Abstract
Yarrowia lipolytica strain is a promising cell factory for the conversion of lignocellulose to biofuels and bioproducts. Despite the inherent robustness of this strain, further improvements to lignocellulose-derived inhibitors toxicity tolerance of Y. lipolytica are also required to achieve industrial application. Here, adaptive laboratory evolution was employed with increasing concentrations of ferulic acid. The adaptive laboratory evolution experiments led to evolve Y. lipolytica strain yl-XYL + *FA*4 with increased tolerance to ferulic acid as compared to the parental strain. Specifically, the evolved strain could tolerate 1.5 g/L ferulic acid, whereas 0.5 g/L ferulic acid could cause about 90% lethality of the parental strain. Transcriptome analysis of the evolved strain revealed several targets underlying toxicity tolerance enhancements. YALI0_E25201g, YALI0_F05984g, YALI0_B18854g, and YALI0_F16731g were among the highest upregulated genes, and the beneficial contributions of these genes were verified via reverse engineering. Recombinant strains with overexpressing each of these four genes obtained enhanced tolerance to ferulic acid as compared to the control strain. Fortunately, recombinant strains with overexpression of YALI0_E25201g, YALI0_B18854g, and YALI0_F16731g individually also obtained enhanced tolerance to vanillic acid. Overall, this work demonstrated a whole strain improvement cycle by “non-rational” metabolic engineering and presented new targets to modify Y. lipolytica for microbial lignocellulose valorization.
Key points
• Adaptive evolution improved the ferulic acid tolerance of Yarrowia lipolytica
• Transcriptome sequence was applied to analyze the ferulic acid tolerate strain
• Three genes were demonstrated for both ferulic acid and vanillic acid tolerance
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Along with rapid global economic development and population growth, issues caused by utilizing fossil resources are attracting increasing attention. Lignocellulosic biomass, one of the most abundant renewable resources on earth, has received widespread attention as it offers an alternative to fossil resources (Jin et al. 2015). In recent years, many researchers have tried to develop technologies for production of biofuels and chemicals from lignocellulosic biomass (such as agro-forestry residues, urban solid wastes) (Ruan et al. 2012, Ruan et al. 2013, Yu et al. 2011). Such a conversion process using lignocellulosic biomass as a substrate can not only produce biofuels and chemicals sustainably, but also reduce the emission of greenhouse-gas.
Structurally, there are three main components in lignocellulosic biomass, which are cellulose, hemicellulose, and lignin (Shishir et al. 2011). Cellulose and hemicellulose can be hydrolyzed to fermentable sugars for further production of biofuels and chemicals. Due to its recalcitrant structure, lignocellulosic biomass must undergo pretreatment to make it easy for the hydrolysis of cellulose and hemicellulose. However, accompanied with the pretreatment of lignocellulose, undesirable compounds, such as weak acids, furans, and phenolics that are toxic to microorganisms, are also generated (Almeida et al. 2010, Klinke et al. 2004, Xu et al. 2019). Weak acids, like formic acid and acetic acid, can render intracellular anion accumulation after entering cells, which will consume ATP to maintain intracellular pH balance (Imai and Ohno 1995, Pampulha and Loureiro-Dias 1990). Furans, such as hydroxymethylfurfural and furfural, can disturb intracellular redox balance, inhibit the activity of enzymes in glycolytic pathway, and prolong the lag phase of microorganisms (Allen et al. 2010, Modig et al. 2002). Phenolics can destroy the integrity and permeability of microbial cell membrane, as well as induce undesirable intracellular reactive oxygen species (ROS) (Heipieper et al. 1994, Larsson et al. 2000). Among the abovementioned three types of inhibitors, phenolics commonly exhibit a higher inhibition effect on microorganisms than weak acids and furans. Ferulic acid (hereafter abbreviated as FA) is one of the main phenolics in lignocellulosic hydrolysate, especially in the hydrolysate of dicotyledons (Mansfield et al. 2012, Xu et al. 2019). According to previous studies, FA can inhibit the growth of many microorganisms even at a low concentration. For example, Larsson and colleagues (Larsson et al. 2000) found that FA at a concentration as low as 0.2 g/L substantially inhibited the cell growth of Saccharomyces cerevisiae.
Although lignocellulose-derived toxins can be removed by detoxifying processes (Cho et al. 2009, Guo et al. 2013), the detoxification operations not only increase the cost but may also cause the loss of fermentable sugars. Using robust microbial strains, which can tolerate multiple inhibitors and utilize sugars in lignocellulosic hydrolysate efficiently, is a more widely applied approach. Adaptive laboratory evolution has been proved an efficient method to obtain high tolerant strains for lignocellulosic feedstock fermentation. For instance, Thompson and colleagues subjected an industrial S. cerevisiae strain to adaptive laboratory evolution in pretreated pine biomass. As a result, two mutants exhibited significantly improved cell growth and fermentation performance in the presence of relevant inhibitors were obtained (Thompson et al. 2016). In another study, Xia and colleagues carried out a long-term adaptive evolution experiment for Corynebacterium glutamicum S9114, and the results indicated that the evolved strain achieved highly increased tolerance to multiple inhibitors (Wang et al. 2018). As an efficient method, adaptive laboratory evolution can also be used to obtain high-yielding strains. For example, Daskalaki and colleagues improved lipid accumulation capacity in oleaginous microorganisms by using evolution-based strategies. The population they obtained was able to accumulate 44% w/w of lipid, which was significantly higher than that of the start strain (Daskalaki et al. 2019).
Yarrowia lipolytica strains are considered as one of the most potential cell factories due to their excellent lipids accumulation capability (Qiao et al. 2017, Xu et al. 2017). Moreover, the composition of lipid from Y. lipolytica strains is similar to that of diesel, making it an ideal substitute for fossil resources-derived diesel. In addition, Y. lipolytica strains have also been applied to produce many other valuable compounds, such as terpenoids (Liu et al. 2019, Yang et al. 2016) and carotinoid (Jacobsen et al. 2020, Larroude et al. 2017). As mentioned above, lignocellulosic biomass is considered as one of the most promising substrates for fermentation. One obstacle impeding the development of large-scale biodiesel production from lignocellulose by Y. lipolytica is the fragile cell growth caused by inhibitors in hydrolysate. For instance, when synthetic medium is applied, 99.30 g/L lipid can be achieved (Qiao et al. 2017). In contrast, only 12.01 g/L lipid was obtained when lignocellulosic hydrolysate was applied as feedstock (Yook et al. 2020). Thus, a robust Y. lipolytica strain, which can tolerate inhibitors in hydrolysate, is essential for microbial lipid production from lignocellulosic biomass. Whereas, few studies have been reported on the construction of robust Y. lipolytica strains, as well as analysis of tolerance mechanism of Y. lipolytica strains.
In this study, efforts have been made to obtain robust Y. lipolytica strains which can tolerate phenolics, especially FA, with an integrated strategy of adaptive laboratory evolution, transcriptome analysis, tests of target genes, and reverse metabolic engineering (Fig. 1). First, a four-stage FA tolerant evolution experiment was carried out with FA concentration gradually increased from 0.5 to 1.5 g/L. Phenotype characterization suggested that evolved strains exhibited regular growth in the presence of 1.5 g/L FA after four stage of laboratory evolutions. Transcriptional analysis of the evolved strains versus the original one revealed that YALI0_E25201g (a gene for pleiotropic drug resistance proteins), YALI0_F05984g (a gene for permease of the major facilitator superfamily), YALI0_B18854g (a gene mapping in the integral component of membrane of GO terms), and YALI0_F16731g (a gene mapping in the integral component of membrane of GO terms) might be responsible for the improved phenotype, which were confirmed by subsequent reverse genetic engineering. This study promotes the understanding of stress tolerance of Y. lipolytica in response to lignocellulose-derived inhibitors, and the mined target genes may be helpful to construct robust strains through genetic engineering.
Schematic representation of the study performed to increase FA tolerance of yl-XYL+. First, the parental strain yl-XYL+ was subjected to adaptive laboratory evolution. During the evolution experiment, the concentration of FA in medium was increased stepwise from 0.5 to 1.5 g/L. Then, the original strain and the evolved strain were subject for transcriptome analysis, and some potential gene targets were selected. Finally, the potential targets involving in FA tolerance were verified by reverse metabolic engineering
Materials and methods
Strains and media
Y. lipolytica XYL+ (hereafter abbreviated as yl-XYL+) used as the parental strain for adaptive laboratory evolution is a xylose-utilizing strain constructed by Ledesma-Amaro and colleagues based on the wild-type strain W29 (ATCC20460) (Ledesma-Amaro et al. 2016) (Table 1). YPX medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L xylose) was used for seed cultivation. YNBXFA medium (1.7 g/L yeast nitrogen base, 5 g/L NH4Cl, 10 g/L xylose, and different amount of FA) was used for adaptive laboratory evolutions. YNBX medium (1.7 g/L yeast nitrogen base, 5 g/L NH4Cl, and 30 g/L xylose) was used for fermentation phenotype tests. SC-S medium (7.9 g/L SC dropout medium and 30 g/L sucrose) was used for recombinant strains’ seed cultivation. SC-S-FA medium (7.9 g/L SC dropout medium, 30 g/L sucrose, and 0.5 g/L FA) and SC-S-VA medium (7.9 g/L SC dropout medium, 30 g/L sucrose, and 1.0 g/L vanillic acid) were used for the test of recombinant strains. YPD medium (10 g/L yeast extract, 20 g/L peptone, and 20 g/L glucose) was applied for the routine culture of yeasts. Additional 20 g/L agar was added for the abovementioned media when solid media were required.
Adaptive laboratory evolution for improving ferulic acid tolerance
YNBXFA medium was used to domesticate yl-XYL+. In order to provide sufficient survival stress, the lethality of FA to Y. lipolytica was evaluated first with an initial Y. lipolytica OD600 of 0.25, 0.5, 0.75, and 1.0. The adaptive laboratory evolution process was divided into four stages with 0.5, 0.75, 1.0, and 1.5 g/L FA added in the medium respectively. In details, yl-XYL+ was inoculated into YNBXFA medium with 0.5 g/L FA in the first stage, and the cultivated cells were transferred into fresh medium every 48 h. Cultivation was carried out at 30 °C with a rotation of 250 rpm. Domestication was stopped when the growths of 3 sequential passages reached similar OD600 after 48-h cultivation. The domesticated cells were spread on YNBXFA plate and ten colonies were picked randomly for phenotype test. The colony with the best performance was selected for the next stage of domestication.
RNA sequencing and quantitative real-time PCR
The target colony from plate was incubated in YPX medium in 250-mL Erlenmeyer flasks at 30 °C with a rotation of 250 rpm. When cells grew up to the logarithmic growth phase, three parallel samples were collected for liquid nitrogen frozen immediately. Then, the frozen samples were milled to power form with a sterile mortar and pestle. The powdered cells were collected in a 1.5-mL centrifuge tube and then 1 mL trizol was added (Zhou et al. 2020). The samples were stored in dry ice and sent out for RNA sequencing. RNA extraction, library construction, and sequencing were performed by Shanghai Shenggong Bioengineering Technology Service Co., Ltd. The sequencing was performed by using an Illumina HiSeqTM 2500 system as described previously (Wei et al. 2017, Wu et al. 2016). RNA-Seq data analysis, including quality control, reads mapping, transcriptome assembly, annotation, gene expression level analysis, and SNP calling, were carried out according to previously published standard procedures (Wei et al. 2017, Wu et al. 2016). Relevant RNA sequencing raw data have been submitted in Genbank with an accession number of PRJNA 675399.
Quantitative real-time PCR (qRT-PCR) was performed to validate the results of RNA-seq. RNA isolation and cDNA synthesis were performed with a PrimeScript™ RT reagent kit and gDNA Eraser (Takara, RR047A) as previously described (Wen et al. 2014). qRT-PCR was conducted in an Applied Biosystems StepOnePlus Real-Time PCR system (Thermo-Fisher Scientific Inc., Waltham, MA, USA). SYBR® Green Premix Ex TaqTM II kits (Takara, Dalian, Liaoning, China) were used under the following reaction conditions: 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles of 95 °C for 3 s, 60 °C for 30 s, and 72 °C for 30 s. All assays were performed in triplicate. All qRT-PCR primers are listed in Table 2.
Reverse engineering of key targets found in evolved strains
Four potential gene targets that might contribute to FA tolerance were chosen based on the RNA-Seq and qRT-PCR results. These targets were in situ verified by gene overexpression with the help of a replicating plasmid. Recombinant plasmids were constructed according to standard procedures with pMCS-CEN1 as a skeleton plasmid (Gao et al. 2016). Using pMCS-CEN1-C20265g as an example, it was constructed by assembling a YALI0_C20265g expression cassette (composed of TEFin promoter, ORF of YALI0_C20265g, and XPR2t terminator) and a SUC cassette (composed of TEFin promoter, ORF of SUC, and Migt terminator) into pMCS-CEN1. The constructed plasmids were transformed into the original yl-XYL+ by electroporation, and then 50 μL transformation mixture was plated onto SC-S solid medium for screening correct recombinant strains. The verified recombinant strains were tested in SC-S-FA and SC-S-VA medium with initial OD600 of 0.5. Three parallel samples were collected at each 24 h for determination of optical density and sugar consumption. Involved strains and plasmids used are summarized in Table 1.
Analytical methods
The concentrations of sugars and phenolics in fermentation broth were analyzed by HPLC equipped with a Bio-rad Aminex HPX-87H column and a refractive index detector. The Aminex HPX-87H column was maintained at 60 °C, and the mobile phase was 0.005 M sulfuric acid with a flow rate of 0.6 mL/min (Chen et al. 2018). Concentrations of monomeric sugars were calculated based on the calibration sugar standards. Cell growth of Y. lipolytica was quantified by measuring the optical density of fermentation broth at 600 nm with a UV-visible spectrophotometer (TU-1810, Beijing Pushen General Instrument Co., Ltd.). The OD600 value was measured before each passage, and the number of generations was calculated with the formula: number of generations = log2(OD600 final/OD600 initial) (Hellgren 2017).
Results
Adaptive laboratory evolution for improving ferulic acid tolerance of Y. lipolytica XYL+
Before performing adaptive laboratory evolution for yl-XYL+ to enhance its FA tolerance, the toxicity of FA to the parental strain was tested. Due to the low solubility of FA in water at fermentation temperature, FA was dissolved in DMSO at a concentration of 100 g/L for further use. Our preliminary experiments showed that DMSO did not inhibit the growth of yl-XYL+ until its concentration was above 1% (data not shown). Thus, the concentration of DMSO used in this study was controlled below 1%. Then, the effect of FA concentration on yl-XYL+ growth was investigated. As shown in supplementary figure S1, 0.5 g/L FA caused about 90% lethality of yl-XYL+, which indicated that this concentration imposed desired pressure to Y. lipolytica XYL+ for evolution. Thus, 0.5 g/L FA was applied to initiate the adaptive laboratory evolution for improving FA tolerance of yl-XYL+.
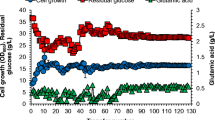
With an appropriate initial FA concentration, the adaptive laboratory evolution for yl-XYL+ was conducted. As shown in Fig. 2, increased cell growth was observed during the evolution process, indicating that the strain was adapted to the FA stress gradually. For instance, in the first stage of evolution, yl-XYL+ grew well in YNBX medium (without FA), in contrast, it could hardly survive in the first 48 h when 0.5 g/L FA was added into the medium. After three transfers, there was no obvious difference between the final OD600 when the strain was cultured with and without 0.5 g/L FA, which indicated that the strain was well adapted to 0.5 g/L FA. Similar domestication processes were also observed in the next three passages. After four stages of adaptive laboratory evolutions, the evolved strains could grow significantly with FA at a concentration of 1.5 g/L, which indicated that this “non-rational” strategy is effective in enhancing the robustness of this Y. lipolytica strain. Although the final OD600 of the evolved strain cultured with 1.5 g/L FA was lower than that of control in the end of stage 4, the cell growth was stable in the last 6 cultivations, which suggested the evolution reached its threshold. Finally, it took 84 days for the whole adaptive laboratory evolution and the obtained cells could survive well with 1.5 g/L FA. To uncover the underlying mechanism of the evolution, a single strain named yl-XYL + *FA*4 was separated from the final evolution stage and selected for further analysis.
Adaptive evolution of yl-XYL+ for FA tolerance. The graph shows the measured OD600 in each subculture during the evolution experiment. The triangles (red) represent the OD600 of yl-XYL+ cultured in YNBX with FA. The squares (black) represent the OD600 of yl-XYL+ cultured in YNBX which was used as control. The vertical gray lines indicate the time where the evolution entered into the next stage. The evolved strains were spread on plate, and ten random colonies were picked for phenotype test at the end of each evolution stage. Domestication had lasted for 86 days with Y. lipolytica transferred for 43 times: the first stage had gone through 7 passages, approximately 27 generations; the second stage had gone through 9 passages, approximately 37 generations; the third stage had gone through 13 passages, approximately 53 generations; the final stage had gone through 14 passages, approximately 57 generations
When the evolved strain yl-XYL + *FA*4 and parental strain yl-XYL+ were cultured on YNBX and YNBXFA solid mediums, it was noticeable that the evolved strain could grow on YNBXFA while the parental strain could not (data not shown), whereas both strains grew well on YNBX solid medium. Then, the morphologies of yl-XYL + *FA*4 and yl-XYL+ were investigated by optical microscopy respectively, and no obvious morphological differences were observed. Furthermore, the fermentation performances of these two strains were compared in YNBXFA and YNBX medium. As shown in Fig. 3, the evolved and parental strains exhibited similar growth profile and xylose consumption when cultured in YNBX liquid medium. However, when cultured in YNBXFA liquid medium, the yl-XYL + *FA*4 strain could grow well, but the parental strain could hardly survive. All the above information indicated that the evolved strain had obtained the ability to tolerate FA compared to the parental strain.
Comparison of the parental strain and the evolved strain by transcriptome analysis
Three parallel RNA samples extracted from yl-XYL+ and yl-XYL + *FA*4 were subjected to transcriptome sequencing to analyze the emerged changes in the evolved strain. As a result, over 40 Gb raw reads were obtained from each cDNA library. In detail, a total of 42,181,222 and 42,259,356 pairs of reads were obtained from RNA of yl-XYL + *FA*4 and yl-XYL+, respectively. Comparing the obtained reads to the reference genome, the proportion of reads in each genome structure, includes exon, intron, and intergenic, was shown in supplementary figure S2. Over 95% of the reads from both samples were mapped to the genome. The average read length was 141 and 142 bases for yl-XYL + *FA*4 samples and yl-XYL+ samples respectively, and the distribution of reads were homogeneous with great randomness of fragmentation (supplementary figure S3). Distributions of genes coverage (the percentage of a gene covered by reads) were shown in supplementary table S1. For each sample of yl-XYL+ and yl-XYL + *FA*4, more than 97% of genes in transcriptomic dataset showed perfect coverage of 90 to 100%.
Gene expression level was directly reflected in the abundance of transcripts. High abundance of transcripts suggested high gene expression level. TPM (Transcripts Per Million) measurement was employed to estimate the expression levels of target genes. In order to obtain genes with significant differences, the screening condition was settled to q value < 0.05 (corrected p value. The smaller q value, the more significant difference in gene expression) and difference multiple (which was also called fold change) > 2.000. Scatterplot provided a global overview of the number of differential genes and the number of up- and downregulated genes (Fig. 4). In particular, the genes significantly over-expressed (upregulated by more than twofold) and significantly under-expressed (decreased by at least 50%) in yl-XYL + *FA*4 was 63 and 333 respectively (supplementary table S2). Besides, all non-synonymous mutations in the evolved strain yl-XYL + *FA*4 based on the transcriptome sequencing have been added to supplementary (supplementary table S3). Further study on the distribution of these difference genes in the annotation function indicated that 69 difference genes were assigned to the KEGG ortholog (KO) system (supplementary figure S4a), 200 difference genes were assigned to KOG (supplementary figure S4b), and 288 genes were categorized into 46 GO functional under three main categories—“Cellular component,” “Biological process,” and “Molecular function” (supplementary figure S4c).
The horizontal and vertical axes are the log2 (TPM) values of yl-XYL+ and yl-XYL + *FA*4 samples respectively. Each dot represents a gene, and the closer to the origin indicating the lower the expression level. The red represents the upregulated genes, green represents the downregulated genes, and black represents a non-differentially expressed genes
To figure out putative genes responsible for high concentration FA tolerance, transcriptional level of significantly changed genes in yl-XYL + *FA*4 was investigated carefully. GO directed acyclic graphs were drawn for visualizing potential genes having most significant differences in the evolved strain (supplementary figure S5). In acyclic graphs, branches represented inclusion relationships, and the range of defined KEGG channels became smaller from top to bottom. It was found that features with significant differences mainly matched with categories of “integral component of membrane,” “xenobiotic transmembrane transporting ATPase activity,” and “drug transport.” Meanwhile, significantly enriched function-gene interaction network diagrams were drawn to better understand the relationship between genes and various functions (supplementary figure S6). Brandt et al. (2019) summarized the inhibitor tolerance mechanisms in S. cerevisiae, including direct mechanisms (such as removing inhibitors by multidrug transporters and enzymatic detoxification), indirect mechanisms (such as producing protectant to repair membranes and proteins), and other mechanisms like regenerating cofactors to aid in the detoxification. Based on inhibitor tolerance mechanisms in S. cerevisiae and the obtained functional-gene interaction network diagram (supplementary figure S6), it was suspected that the good growth of yl-XYL + *FA*4 with 1.5 g/L FA was trigged by changing in multiple “direct and indirect” elements, including these belong to “integral component of membrane,” “xenobiotic transmembrane transporting ATPase activity,” “drug transmembrane transporter activity,” and “drug transport.” In particular, seven most possible target genes were selected for further function demonstration.
Validation of transcriptome sequence results by qRT-PCR
To verify the correctness of the transcriptome profiling, seven potential genes for FA tolerance (YALI0_C20265g, YALI0_B12188g, YALI0_F17996g, YALI0_E25201g, YALI0_F05984g, YALI0_B18854g, and YALI0_F16731g) were re-checked by qRT-PCR. According to function analysis, YALI0_E25201g and YALI0_F17996g encode multidrug resistance-associated proteins, which belong to ATP-binding cassette (ABC) superfamily. They also associate with GO function/term of transmembrane transporter activity, transporter activity, drug transmembrane transporter activity, xenobiotic transmembrane transporting ATPase activity, and xenobiotic transmembrane transporter activity. YALI0_B12188g encodes a multidrug export protein which matches with the plasma membrane function on GO term. YALI0_F05984g, YALI0_B12188g, and YALI0_F16731g mainly involve in organic acid (such as carboxylic acid, oxoacid) metabolism processes. The expression levels of these seven target genes along with a reference gene actin, an endogenous control for normalization, were determined by qRT-PCR. As shown in Fig. 5, qRT-PCR results exhibited similar tendency with the transcriptome results for selected target genes.
Reverse engineering of key targets
Transcriptome and qRT-PCR results suggested the function enhancements of seven genes, which involved with the multidrug resistance, multidrug transport, and other unknown functions, may be responsible for the enhanced FA tolerance ability of yl-XYL + *FA*4. Then, the seven target genes were inserted into a replicable plasmid under a strong constitutive TEFin promoter, and the verified recombinant plasmids were introduced into the parental yl-XYL+ separately, resulting in overexpressing strains yl-XYL + C20265g, yl-XYL + B12188g, yl-XYL + F17996g, yl-XYL + E25201g, yl-XYL + F05984g, yl-XYL + B18854g, and yl-XYL + F16731g. Then, the recombinant strains were cultured with and without 0.5 g/L FA respectively.
The growth profiles of yl-XYL + C20265g, yl-XYL + B12188g, and yl-XYL + F17996g were basically similar to that of the control strain yl-XYL+, or even worse. As well acknowledged that the introduction of replicable plasmids and/or gene overexpression cassettes can bring extensive burden for host cells, which may be the reason for the unchanged or decreased cell growth profiles. As depicted in Fig. 6, yl-XYL + E25201g, yl-XYL + F05984g, yl-XYL + B18854g, and yl-XYL + F16731g exhibited better tolerance to FA than the parental yl-XYL+ when cultured with 0.5 g/L FA. In detail, the final OD600 of these four strains were 8.4, 3.8, 4.0, and 6.8, compared to 3.1 of the control strain.
Reverse engineering of potential target genes. The genes of YALI0_E25201g, YALI0_F05984g, YALI0_B18854g, and YALI0_F16731g were introduced separately into the parental yl-XYL+ via a replicable plasmid driven by a strong constitutive TEFin promoter, resulting in overexpressing strains yl-XYL + E25201g, yl-XYL + F05984g, yl-XYL + B18854g, and yl-XYL + F16731g. a, c, e, g The growth profiles of yl-XYL + E25201g, yl-XYL + F05984g, yl-XYL + B18854g, and yl-XYL + F16731g. b, d, f, h The concentration of sucrose, glucose, and fructose at each moment. Mean values and error bars were calculated using three replicates
Vanillic acid tolerance of recombinant strains
With the aforementioned information, at least four of the seven potential target genes showed relation to the enhanced FA tolerance ability of the evolved strain. Considering the similar structure between FA and vanillic acid, it was anticipated that if the overexpression of these four target genes would endow the parental strain with vanillic acid tolerance ability. Therefore, yl-XYL + E25201g, yl-XYL + F05984g, yl-XYL + B18854g, and yl-XYL + F16731g were cultured in SC-S medium with 1 g/L vanillic acid. As shown in Fig. 7, yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g exhibited enhanced cell growth ability compared with the control strain. In details, the OD600 of yl-XYL+ in SC-S-VA medium reached value of 1.50 at 25 h, then remained almost unchanged in the next 96 h. In contrast, the growth of yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g increased steadily after 25 h, and the final OD600 reached about 7.7, 5.1, and 5.8 respectively (Fig. 7a, c, e). It showed that overexpression of YALI0_E25201g, YALI0_B18854g, and YALI0_F16731g can enhance parental strain’s tolerance to vanillic acid.
Growth curves of recombinant strains yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g in the presence of 1 g/L vanillic acid at SC-S medium. a, c, e The growth profiles of yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g, separately. b, d, f The concentration of sucrose, glucose, and fructose at each moment of yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g, separately. Mean values and error bars were calculated using three replicates
Discussion
Y. lipolytica is a famous microbial lipid-producing strain, whereas when lignocellulosic biomass is applied as the substrate, the lipid production of Y. lipolytica sharply decreases. One of the main reasons for the decreased lipid production with lignocellulosic substrates is the severe cell growth inhibition caused by inhibitors. Enhancing the inhibitor tolerance ability of relevant strains is a feasible method for microbial lipid from lignocellulosic substrates. FA is one of the most common inhibitors in lignocellulosic hydrolysate, especially when alkaline pretreatments were applied to pretreat the lignocellulosic substrates. In this study, a “non-rational” strategy was provided to enhance the FA tolerance ability of Y. lipolytica, which was initiated with adaptive evolution, continued with transcriptome profiles analyzation, and completed with the potential gene targets identification by reversing engineering (Fig. 1). This work provides new ideas for constructing engineered Y. lipolytica that can tolerate inhibitors in lignocellulose hydrolysate.
First, a four-stage adaptive laboratory evolution experiment was successively performed to enhance the FA tolerance of Y. lipolytica for the first time. By continuously transferring the original strain yl-XYL+ in YNBXFA medium (FA concentration was gradually increased from 0.5 to 1.5 g/L) for approximately 3 months, an evolved Y. lipolytica strain which could tolerate 1.5 g/L FA was obtained. As determined, 0.25 g/L FA can inhibit more than 75% cell growth of the original strain yl-XYL+ when it was cultured in YNBX medium. In contrast, the evolved strain yl-XYL + *FA*4 could grow regularly in YNBX medium with 1.5 g/L FA. Similar to phenomena appeared in the adaptive laboratory evolution of Rhodococcus opacus (Kurosawa et al. 2015) and S. cerevisiae (Almario et al. 2013), the time consumed for each evolution stage gradually increased as the evolution proceeded. It was probably because higher concentration of FA caused greater damage to strains, thus it needed longer time to obtain the inhibitor resistance characteristic. In the last stage of the adaptive laboratory evolution experiment, the OD600 of evolved strains cultured with 1.5 g/L FA reached a stable value of about 5.0, which was lower than that of the control strain cultured without FA. This suggested that it was difficult to further improve strains’ tolerance to higher concentrations of FA. Considering the FA concentration in lignocellulosic hydrolysate (e.g., NaOH pretreated rice straw yielded 0.19 g/L FA (Hou et al. 2017), acid pretreated corn stover yielded 0.38 g/L FA (López et al. 2004); tolerance to 1.5 g/L FA is enough to make the evolved strain resist the FA in lignocellulosic hydrolysate, and thus the evolution experiment was terminated. According to previous studies, there are mainly two ways for selecting samples for the next round of evolution in adaptive laboratory evolution experiments: selecting a mixture of evolved strains and selecting one colony (Dragosits and Mattanovich 2013; Sandberg et al. 2019). In this study, only one colony with an excellent phenotype was selected for the next round of evolution during the experiment. Based on the adaptive laboratory evolution hypothesis that strains with an excellent phenotype may carry more favorable mutations, we believe this operation can enable more advantageous mutations to be inherited, ensuring the effectiveness of domestication. Moreover, this operation can also make sure that there is a clear genetic relationship between the selected strains at each stage, which will make it easier to analyze the genetic backgrounds of evolved strains.
To explore the mechanisms of enhanced FA tolerance of the evolved strain, transcriptomic profiles of the evolved strain and the parental strain were analyzed. The expression levels of genes in the PPP pathway and TCA cycle were similar in these two strains, indicating that the hexose metabolism was not affected by the adaptive laboratory evolution. Some genes involved in amino acids metabolism changed in the evolved strain. In detail, five genes involved in the biosynthesis of isoleucine, lysine, glutamate, and glycine were upregulated varying from 2.621-fold to 3.010-fold. A gene involved in the serine to pyruvate pathway was upregulated, implying that the intracellular serine concentration maybe slightly decreased. Reactive oxygen species (ROS) accumulation is commonly considered the main reason for the inhibition of phenolics because excess intracellular ROS will damage DNA and inhibit protein and RNA synthesis. Many researches have proved that glutathione and catalase could reduce the accumulation of ROS (Qi et al. 2015, Qiu et al. 2015). Whereas, three genes related to glutathione transport and synthesis (YALI0_B20416g, 0.495-fold; YALI0_A06743g, 0.487-fold; YALI0_E02310g, 0.404-fold), and two genes related to catalase (YALI0_E34265g, 0.131-fold and YALI0_E34749g, 0.080-fold) (Ask et al. 2013, Chen et al. 2019) were downregulated in the evolved strain. Therefore, there must be other alterations contributing to the FA tolerance of the evolved strain.
The constant FA concentration in medium excluded the possibility that Y. lipolytica could degrade FA. According to the reported inhibitor resistance mechanism in S. cerevisiae (Brandt et al. 2019), the expression changes of genes related to multidrug transport and multidrug resistance were investigated. Based on significant GO-directed acyclic graphs of biological processes (supplementary figure S5) and significantly enriched function-gene interaction network diagram (supplementary figure S6), five genes involved in drug resistance and drug transport were significantly upregulated. They were YALI0_C20265g, YALI0_E25201g, YALI0_F17996g, YALI0_F05984g, and YALI0_B12188g. The growths of yl-XYL + C20265g, yl-XYL + B12188g, and yl-XYL + F17996g were similar to that of the control strain, or even worse when cultured with 0.5 g/L FA, which may be because the extensive pressure brought by overexpression of those genes was greater than the enhanced FA resistance in those strains. Whereas, the maximum OD600 of recombinant strains yl-XYL + E25201g and yl-XYL + F05984g in SC-S-FA medium were 2.70-fold and 1.23-fold higher compared with that of the control strain, respectively (Fig. 6). YALI0_F05984g is a homologous gene of Tna1p in S. cerevisiae, but no researches have showed Tna1p was related to phenolics tolerance. For YALI0_E25201g, no homologous gene was not reported, let alone its function. According to previous studies, phenolic compounds can embed in, or even break and pass through the cell membrane, disrupting cell’s normal metabolism due to their lipophilic properties (Mansfield et al. 2012, Xu et al. 2019). Many genes matching with GO classified of integral component of membrane (such as YALI0_B18854g and YALI0_F16731g) in the evolved strain were upregulated. This may be one of the reasons for the enhanced tolerance to FA of the evolved strain. Reverse metabolic engineering validated that overexpression of YALI0_B18854g and YALI0_F16731g did enhance the parental strain’s tolerance to FA (Fig. 6).
The recombinant strains yl-XYL + E25201g, yl-XYL + B18854g, and yl-XYL + F16731g also obtained enhanced vanillic acid tolerance (Fig. 7), which indicated that YALI0_B18854g and YALI0_F16731g maybe played a key role in resisting to multiple phenolic acid inhibitors in Y. lipolytica. YALI0_B18854g encodes alpha/beta hydrolase, and its homologous gene MGL2 was found in S. cerevisiae. YALI0_F16731g’s function was unknown but its homologous gene Lpx1 was found in S. cerevisiae. There have been no direct evidences suggest that MGL2 and Lpx1 are related to phenolics tolerance in previous studies. Whereas, Wu and colleagues found that disruption of an unknown alpha/beta hydrolase made Kluyveromyces marxianus more susceptible to lignocellulose-derived inhibitors (Wu et al. 2020). This may partly explain the enhanced tolerance to FA and vanillic acid of yl-XYL + B18854g and yl-XYL + F16731g. Interestingly, not all genes mapping integral membrane protein were upregulated, such as YALI0_F08349g (0.429-fold), suggesting the complexity of membrane protein functions and the necessity of re-exploring these proteins.
Overall, adaptive laboratory evolution was first applied to improve FA tolerance of Y. lipolytica yl-XYL+. As a result, an excellent strain yl-XYL + *FA*4, which can survive well with 1.5 g/L FA, was obtained. The results of transcriptomic analysis and reverse metabolic engineering showed that the overexpression of two multidrug resistance-associated proteins (YALI0_E25201g and YALI0_F05984g) and two integral membrane proteins (YALI0_B18854g and YALI0_F16731g) in the evolved strain contributed to the FA tolerance improvement. Moreover, three of the four target genes were also demonstrated playing an important role in VA resistance. Efforts have been made to find out the reason why these four targets were upregulated by sequencing the promoter regions of the four targets, but the results suggested that there were no mutations in the promoter regions of these four targets (supplementary table S4). The specific mechanisms why these targets were upregulated remain to be further studied, and the whole-genome sequencing can provide more information about the evolved strain. Maybe, the expressional level changes of these four targets were inspired by the mutation of some transcriptional regulatory factors. Our work paves the way for using non-conventional yeast of Y. lipolytica as chassis cell to convert lignocellulosic hydrolysates effectively into biofuels and other chemicals.
Data availability
The datasets of RNA sequencing raw data generated in this study have been submitted in Genbank with an accession number of PRJNA 675399 (https://dataview.ncbi.nlm.nih.gov/object/PRJNA675399).
References
Allen SA, Clark W, Mccaffery JM, Zhen C, Gorsich SW (2010) Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol Biofuels 3:1–10. https://doi.org/10.1186/1754-6834-3-2
Almario M, Reyes L, Kao K (2013) Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng 110. 2616-2623. doi:https://doi.org/10.1002/bit.24938
Almeida JR, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF (2010) Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J Chem Technol Biotechnol 82:340–349. https://doi.org/10.1002/jctb.1676
Ask M, Mapelli V, Hck H, Olsson L, Bettiga M (2013) Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials. Microb Cell Factories 12:87. https://doi.org/10.1186/1475-2859-12-87
Brandt BA, Jansen T, Görgens JF, van Zyl WH (2019) Overcoming lignocellulose-derived microbial inhibitors: advancing the Saccharomyces cerevisiae resistance toolbox. Biofuels Bioprod Biorefin 13:1520–1536. https://doi.org/10.1002/bbb.2042
Chen H, Li J, Wan C, Fang Q, Zhao X (2019) Improvement of inhibitor tolerance in Saccharomyces cerevisiae by overexpression of the quinone oxidoreductase family gene YCR102C. FEMS Yeast Res 19:55. https://doi.org/10.1093/femsyr/foz055
Chen X, Zhai R, Shi K, Yuan Y, Dale BE, Gao Z, Jin M (2018) Mixing alkali pretreated and acid pretreated biomass for cellulosic ethanol production featuring reduced chemical use and decreased inhibitory effect. Ind Crop Prod 124:719–725. https://doi.org/10.1016/j.indcrop.2018.08.056
Cho DH, Lee YJ, Um Y, Sang BI, Kim YH (2009) Detoxification of model phenolic compounds in lignocellulosic hydrolysates with peroxidase for butanol production from Clostridium beijerinckii. Appl Microbiol Biotechnol 83:1035–1043. https://doi.org/10.1007/s00253-009-1925-8
Daskalaki A, Perdikouli N, Aggeli D, Aggelis G (2019) Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl Microbiol Biotechnol 103:8585–8596. https://doi.org/10.1007/s00253-019-10088-7
Dragosits M, Mattanovich D (2013) Adaptive laboratory evolution–principles and applications for biotechnology. Microb Cell Factories 12:64. https://doi.org/10.1186/1475-2859-12-64
Gao S, Tong Y, Wen Z, Zhu L, Ge M, Chen D, Jiang Y, Yang S (2016) Multiplex gene editing of the Yarrowia lipolytica genome using the CRISPR-Cas9 system. J Ind Microbiol Biotechnol 43:1085–1093. https://doi.org/10.1007/s10295-016-1789-8
Guo X, Cavka A, Jönsson LJ, Hong F (2013) Comparison of methods for detoxification of spruce hydrolysate for bacterial cellulose production. Microb Cell Factories 12:93. https://doi.org/10.1186/1475-2859-12-93
Heipieper HJ, Weber FJ, Sikkema J, Keweloh H, de Bont JA (1994) Mechanisms of resistance of whole cells to toxic organic solvents. Trends Biotechnol 12:409–415. https://doi.org/10.1016/0167-7799(94)90029-9
Hellgren J (2017) Adaptive evolution of Yarrowia lipolytica for osmotic and saline tolerance. Dissertation, Chalmers University Of Technology
Hou J, Ding C, Qiu Z, Zhang Q, Xiang WN (2017) Inhibition efficiency evaluation of lignocellulose-derived compounds for bioethanol production. J Clean Prod 165:1107–1114. https://doi.org/10.1016/j.jclepro.2017.07.204
Imai T, Ohno T (1995) The relationship between viability and intracellular pH in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 61:3604–3608. https://doi.org/10.1002/bit.260480111
Jacobsen IH, Ledesma-Amaro R, Martinez JL (2020) Recombinant β-Carotene production by Yarrowia lipolytica – assessing the potential of micro-scale fermentation analysis in cell factory design and bioreaction optimization. Front Bioeng Biotech 8. 29. doi:https://doi.org/10.3389/fbioe.2020.00029
Jin M, Slininger PJ, Dien BS, Waghmode S, Moser BR, Orjuela A, Sousa LC, Balan V (2015) Microbial lipid-based lignocellulosic biorefinery: feasibility and challenges. Trends Biotechnol 33:43–54. https://doi.org/10.1016/j.tibtech.2014.11.005
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. https://doi.org/10.1007/s00253-004-1642-2
Kurosawa K, Laser J, Sinskey AJ (2015) Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels 8:76. https://doi.org/10.1186/s13068-015-0258-3
Larroude M, Celinska E, Back A, Thomas S, Nicaud JM, Ledesma-Amaro R (2017) A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol Bioeng 115(2):464–472. https://doi.org/10.1002/bit.26473
Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant NO, Jönsson LJ (2000) Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl Biochem Biotechnol 84-86:617–632. https://doi.org/10.1385/abab:84-86:1-9:617
Ledesma-Amaro R, Lazar Z, Rakicka M, Guo Z, Fouchard F, Coq CL, Nicaud JM (2016) Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metab Eng 38:115–124. https://doi.org/10.1016/j.ymben.2016.07.001
Liu Y, Jiang X, Cui Z, Wang Z, Hou J (2019) Engineering the oleaginous yeast Yarrowia lipolytica for production of α-farnesene. Biotechnol Biofuels 12:296. https://doi.org/10.1186/s13068-019-1636-z
López MJ, Nichols NN, Dien BS, Moreno J, Bothast RJ (2004) Isolation of microorganisms for biological detoxification of lignocellulosic hydrolysates. Appl Microbiol Biotechnol 64:125–131. https://doi.org/10.1007/s00253-003-1401-9
Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7:1579–1589. https://doi.org/10.1038/nprot.2012.064
Modig T, Lidén G, Taherzadeh MJ (2002) Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem J 363:769–776. https://doi.org/10.1042/0264-6021:3630769
Pampulha ME, Loureiro-Dias MC (1990) Activity of glycolytic enzymes of Saccharomyces cerevisiae in the presence of acetic acid. Appl Microbiol Biotechnol 34:375–380. https://doi.org/10.1007/BF00170063
Qi K, Xia X, Zhong J (2015) Enhanced anti-oxidative activity and lignocellulosic ethanol production by biotin addition to medium in Pichia guilliermondii fermentation. Bioresour Technol 189:36–43. https://doi.org/10.1016/j.biortech.2015.02.089
Qiao K, Wasylenko TM, Zhou K, Xu P, Stephanopoulos G (2017) Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol 35:173–177. https://doi.org/10.1038/nbt.3763
Qiu Z, Deng Z, Tan H, Zhou S, Cao L (2015) Engineering the robustness of Saccharomyces cerevisiae by introducing bifunctional glutathione synthase gene. J Ind Microbiol Biotechnol 42:537–542. https://doi.org/10.1007/s10295-014-1573-6
Ruan Z, Zanotti M, Wang X, Ducey C, Liu Y (2012) Evaluation of lipid accumulation from lignocellulosic sugars by Mortierella isabellina for biodiesel production. Bioresour Technol 110:198–205. https://doi.org/10.1016/j.biortech.2012.01.053
Ruan Z, Zanotti M, Zhong Y, Liao W, Ducey C, Liu Y (2013) Co-hydrolysis of lignocellulosic biomass for microbial lipid accumulation. Biotechnol Bioeng 110:1039–1049. https://doi.org/10.1002/bit.24773
Sandberg TE, Salazar MJ, Weng LL, Palsson BO, Feist AM (2019) The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab Eng 56:1–16. https://doi.org/10.1016/j.ymben.2019.08.004
Shen H, Zhang X, Gong Z, Wang Y, Yu X, Yang X, Zhao Z (2017) Compositional profiles of Rhodosporidium toruloides cells under nutrient limitation. Appl Microbiol Biotechnol 101:3801–3809. https://doi.org/10.1007/s00253-017-8157-0
Shishir PSC, Gregg TB, Michael EH, Bruce ED (2011) Deconstruction of lignocellulosic biomass to fuels and Cchemicals. Annu Rev Chem Biomol Eng 2:121–145. https://doi.org/10.1146/annurev-chembioeng-061010-114205
Thompson OA, Hawkins GM, Gorsich SW, Doran-Peterson J (2016) Phenotypic characterization and comparative transcriptomics of evolved Saccharomyces cerevisiae strains with improved tolerance to lignocellulosic derived inhibitors. Biotechnol Biofuels 9:200. https://doi.org/10.1186/s13068-016-0614-y
Wang X, Khushk I, Xiao Y, Gao Q, Bao J (2018) Tolerance improvement of Corynebacterium glutamicum on lignocellulose derived inhibitors by adaptive evolution. Appl Microbiol Biotechnol 102:377–388. https://doi.org/10.1007/s00253-017-8627-4
Wei L, Cao L, Miao Y, Wu S, Xu S, Wang R, Du J, Liang A, Fu Y (2017) Transcriptome analysis of Spodoptera frugiperda 9 (Sf9) cells infected with baculovirus, AcMNPV or AcMNPV-BmK IT. Biotechnol Lett 39:1129–1139. https://doi.org/10.1007/s10529-017-2356-8
Wen Z, Wu M, Lin Y, Yang L, Lin J, Cen P (2014) Artificial symbiosis for acetone-butanol-ethanol (ABE) fermentation from alkali extracted deshelled corn cobs by co-culture of Clostridium beijerinckii and Clostridium cellulovorans. Microb Cell Factories 13:1–11. https://doi.org/10.1186/s12934-014-0092-5
Wu D, Wang D, Hong J (2020) Effect of a novel alpha/beta hydrolase domain protein on tolerance of K. marxianus to lignocellulosic biomass derived inhibitors. Front Bioeng Biotechnol 8:844. https://doi.org/10.3389/fbioe.2020.00844
Wu Y, Wang T, Wang K, Liang Q, Bai Z, Liu Q, Pan Y, Jiang B, Zhang L (2016) Comparative analysis of the chrysanthemum leaf transcript profiling in response to salt stress. PLoS One 11:e0159721. https://doi.org/10.1371/journal.pone.0159721
Xu J, Liu N, Qiao K, Vogg S, Stephanopoulos G (2017) Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation. Proc Natl Acad Sci U S A 114:E5308–E5316. https://doi.org/10.1073/pnas.1703321114
Xu Z, Lei P, Zhai R, Wen Z, Jin M (2019) Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol Biofuels 12:32. https://doi.org/10.1186/s13068-019-1376-0
Yang X, Nambou K, Wei L, Hua Q (2016) Heterologous production of α-farnesene in metabolically engineered strains of Yarrowia lipolytica. Bioresour Technol 216:1040–1048. https://doi.org/10.1016/j.biortech.2016.06.028
Yook SD, Kim J, Gong G, Ko JK, Um Y, Han SO, Lee SM (2020) High-yield lipid production from lignocellulosic biomass using engineered xylose-utilizing Yarrowia lipolytica. GCB Bioenergy 12:670–679. https://doi.org/10.1111/gcbb.12699
Yu X, Zheng Y, Dorgan KM, Chen S (2011) Oil production by oleaginous yeasts using the hydrolysate from pretreatment of wheat straw with dilute sulfuric acid. Bioresour Technol 102:6134–6140. https://doi.org/10.1016/j.biortech.2011.02.081
Zhou L, Wen Z, Wang Z, Zhang Y, Ledesma-Amaro R, Jin M (2020) Evolutionary Engineering Improved d-Glucose/Xylose Cofermentation of Yarrowia lipolytica. Ind Eng Chem Res 59:17113–17123. https://doi.org/10.1021/acs.iecr.0c00896
Acknowledgements
We would like to thank Dr. Rodrigo Ledesma-Amaro for providing Y. lipolytica yl-XYL+.
Funding
This work was supported by “Natural Science Foundation of Jiangsu Province,” Grant No. BK20170037 and BK20170829; “National Key R&D Program of China,” Grant No. 2016YFE0105400; “National Natural Science Foundation of China,” Grant Nos. 21606132 and 21706133.
Author information
Authors and Affiliations
Contributions
JM and WZ conceived and designed research. WZ, ZL, WZ, ZY, LM, and ZH performed the experiments; WZ, ZL, XZ, and WZ analyzed the data; JM, XZ, and WZ coordinated and supervised this study; WZ drafted the manuscript. ZL, XZ, and JM revised the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals that were performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Z., Zhou, L., Lu, M. et al. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl Microbiol Biotechnol 105, 1745–1758 (2021). https://doi.org/10.1007/s00253-021-11130-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-021-11130-3