow in PU coatings as the only carbon source, although, to a different extent, reflecting a common biochemical background but also specific particularities. Therefore, the slightly different but significantly better performance of some communities seems to be based on specific taxa with better PU biodegradation abilities.
The direct participation of esterases in PU biodegradation has been well documented (Amobonye et al. 2021; Magnin et al. 2020). Genetic evidence showed that in a Pseudomonas aeruginosa mutant, growth impairment in PU-diol and reduced biodegradation were associated with reduced esterase activity (Mukherjee et al. 2011). In other work, three recombinant polyester hydrolases with esterase-lipase activity, cloned from two Thermobifida species, hydrolyzed Impranil DLN, a PS-PU coating, and caused significant weight losses and surface cracks on the thermoplastic PS-PU Elastollan B85A-10 and C85A-10, as a result of ester bonds cleavage (Schmidt et al. 2017). However, less evidence has been published about the action of ureases/urethanases in PU biodegradation. Recently, from a collection of 50 hydrolases, a combination of amidase E4143 and esterase E3576 degraded a polycaprolactone polyol-based PU by inducing deep cracks on the polymer surface more effectively than the esterase alone (Magnin et al. 2019). As part of the enzymatic mechanisms, the BP communities may display for degrading PolyLack, esterase, and urease activities were assayed in the extracellular fractions of the BP2 and BP7 communities selected for their different activity over the urethane groups (signals at 1532/1223 cm−1) (Fig. 4). Similarly, as we previously observed (Pérez-Lara et al. 2016; Vargas-Suárez et al. 2019), higher esterase and lower urease activities were detected at early cultivation times than at later stages. Interestingly, the BP2 community, which reduces the urethane signals more effectively, was the one that showed the highest esterase activity at early cultivation time and urease activity at later. However, the last one was not statistically different from those displayed by the other communities. From these results, we hypothesized that the different behavior shown by esterase and urease activities should be related to the requirements of the biochemical processes for attacking PU. Currently, we are addressing this subject in our laboratory. Based on these data, it is feasible that the extracellular esterase and urease activities exhibited by the BP1, BP2, and BP7 communities degrade the PE-PU-A present in PolyLack.
The microbial composition analysis of the BP communities studied in this work (Table 1) revealed that Proteobacteria and Actinobacteria followed by a small proportion of Bacteroidetes populate them, similarly to the reported profiles of diverse municipal landfills (Sekhohola-Dlamini and Tekere 2020). Six from the eight genera assigned, Acinetobacter, Advenella, Bordetella, Microbacterium, Paracoccus, and Pseudomonas, have already been reported as degraders of diverse xenobiotics (Lee et al. 2018; Li et al. 2020; Zhao et al. 2012; Yadav et al. 2020; Wang et al. 2007), whereas the other two, Castellaniella and Populibacterium, have not yet been. From them, Pseudomonas has been reported in PU biodegradation (Howard et al. 2012), Acinetobacter in acrylics (Kawai 1993) and PU biodegradation (Wilkes and Aristilde 2017), whereas Paracoccus has been identified as a member of several PU-degrading communities (Faccia et al. 2021; Gaytán et al. 2020; Vargas-Suárez et al. 2019). Thus, Advenella, Bordetella, Castellaniella, Microbacterium, and Populibacterium are potentially PU-degrading genera. The BP communities were isolated from decomposing foams collected at a municipal landfill, where hundreds of pollutants of diverse chemical compositions are dumped, and their presence exerts selective pressure on microbial composition. Therefore, many of the specimens that could not be identified at the genera level might display some xenobiotic degradation capacity.
When comparing the microbial composition of BP1 and BP7 communities grown in MM-PolyLack to that of BP1h and BP7h, selected from the same PU foam samples but cultured in MM-Bayhydrol a polyacrylic coating, and in MM-NeoRez a PS-PU coating, respectively (Vargas-Suárez et al. 2019), some differences were observed. In the BP1 community, one Acinetobacter, three Microbacterium, and five Rhizobiales were identified, while in BP1h, only two Acinetobacter could be isolated. Similarly, in BP7, two Advenella, two Paracoccus, and one Rhizobiales were identified, whereas from BP7h Acinetobacter, Bacillus, Hydrogenophaga, Microbacterium, and Paracoccus were isolated. These differences can be based on the different PU coatings used in the analyses. It is known that the type of nutrients available in an environment drives different microbial community structures (Mello et al. 2016; Zuo et al. 2020). Nevertheless, the different approaches used in these studies, i.e., culture-free bacterial identification (DGGE) in BP communities compared to strain isolation by dilution and streaking in a rich medium for BPh communities, cannot be ruled out as the reason for these differences. On the other hand, more considerable microbial composition differences were observed between the BP communities analyzed in this work and the BP8 community cultured in identical conditions but selected from another foam, representing another microenvironment. In the BP8 work, a Hi-C metagenomic sequencing analysis was performed though (Gaytán et al. 2020). From 16 genera identified in BP8, only Bordetella and Paracoccus were shared with BP2 and Paracoccus with BP7. Since these communities were cultured under identical conditions in MM-PolyLack, their different microbial composition reflects the diversity of the indigenous landfill bacteria, driven by the selection pressure imposed in the different microhabitats where the foam samples were collected. Studies showing that ecological factors influence the microbial diversity of municipal solid waste landfills have been published (Sekhohola-Dlamini and Tekere 2020).
While several BP communities shared some taxa such as Microbacterium, Populibacterium, Advenella, Paracoccus, Micrococcales, and Rhizobiales, others had unique members (Fig. 6). The taxonomic redundancy detected could explain the shared capability of the BP communities to thrive in PolyLack. Considering that the closer the phylogenetic distance between microorganisms, the greater the degree of metabolic overlapping (Hester et al. 2019), it can be deduced that the communities sharing phylogenetically related taxa would exhibit similar metabolic pathways or key biochemical reactions to biodegrade PE-PU-A and xenobiotics, i.e., functional redundancy. However, the unique microbial members could provide a community with better performance, exerting their influence directly or through syntrophic interactions. The exclusive presence of members of the Weeksellaceae family, from the Bacteroidetes phylum, existing only in the BP4/BP5 group, which showed the best growth in PolyLack (Fig. 2), suggests that their metabolic activity could be the basis for this difference. The Weeksellaceae family includes the genus Chryseobacterium, which was identified in the metagenome of the BP8 community also thriving in PolyLack (Gaytán et al. 2020). Our laboratory is currently addressing this possibility.
BP1, BP2, BP4, BP5, and BP7s are landfill microbial communities isolated from deteriorated PU foams collected at different microhabitats, which thrive in a PE-PU-A coating-containing medium as the sole carbon source. These communities degrade the PE-PU-A copolymer by cleaving different functional groups, including ester and the more recalcitrant urethane groups, presumably mediated by esterases and ureases, active during the cultivation period. Proteobacteria was the most abundant phylum in the BP communities, with Micrococcales and Rhizobiales as the more redundant orders. Data revealed several already reported xenobiotics- and PU-degrading genera (Acinetobacter, Paracoccus, and Pseudomonas) and novel, potentially PU-degrading genera (Advenella, Bordetella, Castellaniella, Microbacterium, and Populibacterium). Partial taxonomic redundancy, observed in the BP communities, sustains functional redundancy concerning the polyether-polyurethane-acrylic polymer biodegradation. However, unique microbial members could lead to more efficient PU-degrading communities providing distinctive and more effective enzymatic activities. The BP communities are valuable biological materials, either as a whole or selecting some of their members for assembling dedicated consortia, for using them to treat PU waste, or for xenobiotics bioremediation.
Data availability
Data sharing does not apply to this article as no datasets were generated or analyzed during the current study.
References
Akindoyo JO, Beg M, Ghazali S, Islam M, Jeyaratnam N, Yuvaraj A (2016) Polyurethane types, synthesis and applications–a review. RSC Adv 6:114453–114482
Ali SS, Elsamahy T, Koutra E, Kornaros M, El-Sheekh M, Abdelkarima EA, Zhu D, Sun J (2021) Degradation of conventional plastic wastes in the environment: a review on current status of knowledge and future perspectives of disposal. Sci Total Environ 771:144719
Amobonye A, Bhagwat P, Singh S, Pillai S (2021) Plastic biodegradation: frontline microbes and their enzymes. Sci Total Environ 759:143536
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current protocols in molecular biology. Wiley , USA
Aziz A, Agamuthu P, Alaribeb FO, Fauziah SH (2018) Biodegradation of benzo[a]pyrene by bacterial consortium isolated from mangrove sediment. Environ Technol 39:527–535
Bradford MM (1976) Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Brown MB, Forsythe AB (1974) Robust tests for the equality of variances. J Am Stat Assoc 69:364–367
Cosgrove L, McGeechan PL, Robson GD, Handley PS (2007) Fungal communities associated with degradation of polyester polyurethane in soil. Appl Environ Microbiol 73:5817–5824
Cregut M, Bedas M, Assaf A, Durand-Thouand MJ, Thouand G (2014) Applying Raman spectroscopy to the assessment of the biodegradation of industrial polyurethanes wastes. Environ Sci Pollut Res Int 21:9538–9544
Das S, Pandey P, Mohanty S, Kumar S, Nayak SK (2017) Evaluation of biodegradability of green polyurethane/nanosilica composite synthesized from transesterified castor oil and palm oil based isocyanate. Int Biodeterior Biodegrad 117:278–288
Dong W, Liu K, Wang F, Xin F, Zhang W, Zhang M, Wu H, Ma J, Jiang M (2017) The metabolic pathway of metamifop degradation by consortium ME-1 and its bacterial community structure. Biodegradation 28:181–194
Faccia PA, Pardini FM, Agnello AC, Amalvy JI, Del Panno MT (2021) Degradability of poly(ether-urethanes) and poly(ether-urethane)/acrylic hybrids by bacterial consortia of soil. Int Biodeterior Biodegrad 160:105205
Fernandes IP, Barbosa M, Amaral JS, Pinto V, Rodrigues JL, Ferreira MJ, Barreiro MF (2016) Biobased additives as biodegradability enhancers with application in TPU-based footwear components. J Renew Mater 4:47–56
Filip Z (1978) Decomposition of polyurethane in garbage landfill leakage water and by soil microorganisms. Appl Microbiol Biotechnol 5:225–231
Gaytán I, Sánchez-Reyes A, Burelo M, Vargas-Suárez M, Liachko I, Press M, Sullivan S, Cruz-Gómez MJ, Loza-Tavera H (2020) Degradation of recalcitrant polyurethane and xenobiotic additives by a selected landfill microbial community and its biodegradative potential revealed by proximity ligation-based metagenomic analysis. Front Microbiol 10:2986
Ge J, Zhong W, Guo Z, Li W, Sakai K (2000) Biodegradable polyurethane materials from bark and starch. I. Highly resilient foams. J Appl Polym Sci 77:2575–2580
Hester ER, Jetten MSM, Welte CU, Lücker S (2019) Metabolic overlap in environmentally diverse microbial communities. Front Genet 10:989
Howard GT, Norton WN, Burks T (2012) Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme. Biodegradation 23:561–573
Kawai F (1993) Bacterial degradation of acrylic oligomers and polymers. Appl Microbiol Biotechnol 39:382–385
Leadbeater DR, Oates NC, Bennett JP, Li Y, Dowle AA, Taylor JD, Sanchez Alponti J, Setchfield AT, Alessi AM, Helgason T, McQueen-Mason SJ, Bruce NC (2021) Mechanistic strategies of microbial communities regulating lignocellulose deconstruction in a UK salt marsh. Microbiome 9:48
Lee Y, Jeong SE, Hur M, Ko S, Jeon CO (2018) Construction and evaluation of a Korean native microbial consortium for the bioremediation of diesel fuel-contaminated soil in Korea. Front Microbiol 9:2594
Li C-M, Wu H-Z, Wang Y-X, Zhu S, Wei C-H (2020) Enhancement of phenol biodegradation: metabolic division of labor in coculture of Stenotrophomonas sp. N5 and Advenella sp. B9. J Hazard Mater 400:123214
Lucas N, Bienaime C, Belloy C, Queneudec M, Silvestre F, Nava-Saucedo JE (2008) Polymer biodegradation: mechanisms and estimation techniques. Chemosphere 73:429–442
Magnin A, Pollet E, Perrin R, Ullmann C, Persillon C, Phalip V, Avérous L (2019) Enzymatic recycling of thermoplastic polyurethanes: synergistic effect of an esterase and an amidase and recovery of building blocks. Waste Manag 85:141–150
Magnin A, Pollet E, Phalip V, Avérous L (2020) Evaluation of biological degradation of polyurethanes. Biotechnol Adv 39:107457
Massey F (1951) The Kolmogorov-Smirnov test for goodness of fit. J Am Stat Assoc 46:68–78
McCarthy SJ, Meijs GF, Mitchell N, Gunatillake PA, Heath G, Brandwood A, Schindhelm K (1997) In-vivo degradation of polyurethanes: transmission-FTIR microscopic characterization of polyurethanes sectioned by cryomicrotomy. Biomaterials 18:1387–1409
Mello BL, Alessi AM, McQueen-Mason S, Bruce NC, Polikarpov I (2016) Nutrient availability shapes the microbial community structure in sugarcane bagasse compost-derived consortia. Sci Rep 6:38781
Mukherjee K, Tribedi P, Chowdhury A, Ray T, Joardar A, Giri S, Sil AK (2011) Isolation of a Pseudomonas aeruginosa strain from soil that can degrade polyurethane diol. Biodegradation 22:377–388
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Nakajima-Kambe T, Onuma F, Kimpara N, Nakahara T (1995) Isolation and characterization of a bacterium which utilizes polyester polyurethane as a sole carbon and nitrogen source. FEMS Microbiol Lett 129:39–42
Obafemi YD, Taiwo OS, Omodara OJ, Dahunsi OS, Oranusi S (2018) Biodegradation of crude petroleum by bacterial consortia from oil-contaminated soils in Ota, Ogun State, South-Western, Nigeria. Environ Technol Innov 12:230–242
Obruca S, Marova I, Vojtova L (2011) Biodegradation of polyether-polyol-based polyurethane elastomeric films: influence of partial replacement of polyether polyol by biopolymers of renewable origin. Environ Technol 32:1043–1052
Pérez-Lara LF, Vargas-Suárez M, López-Castillo NN, Cruz-Gómez MJ, Loza-Tavera H (2016) Preliminary study on the biodegradation of adipate/phthalate polyester polyurethanes of commercial-type by Alicycliphilus sp. BQ8. J Appl Polym Sci 133:42992
PlasticsEurope (2020) Plastics - the Facts 2020. https://www.plasticseurope.org/es/resources/market-data. Accessed 24 March 2021
Raimundo I, Silva R, Meunier L, Valente SM, Lago-Lestón A, Keller-Costa T, Costa R (2021) Functional metagenomics reveals differential chitin degradation and utilization features across free-living and host-associated marine microbiomes. Microbiome 9:43
Roy R, Mukherjee G, Das Gupta A, Tribedi P, Sil AK (2021) Isolation of a soil bacterium for remediation of polyurethane and low-density polyethylene: a promising tool towards sustainable cleanup of the environment. 3 Biotech 11:29
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schmidt J, Wei R, Oeser T, Dedavid e Silva LA, Breite D, Schulze A, Zimmermann W (2017) Degradation of polyester polyurethane by bacterial polyester hydrolases. Polymers (Basel) 9:65
Sekhohola-Dlamini L, Tekere M (2020) Microbiology of municipal solid waste landfills: a review of microbial dynamics and ecological influences in waste bioprocessing. Biodegradation 31:1–21
Shah AA, Hasan F, Akhter JI, Hameed A, Ahmed S (2008) Degradation of polyurethane by novel bacterial consortium isolated from soil. Ann Microbiol 58:381–386
Shah Z, Gulzar M, Hasan F, Shah AA (2016) Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polym Degrad Stab 134:349–356
StatSoft, Inc. (2004) STATISTICA (data analysis software system), version 7. http://www.statsoft.com/Products/STATISTICA-Features. Accesed 23 February 2020
Syranidou E, Karkanorachaki K, Amorotti F, Avgeropoulos A, Kolvenbach B, Zhou N-Y, Fava F, Corvini PF-X, Kalogerakis N (2019) Biodegradation of mixture of plastic films by tailored marine consortia. J Hazard Mater 375:33–42
Vargas-Suárez M, Fernández-Cruz V, Loza-Tavera H (2019) Biodegradation of polyacrylic and polyester polyurethane coatings by enriched microbial communities. Appl Microbiol Biotechnol 103:3225–3236
Wang F, Grundmann S, Schmid M, Dörfler U, Roherer S, Charles Munch J, Hartmann A, Jiang X, Schroll R (2007) Isolation and characterization of 1,2,4-trichlorobenzene mineralizing Bordetella sp. and its bioremediation potential in soil. Chemosphere 67:896–902
Wilkes RA, Aristilde L (2017) Degradation and metabolism of synthetic plastics and associated products by Pseudomonas sp.: capabilities and challenges. J Appl Microbiol 123:582–593
Yadav S, Sharma A, Khan MA, Sharma R, Celin M, Malik A, Sharma S (2020) Enhancing hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) remediation through water-dispersible Microbacterium esteraromaticum granules. J Environ Manage 264:110446
Zafar U, Houlden A, Robson GD (2013) Fungal communities associated with the biodegradation of polyester polyurethane buried under compost at different temperatures. Appl Environ Microbiol 79:7313–7324
Zhao C, Wen D, Zhang Y, Zhang J, Tang X (2012) Experimental and mathematical methodology on the optimization of bacterial consortium for the simultaneous degradation of three nitrogen heterocyclic compounds. Environ Sci Technol 46:6205–6213
Zuo Y, He C, He X, Li X, Xue Z, Li X, Wang S (2020) Plant cover of Ammopiptanthus mongolicus and soil factors shape soil microbial community and catabolic functional diversity in the arid desert in Northwest China. Appl Soil Ecol 147:103389
Acknowledgements
AS-G acknowledges Consejo Nacional de Ciencia y Tecnología (CONACYT) for her Master Science scholarship and DGAPA-PAPIIT, UNAM, for a partial scholarship at the end of her studies. LD-M acknowledges CONACYT for her scholarship for a postdoctoral position (grant 290847) at Facultad de Química, Universidad Nacional Autónoma de México. We thank to Carolina Peña-Montes for her technical assistance with DGGE setup experiments, Alejandro Camacho for his assistance in the bacterial community deposit into Cepario Facultad de Química, UNAM, World Data Centre for Microorganisms CFQ100 and Marisela Gutiérrez in her technical support for the FTIR experiments at Unidad de Servicios de Apoyo a la Investigación y a la Industria, Facultad de Química, UNAM. We also thank the supercomputing resources and services of the Dirección General de Cómputo y de Tecnologías de Información y Comunicación (DGTIC) from Universidad Nacional Autónoma de México, through the project: LANCAD-UNAM-DGTIC-371.
Funding
This work was supported by Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica, Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México grants IN223317 and IN227620; Programa de Apoyo a la Investigación y el Posgrado, Facultad de Química, Universidad Nacional Autónoma de México, grant 5000–9117; and CONACYT Proyecto FORDECYT-PRONACES 101737 (Ciencia de Frontera 2019).
Author information
Authors and Affiliations
Contributions
A.S.-G. and H.L.-T. conceived the project and designed the experiments. A.S.-G., L.D.-M., and M.V.-S. performed the experiments. M.Q.-B. provided experimental support for the DGGE analysis. A.S.-R. performed the taxonomic and statistical analysis. M.V.-S., L.D.-M., and A.S.-R. prepared the figures. M.V.-S. and H.L.-T. wrote the original draft of the manuscript. All the authors analyzed the data, edited the manuscript, read, and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies performed on humans or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vargas-Suárez, M., Savín-Gámez, A., Domínguez-Malfavón, L. et al. Exploring the polyurethanolytic activity and microbial composition of landfill microbial communities. Appl Microbiol Biotechnol 105, 7969–7980 (2021). https://doi.org/10.1007/s00253-021-11571-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11571-w