Abstract
Although a variety of whole-cell biosensors and biosorbents have been developed for detection and removal of heavy metal contaminants, few whole cells can be applied to both monitoring and remediation of copper pollution in water. In this study, a modified plasmid was constructed by incorporating a copper-sensing element and a copper-adsorbing element into a temperature-inducible plasmid, pBV220. This plasmid was subsequently transformed into an engineered Escherichia coli strain lacking copA and cueO. This dual-functional E. coli cell selectively responded to copper ions with a linear detection range of 0.01–25 μM at 37 °C and could express surface-displayed CueR when treated at 42 °C without any costly chemical inducers. The display of CueR on the cell surface specifically enhanced its copper adsorption capacity and rapidly removed copper ions from aqueous solutions. In addition, the CueR surface-displayed cells could be regenerated by adsorption-desorption cycles via pH regulation. Moreover, by simply using two different temperatures, the detection or adsorption of copper using this dual-functional whole cell was achieved without any cross-interference. Most importantly, it provided highly sensitive, accurate quantification, and effective removal of copper in real environmental water samples. Thus, this E. coli cell can be used for large-scale detection and remediation of copper pollutants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
While Copper (Cu) is one of the most ubiquitous trace elements essential for all living organisms, chronic exposure to excess copper is involved in the development of various human diseases (Cox and Moore 2002). Continuous discharge of copper-containing industrial effluent into the environment is greatly harmful to human health and ecosystems. Therefore, extensive monitoring and efficient remediation of copper should be of vital importance. Conventionally, copper detection and removal in wastewater have primarily relied on physicochemical technologies (Blake et al. 2014; Sabermahani and Taher 2014). Although they are accurate and effective, most of these methods require expensive instruments that are difficult to operate. Thus, the application of biotechnology has become an attractive alternative because the methods are usually low cost and eco-friendly. Various whole-cell biosensors have been developed for the detection of different heavy metals. The sensing of metals by whole-cell biosensors is typically based on heavy metal resistance/homeostasis mechanisms. For example, E. coli has evolved several copper efflux systems to survive when exposed to toxic levels of copper (Rensing and Grass 2003). Among these, CueR acts as the key transcription regulator of the copper tolerance genes, copA and cueO (Outten et al. 2000). As a P-type ATPase located on the cytoplasmic membrane, CopA was proposed to export Cu(I) from the cytoplasm to the periplasm (Rensing et al. 2000). CueO is a multi-copper oxidase that converts periplasmic Cu(I) to the less toxic form Cu(II) (Grass and Rensing 2001). Recently, whole-cell biosensors harboring a vector with simple fusion of the copA promoter (PcopA) and a fluorescent protein-encoding gene were constructed to detect bioavailable copper ions (Kang et al. 2018; Li et al. 2014) and the E. coli was engineered to remove copper by adsorption through surface display of a copper-binding peptide (Ravikumar et al. 2011).
Biodetection and bioremediation of heavy metals were usually achieved by utilizing at least two different engineered whole-cell strains that could detect or remove metal ions respectively (Wei et al. 2014). Currently, few bacterial cells have been reported that can do both. One exception is the carboxylesterase E2 surface-displayed E. coli cell constructed by Yin and coworkers (Yin et al. 2016). Furthermore, overexpression of metal binding proteins/peptides that enhance whole-cell bioadsorption capacity is primarily induced by chemical reagents, such as isopropyl β-d-thiogalactoside (IPTG), or arabinose, which would be extremely costly when treating large volumes of wastewater.
In this study, we developed a dual-functional E. coli cell that can detect and remove copper ions. This whole cell responds to copper ions by emitting green fluorescence at their optimal temperature of 37 °C. When the temperature switches to 42 °C, the copper-binding protein CueR will be displayed on the outer membrane of the E. coli cell without any chemical inducers, resulting in enhanced adsorption of copper ions. In addition, the bound copper ions can be desorbed by modulating the pH value, and the whole cell can be used for repeated adsorption. Thus, this E. coli cell performs as a biosensor at 37 °C and becomes adsorbent at 42 °C. This technology allows for sensitive, accurate measurement, and efficient removal of copper in various field water samples and may provide a useful strategy for large-scale whole-cell detection and remediation of heavy metal pollutants.
Materials and methods
Construction of dual-functional E. coli cell for biodetection and bioadsorption of copper
Three DNA fragments encoding the N-terminal amino acids (1–29) of prolipoprotein (Lpp) (EcoGene accession number: EG10544), residues 45–159 of the outer membrane protein OmpA (EcoGene accession number: EG10669), and the entirety of CueR (EcoGene accession number: EG13256) were amplified by PCR from E. coli JM109 genomic DNA using the primers Lpp-1/Lpp-2, OmpA-1/OmpA-2, and CueR-1/CueR-2 (Table S1 in the Supplementary Material), respectively. The primers Lpp-1 and CueR-2 were used to join the DNA sequences by recombinant PCR. The obtained PCR product lpp-ompA-cueR was digested with EcoRI and SalI, and subsequently cloned into the same restriction sites on pBV220 to generate the plasmid pBV220-lpp-ompA-cueR. Next, the copper-sensing element PcopA-egfp, consisting of the upstream promoter region of copA (− 227 to − 1 bp, PcopA) (EcoGene accession number: EG13246) and egfp (encoding enhanced green fluorescent protein), was amplified as described above using the PcopA-1/PcopA-2, eGFP-1/eGFP-2 primers, respectively. This element was then inserted into the site following the rrnB transcriptional terminator of pBV220-lpp-ompA-cueR, which had been pre-linearized by inverse PCR amplification with pBV220-1 and pBV220-2 primers. This restriction site-free cloning was achieved using ClonFast kit based on seamless cloning technology (Messerschmidt et al. 2016), resulting in the dual-functional plasmid, pBV220-lpp-ompA-cueR-PcopA-egfp (abbreviated as pBV-LOCG). Another two plasmids only expressing intracellular CueR (pBV-CG, pBV220-cueR-PcopA-egfp) or the membrane fusion protein Lpp-OmpA (pBV-LOG, pBV220-lpp-ompA-PcopA-egfp) were also constructed similarly.
E. coli strains lacking copA and/or cueO (EcoGene accession number: EG12318) (ΔcopA or ΔcueO single mutant, and ΔcopA/cueO double mutant) were genetically engineered from a wild-type E. coli strain (MC4100) following the gene-knockout procedures described by Datsenko and Wanner (Datsenko and Wanner 2000). The primers for gene deletion (CopA-D1/CopA-D2, CueO-D1/CueO-D2) are listed in Table S1. The final dual-functional E. coli cell (pBV-LOCG/ΔcopA/cueO) was obtained by transforming pBV-LOCG into E. coli ΔcopA/cueO, which has been deposited at China General Microbiological Culture Collection Center (CGMCC No. 17078).
Fluorescence assay of eGFP and SDS-PAGE/Western blotting analysis of surface-displayed CueR
The pBV-LOCG/ΔcopA/cueO was grown in M9S medium (M9 minimal medium supplemented with 0.2% glucose, 5 μg/mL thiamin, and the 20 amino acids (10 μg/mL each)) at 37 °C to OD600 = 0.5. The culture was equally divided into four tubes and then treated with or without 10 μM Cu2+ at 37 °C or 42 °C. After 4 h, the cells were harvested and resuspended in Tris buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.5) to OD600 = 10. The fluorescence intensity of eGFP was measured using a fluorescence spectrophotometer (λem = 510/λex = 480 nm). The expression of surface-displayed CueR was detected by SDS-PAGE and Western blot using an anti-CueR monoclonal antibody (produced by GenScript Biotech Corp.). To verify the surface display of CueR, cells were incubated with trypsin (100 μg/mL final) at 37 °C for 1 h. The reaction was quenched by washing cells twice with Tris buffer, and then CueR was detected by SDS-PAGE and Western blot.
Biodetection of copper ions using dual-functional E. coli cell
pBV-LOCG was transformed into E. coli MC4100, ΔcopA, ΔcueO, and ΔcopA/cueO. The cells were grown in M9S medium at 37 °C until the OD600 reached 0.5. Cu2+ solution (10−10 to 10−1 M) was added to the medium. After 4 h of incubation at 37 °C, cells were harvested and resuspended in Tris buffer before measurement of eGFP signal. The linear regression curve showing copper detection by pBV-LOCG/ΔcopA/cueO was generated following the same procedures, while induced by Cu2+ solution at final concentrations of 0.001, 0.005, 0.01, 0.025, 0.1, 0.5, 1, 5, 10, 25, and 50 μM.
To examine the copper detecting selectivity of pBV-LOCG/ΔcopA/cueO, final concentrations of 5 and 25 μM Cu2+, Pb2+, Hg2+, Fe3+, Zn2+, Ni2+, Co2+, Cr3+, Cd2+, and Ag+ aqueous solutions were added to cell cultures (OD600 = 0.5), respectively. To explore possible interference of other ions on the copper detecting selectivity, Cu2+ (5 μM) with each of the other 9 metals (25 μM) were simultaneously added to the bacterial solutions. Cells were incubated at 37 °C for 4 h, harvested, and resuspended in Tris buffer for measurement of eGFP signal.
Bioadsorption of copper ions using dual-functional E. coli cell
E. coli cells were grown in M9S medium at 37 °C to OD600 = 0.5, and the surface-displayed CueR was induced by switching the temperature to 42 °C. After 4 h, the cells were harvested and washed with deionized water at least three times. The cells were resuspended in water solutions (OD600 = 10) containing 5, 25, and 125 μM Cu2+ and incubated at 25 °C for 10 min. Copper-bound cells were harvested by centrifugation, lysed by microwave digestion, and analyzed for copper content using inductively coupled plasma mass spectrometry (ICP-MS). Copper adsorption capacity (μmol/g) was defined as the amount of copper bound per gram (dry weight) of bacterial cells, which can be measured after lyophilization. To test the copper adsorption selectivity of pBV-LOCG/ΔcopA/cueO, surface-displayed CueR was induced at 42 °C or not induced at 37 °C for 4 h. The induced and uninduced cells were simultaneously treated with 25 μM Cu2+, Ag+, Zn2+, Cd2+, Ni2+, Cr3+, Hg2+, Fe3+, Co2+, or Pb2+. To explore any effects of other metals on copper adsorption, 25 μM Cu2+ and 125 μM Cu2+, Ag+, Zn2+, Cd2+, Ni2+, Cr3+, Hg2+, Fe3+, Co2+, or Pb2+ were jointly added to the bacterial solutions, and the cells were incubated at 25 °C for 10 min prior to measuring copper adsorption capacity.
Desorption of copper ions and regeneration of dual-functional E. coli cell
To evaluate the regeneration ability of the dual-functional E. coli cell for copper adsorption, cells induced with surface-displayed CueR (OD600 = 10) were treated with 1 mM Cu2+ and then washed twice with deionized water to remove unbound copper ions. The copper-treated cells were resuspended in Tris buffers with pH values of 2, 3, 4, 5, 6, and 7 and incubated at 25 °C for 1 h. After 10 min of centrifugation, the total copper content in each cell pellet was measured by ICP-MS. After copper desorption at the optimal pH, cells were treated with 1 mM Cu2+ for repeat analysis of the copper adsorption capacity using ICP-MS. At least 5 cycles of adsorption-desorption were performed.
Detection and remediation of copper in field water samples using dual-functional E. coli cell
Water samples (117 in total) were randomly collected from rivers throughout three industrial districts in Wenzhou (Southeast China). The samples were centrifuged (12,000 rpm, 10 min) to remove any sediment, and the supernatants were transferred to new tubes for independent copper detection by the dual-functional E. coli cell and ICP-MS. For the whole-cell assay, the samples were adjusted to pH ≈ 2 using HNO3 (Merck) at a final concentration of 10–15 mM. The cells were pre-incubated in M9S medium at 37 °C until reaching OD600 = 0.5 and harvested into half of the original volume using freshly prepared × 2 M9S medium. The acidified field sample (1 mL) was mixed with 1 mL of the cell suspension. The mixture was incubated at 37 °C for 4 h before measuring eGFP signal. The total copper content in the samples was determined based on linear standard curves obtained from acidified copper-free (< 0.1 μg/L) field water spiked with Cu2+ solution (final concentrations of 0.01, 0.025, 0.1, 0.5, 1, 5, 10, and 25 μM). To remove copper in these water samples, E. coli cells with surface-displayed CueR were prepared at 42 °C and directly resuspended in the water samples (50 mL, OD600 = 10). After incubated at 25 °C for 10 min, the cells were removed by centrifugation (12,000 rpm, 10 min). Following the same procedures as described above, the total copper contents of the treated water samples (supernatant fraction) were quantified again using the whole cell and ICP-MS.
Results
Characterization of dual-functional E. coli cell under different conditions
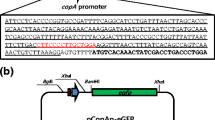
Few bacteria-based whole cells can be used for simultaneous detection and remediation of copper. Therefore, we aimed to develop a dual-functional bacterial cell by combining copper-sensing and copper-adsorbing elements into a single vector. To avoid interference between detection and adsorption, the copper-sensing and copper-adsorbing elements must be controlled by different conditions. Thus, pBV220 was utilized to construct the dual-functional plasmid due to its strong temperature-inducible promoter, PLPR, which can be induced for high-yield protein overexpression at 42 °C (Hu et al. 1993). The copper-binding element lpp-ompA-cueR and the copper-sensing fragment PcopA-egfp were inserted in opposite directions into pBV200 (Fig. 1a). Furthermore, the transcriptional termination region of rrnB gene located between the two elements ensured that the transcriptional expression of one element would not activate the other. The constructed dual-functional vector pBV-LOCG was transformed to E. coli ΔcopA/cueO, which was the most sensitive host cell for copper detection (Fig. 2a). The dual-functional whole cell, pBV-LOCG/ΔcopA/cueO, was subsequently analyzed. After 4 h of treatment with 10 μM Cu2+ at 37 °C, fluorescence measurements and SDS-PAGE analyses showed the E. coli strongly responded to copper by producing eGFP (27 kDa); no significant eGFP was detected in the absence of copper (Fig. 1b and c). At other temperatures, the whole cell also responded to copper. The time course results showed a sharper increase in fluorescence intensity during the first 4 h of incubation at 37 °C as compared to other temperatures (Fig. S1), indicating that the optimal copper detection occurs at 4 h of incubation at 37 °C. As shown in Fig. S2, fluorescence intensity was highest when the cell density reached OD600 = 0.5 before copper was added. Therefore, samples were mixed with a bacterial solution at this density in the following experiments. To induce expression of the copper adsorbing protein Lpp-OmpA-CueR, the cells were incubated at 42 °C for 4 h; this 31-kDa protein was observed by SDS-PAGE (Fig. 1b), but was not induced at other temperatures (Fig. S3), suggesting that 42 °C was the optimal temperature for overexpression of surface-displayed CueR. Because the molecular weight of Lpp-OmpA-CueR is similar to that of eGFP, they cannot be easily differentiated by SDS-PAGE. Therefore, surface-displayed CueR was further verified by Western blot using anti-CueR antibodies, which showed results consistent with the SDS-PAGE analyses. To further ascertain the cell surface localization of CueR, we conducted a protease accessibility experiment. The protease cannot readily permeate through the membrane, thus only proteins exposed on the cell surface are digested. After 1 h of incubation with trypsin, intracellular CueR was resistant to protease digestion (Fig. 1d). In contrast, surface-displayed CueR was almost degraded under the same condition, suggesting that CueR was localized on the surface at 42 °C. The above results collectively illustrate that the dual-functional whole cell detects copper and becomes copper adsorbent under temperature control.
a Schematic diagram of the dual-functional plasmid, pBV-LOCG. b SDS-PAGE gel (top) and Western blot (bottom) of surface-displayed CueR induced by pBV-LOCG/ΔcopA/cueO under different conditions. c Fluorescence measurements of eGFP expressed by pBV-LOCG/ΔcopA/cueO under different conditions. d SDS-PAGE gel (top) and Western blot (bottom) showing a trypsin accessibility analysis of E. coli cell with intracellular and surface-displayed CueR. Cells were incubated with 100 μg/mL trypsin at 37 °C for 1 h. The results are representative of three independent experiments
a eGFP intensity in response to increasing concentrations of copper ions for wild type, ΔcopA, ΔcueO, and ΔcopA/cueO E. coli cells transformed with pBV-LOCG. b Determination of the linear detection range of pBV-LOCG/ΔcopA/cueO towards copper concentration (0.01–25 μM, R2 = 0.996). c The eGFP responses of pBV-LOCG/ΔcopA/cueO to different metals. d Effect of other heavy metals on the copper detection ability of pBV-LOCG/ΔcopA/cueO. Data are the means of three independent experiments; error bars represent the standard deviation
Biodetection of copper ions using dual-functional E. coli cell
Although many bioreporters for heavy metal detection have been constructed, they have not been widely used to analyze authentic environmental samples, partly due to their insensitivity. To address this, we removed the copper efflux transporter CopA to test whether this would increase the sensitivity of the E. coli bioreporter. We also tested whether the deletion of another copper tolerance gene, cueO, would further improve the copper sensitivity. Thus, we constructed ΔcopA, ΔcueO, and ΔcopA/cueO E. coli strains. As shown in Fig. 2A, E. coli MC4100 did not respond to copper ions until the concentration reached 100 μM. For ΔcueO, there was no significant sensitivity improvement, though its eGFP intensity was slightly higher than that of MC4100. Consistent with a previous study (Kang et al. 2018), ΔcopA was much more sensitive and responded to copper ions as low as 10 nM. Interestingly, the eGFP intensity of ΔcopA/cueO was higher than that of ΔcopA within the copper concentration range of 0.01–100 μM, suggesting that removal of cueO further enhanced the sensitivity of the copper bioreporter. Hence, the ΔcopA/cueO harboring the pBV-LOCG plasmid was used as the dual-functional whole cell. As presented in Fig. 2b, the linear range for copper detection of the bioreporter was 0.01–25 μM (0.64–1600 μg/L; R2 = 0.996) and the detection limit (LOD) was 6.82 nM or 0.44 μg/L (signal-to-noise ratio = 3), which was a wider linear range and more sensitive LOD than previously reported whole-cell bioreporters and comparable to some nanomaterial-based biosensors (Kanellis 2018) or conventional physicochemical methods such as atomic absorption spectroscopy (AAS) (Zaman et al. 2018). Because of the complexity of environmental samples, which may contain various types of metal ions, we evaluated the copper selectivity of the whole cell. The eGFP signal was significantly induced by copper but not by the other metals, except silver (Fig. 2c). Furthermore, no significant change of the eGFP intensity was observed even when 5-fold excess of other metals (including silver) was added together with 5 μM Cu2+ (Fig. 2d), implying that the whole cell selectively responds to copper over silver if both are present. Given that there are usually negligible amounts of silver in environmental samples, these results suggest that copper detection using this whole cell would not be affected by other metals (including silver).
Bioadsorption of copper ions using dual-functional E. coli cell
Next, we investigated the copper adsorption capacities of the dual-functional E. coli cell. After 4 h of incubation at 42 °C, surface-displayed CueR was induced in the MC4100, ΔcopA, ΔcueO, and ΔcopA/cueO strains. After treatment with 5, 25, and 125 μM Cu2+ solutions, we found the adsorption capacity of pBV-LOCG/ΔcopA/cueO was higher than the other whole cells at each Cu2+ concentration (Fig. 3a). This result should be attributed to the deletion of copA and cueO, which were previously proposed to enhance intracellular copper accumulation (Rensing et al. 2000). In combination with the copper detection results in Fig. 2a, pBV-LOCG/ΔcopA/cueO was determined to be the optimal dual-functional E. coli cell for copper detection and adsorption.
a The copper adsorption capacities of wild type, ΔcopA, ΔcueO, and ΔcopA/cueO E. coli cells transformed with pBV-LOCG after treatment at 42 °C. b The copper adsorption capacity of ΔcopA/cueO transformed with pBV-LOCG or pBV-LOG after treatment at 37 °C or 42 °C. c The ratios of metal binding capacities of pBV-LOCG/ΔcopA/cueO (capacity at 42 °C/capacity at 37 °C). d Effect of other heavy metals on the copper adsorption of pBV-LOCG/ΔcopA/cueO. Data are the means of three independent experiments; error bars represent the standard deviation
To verify whether the enhanced copper adsorption capacity was due to the expression of surface-displayed CueR, we compared it with uninduced cells and cells expressing only Lpp-OmpA. After incubation with Cu2+ solutions, pBV-LOCG/ΔcopA/cueO pre-treated at 42 °C adsorbed approximately 3.8–5.9-fold more copper than the uninduced cells. The expression of Lpp-OmpA on the cell surface did not significantly increase copper adsorption capacity (Fig. 3b). Furthermore, a water sample prepared with 25 μM Cu2+, which is close to the World Health Organization recommended allowable copper content in drinking water (1.5 mg/L or 23.4 μM) (Bilal et al. 2013), was used to verify the copper removal capability of the whole cell. As shown in Fig. S4, the copper concentration of the water sample decreased by 22.5%, 51.7%, 78.3%, and 91.5% after 10 min of incubation with the whole cell pre-treated at 42 °C (OD600 = 2, 5, 10, 20) (Fig. S4a), whereas the same amount of 37 °C treated whole cell removed only 11.2%, 15.5%, 28.3%, and 32.6% of the total copper, respectively (Fig. S4b). The maximum copper adsorption capacity of pBV-LOCG/ΔcopA/cueO was approximately 358 μmol/g cells based on the dose-dependent saturation curve (Fig. S5), which was comparable with reported bioadsorption or physicochemical techniques for copper remediation (Bilal et al. 2013; Fu and Wang 2011). These results demonstrate that the display of CueR on the cell surface largely enhances the copper adsorption capability and results in efficient removal of copper ions from aqueous solutions.
For metal ions bioadsorption using microorganisms, the accumulation of specific metal ions from the mixture is generally expected, which may facilitate the recovery and recycling of the desired metal. Therefore, the selectivity of pBV-LOCG/ΔcopA/cueO to adsorb copper was further examined. Most microorganisms have intrinsic abilities to adsorb diverse metal ions, thus the ratio of metal adsorption capacity of induced cells to that of uninduced cells was used to assess the copper adsorption selectivity. As shown in Fig. 3d, the ratio of copper adsorption of the 42 °C treated cells to that of the 37 °C treated ones was much higher compared to the other metals. Induction of CueR on the cell surface did not significantly increase adsorption of the other metals (except silver). These results are in agreement with a previous study suggesting that CueR is a copper- and silver-binding protein (Stoyanov et al. 2001). The influences of the other metal ions on copper adsorption were also tested. The adsorption capacity of pBV-LOCG/ΔcopA/cueO was unaffected in the presence of 5-fold excess of other metal ions, including Ag+, Zn2+, Cd2+, Ni2+, Cr3+, Hg2+, Fe3+, Co2+, or Pb2+ (Fig. 3e).
Desorption of copper ions and regeneration of dual-functional E. coli cell
To be used in wastewater treatment, it will be necessary to regenerate the biosorbent to ensure a low-cost process and continuous biomass supply. Therefore, the adsorption process should be followed by contaminant desorption. Metal stripping from biomass can usually be achieved with inexpensive acids or chelators, such as HCl and EDTA (Singh et al. 2008). Thus, desorption of copper-bound dual-functional E. coli cells was performed in Tris buffers of varying pH. As shown in Fig. S6, only 10.9–13.7% was Cu2+ released from the dual-functional E. coli cell at pH 6.0 and 7.0, while almost 89.7% Cu2+ desorbed when the pH was decreased to 3.0. Since biomass would denature after strong acid treatment (pH < 2.0), preventing its reuse, pH 3.0 was used to desorb copper and regenerate the dual-functional E. coli cell. Compared to a freshly prepared E. coli cell, no significant loss in adsorption capacity was observed in the regenerated bacteria during the first three repeated adsorption-desorption cycles (> 90%), and 69.2% binding capacity remained at the end of the fifth cycle (Fig. S7). These results demonstrate that the dual-functional E. coli cell has good regeneration ability for continued copper bioadsorption.
Application of dual-functional E. coli cell in detection and remediation of copper in environmental water samples
To assess the applicability of pBV-LOCG/ΔcopA/cueO for environmentally relevant samples, it was tested at large scale on 117 water samples collected from places where contaminant levels are usually very high. Because copper exists in a variety of physicochemical forms or speciations in environmental aquatic samples (Wang et al. 2016) and the bacterial biosensor only detects freely diffusible (bioavailable) metal ions, acid was routinely added to ensure the samples were dissolved and to prevent complex formation (Thompson et al. 2013). In this study, samples were acidified to pH of 2 using nitric acid and then neutralized to pH ≈ 7 when mixed with × 2 M9S medium. The total copper content in each sample was analyzed by the dual-functional whole cell and ICP-MS for comparison. The correlation coefficient between copper concentrations measured by the whole cell and ICP-MS was 0.941, and the average accuracy of the whole cell for copper quantification was 93.8 ± 30.1% as compared to ICP-MS (Fig. 4a). Among the 117 tested samples, 11 samples were identified to be hazardous for potable water (copper concentration above 1.5 mg/L) by ICP-MS. Only two samples (1.7%) were false negatively determined to be safe, and no false positives were identified by whole cell (Fig. 4a). Thus, this E. coli cell is a reliable tool for high-throughput screening of copper in environmental samples.
Comparative analysis between the copper detection of 117 field water samples using the dual-functional E. coli cell and ICP-MS. The copper concentrations of untreated (a) or treated (b) samples determined by both methods were fit to two correlation curves. The accuracy was defined as the percentage ratio of the copper concentration analyzed by the whole cell to ICP-MS (n = 117). The ratios of copper contents obtained from the whole-cell assay (d) or ICP-MS analysis (c) in the treated sample to the matching untreated samples were plotted as percentages with dashed lines representing the average values (n = 117)
To remediate copper in the water samples, they were treated with the 42 °C pre-treated whole cell for 10 min. The copper content of treated samples was requantified by whole cell and ICP-MS. After treatment, the average copper concentration of all samples decreased to about 1/4 (25.2 ± 9.2%) of the original content, as determined by the whole-cell assay (Fig. 4d), which was similar to the ICP-MS results (21.5 ± 7.1%) (Fig. 4c). All of the 11 over-limit samples were detoxified to safe levels as analyzed by both methods. In the treated samples, close correlation (R2 = 0.964) and high accuracy (113.2 ± 51.7%) for copper detection were observed for the two methods (Fig. 4b). These results demonstrate the effectiveness of the dual-functional whole cell to remove copper in water and proved its authenticity and reliability for copper detection in environmental samples either prior to or after bioremediation. The overall strategy and technical roadmap for biodetection and bioremediation of copper ions using the dual-functional E. coli cell were illustrated in Fig. 5.
Discussion
During recent decades, continuous efforts have been directed towards the development and optimization of bacteria-based whole cells for monitoring and removal of heavy metal contaminants. For detection, improvements in specificity and sensitivity were always the priority. For example, some bioreporters with broad selectivity were modified to respond to one single metal ion by altering amino acid sequences of the native regulatory protein (Hakkila et al. 2011). To increase sensitivity, whole-cell bioreporters were usually engineered for overexpression of metal uptake systems to enhance contaminant import (Selifonova et al. 1993) or efflux transporters were deleted to decrease the export of detectable ions from the cell (Hynninen et al. 2010). Deletion of the copper efflux pump copA dramatically enhanced the sensitivity to copper in an earlier study (Kang et al. 2018). Beyond this, we constructed a ΔcopA/cueO double mutant as the host cell. Although CueR was found to exhibit very high sensitivity to free Cu(I) (Changela et al. 2003), CueO-mediated oxidation of Cu(I) to Cu(II) (Grass and Rensing 2001) would attenuate the copper-sensing capability. Thereby, the deletion of cueO could hinder Cu(I) oxidation, which would further increase the copper responses of bioreporters (Fig. 2a). As another advantage, the deletion of cueO could also increase the whole-cell accumulation of copper (Fig. 3a). Under the aerobic condition, the majority of copper ions may exist in water in the divalent copper (II) valence state, to which CueR cannot readily bind. However, the NADH-linked cupric reductase activity from the E. coli respiratory chain may promote the reduction of intracellular Cu(II) to Cu(I) (Rapisarda et al. 1999), which would enable surface-displayed CueR to adsorb copper in aqueous solution. This scenario reasonably explains the enhanced accumulation of membrane-bound Cu(I) when CueO-mediated copper oxidation was eliminated.
It should be noted that the test medium also exerts an effect on the sensitivity and accuracy of whole-cell bioassay. Compared to defined minimal medium, the rich medium was considered to promote the biosynthesis of reporter protein and enhance the response of bioreporters due to nutrients abundance. Nevertheless, the complex components of the rich medium may bind metal ions and influence metal speciation, resulting in diminished bioavailability of these metals. The limit of whole-cell copper detection in M9 minimal medium was determined to be much lower than that in LB medium (Li et al. 2014). Consistently, the whole cell constructed in this study was able to detect as low as 10 nM copper with a broad linear detection range in this medium. On the other hand, to overcome its drawback, the M9 minimal medium was supplemented with glucose, thiamin, and 20 amino acids (defined as M9S medium), which was found to greatly enhance the expression of eGFP and induction surface-displayed CueR (results not shown). Furthermore, the phosphate buffer system in M9S medium provides very strong buffering capability and could neutralize acidified water samples of pH 2 to approximately 7 without requiring additional alkaline reagents, such as NaOH or Na4P2O7 (Trang et al. 2005).
With the increasing heavy metal pollution, the development of biosensors able to be applied on site to monitor environmental contaminants is an urgent demand. Researchers have attempted to integrate biosensors into miniaturized devices for easier field testing (Roggo and van der Meer 2017). Given the advantage of bioadsorbent-based remediation, it will be an effective approach to establish combinational whole-cell-based systems for real-time monitoring and continuous treatment of metal pollutants. This dual-functional E. coli cell described herein enables sensitive, accurate detection of copper, and efficiently removes copper in various field samples by simply controlling the temperature. Moreover, it can provide reliable feedback on the metal concentrations of waste effluents which have been treated. Although further research efforts are needed before bacterial cells can be widely applied, our work may provide the feasibility of online biodetection and bioremediation of heavy metal pollutants using this dual-functional whole cell.
References
Bilal M, Shah JA, Ashfaq T, Gardazi SM, Tahir AA, Pervez A, Haroon H, Mahmood Q (2013) Waste biomass adsorbents for copper removal from industrial wastewater--a review. J Hazard Mater 263(Pt 2):322–333. https://doi.org/10.1016/j.jhazmat.2013.07.071
Blake D, Nar M, D'Souza NA, Glenn JB, Klaine SJ, Roberts AP (2014) Treatment with coated layer double hydroxide clays decreases the toxicity of copper-contaminated water. Arch Environ Contam Toxicol 66(4):549–556. https://doi.org/10.1007/s00244-013-9986-1
Changela A, Chen K, Xue Y, Holschen J, Outten CE, O’Halloran TV, Mondragon A (2003) Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science (New York, NY) 301(5638):1383–1387. https://doi.org/10.1126/science.1085950
Cox DW, Moore SD (2002) Copper transporting P-type ATPases and human disease. J Bioenerg Biomembr 34(5):333–338
Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97(12):6640–6645. https://doi.org/10.1073/pnas.120163297
Fu F, Wang Q (2011) Removal of heavy metal ions from wastewaters: a review. J Environ Manag 92(3):407–418. https://doi.org/10.1016/j.jenvman.2010.11.011
Grass G, Rensing C (2001) Genes involved in copper homeostasis in Escherichia coli. J Bacteriol 183(6):2145–2147. https://doi.org/10.1128/jb.183.6.2145-2147.2001
Hakkila KM, Nikander PA, Junttila SM, Lamminmaki UJ, Virta MP (2011) Cd-specific mutants of mercury-sensing regulatory protein MerR, generated by directed evolution. Appl Environ Microbiol 77(17):6215–6224. https://doi.org/10.1128/aem.00662-11
Hu B, Li J, Yu W, Fang J (1993) Cloning of human prourokinase cDNA without the signal peptide and expression in Escherichia coli. Chin J Biotechnol 9(2):95–101
Hynninen A, Tonismann K, Virta M (2010) Improving the sensitivity of bacterial bioreporters for heavy metals. Bioeng Bugs 1(2):132–138. https://doi.org/10.4161/bbug.1.2.10902
Kanellis VG (2018) Sensitivity limits of biosensors used for the detection of metals in drinking water. Biophys Rev 10(5):1415–1426. https://doi.org/10.1007/s12551-018-0457-9
Kang Y, Lee W, Kim S, Jang G, Kim BG, Yoon Y (2018) Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering. Appl Microbiol Biotechnol 102(3):1513–1521. https://doi.org/10.1007/s00253-017-8677-7
Li PS, Peng ZW, Su J, Tao HC (2014) Construction and optimization of a Pseudomonas putida whole-cell bioreporter for detection of bioavailable copper. Biotechnol Lett 36(4):761–766. https://doi.org/10.1007/s10529-013-1420-2
Messerschmidt K, Hochrein L, Dehm D, Schulz K, Mueller-Roeber B (2016) Characterizing seamless ligation cloning extract for synthetic biological applications. Anal Biochem 509:24–32. https://doi.org/10.1016/j.ab.2016.05.029
Outten FW, Outten CE, Hale J, O’Halloran TV (2000) Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, cueR. J Biol Chem 275(40):31024–31029. https://doi.org/10.1074/jbc.M006508200
Rapisarda VA, Montelongo LR, Farias RN, Massa EM (1999) Characterization of an NADH-linked cupric reductase activity from the Escherichia coli respiratory chain. Arch Biochem Biophys 370(2):143–150. https://doi.org/10.1006/abbi.1999.1398
Ravikumar S, Yoo IK, Lee SY, Hong SH (2011) Construction of copper removing bacteria through the integration of two-component system and cell surface display. Appl Biochem Biotechnol 165(7-8):1674–1681. https://doi.org/10.1007/s12010-011-9386-9
Rensing C, Grass G (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27(2-3):197–213. https://doi.org/10.1016/s0168-6445(03)00049-4
Rensing C, Fan B, Sharma R, Mitra B, Rosen BP (2000) CopA: An Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci U S A 97(2):652–656
Roggo C, van der Meer JR (2017) Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr Opin Biotechnol 45:24–33. https://doi.org/10.1016/j.copbio.2016.11.023
Sabermahani F, Taher MA (2014) Determination of ultra trace amounts of copper by a multi-injection technique of electrothermal atomic absorption spectrometry after using solid-phase extraction. J AOAC Int 97(6):1713–1718
Selifonova O, Burlage R, Barkay T (1993) Bioluminescent sensors for detection of bioavailable Hg(II) in the environment. Appl Environ Microbiol 59(9):3083–3090
Singh A, Kumar D, Gaur JP (2008) Removal of Cu(II) and Pb(II) by Pithophora oedogonia: sorption, desorption and repeated use of the biomass. J Hazard Mater 152(3):1011–1019. https://doi.org/10.1016/j.jhazmat.2007.07.07
Stoyanov JV, Hobman JL, Brown NL (2001) CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol 39(2):502–511
Thompson CM, Ellwood MJ, Wille M (2013) A solvent extraction technique for the isotopic measurement of dissolved copper in seawater. Anal Chim Acta 775:106–113. https://doi.org/10.1016/j.aca.2013.03.020
Trang PT, Berg M, Viet PH, Van Mui N, Van Der Meer JR (2005) Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ Sci Technol 39(19):7625–7630
Wang D, Gao Y, Larsson K, Lin W (2016) Speciation of dissolved copper in human impacted freshwater and saltwater lakes. Environ Sci Pollut Res Int 23(11):10832–10840. https://doi.org/10.1007/s11356-016-6140-4
Wei W, Liu X, Sun P, Wang X, Zhu H, Hong M, Mao ZW, Zhao J (2014) Simple whole-cell biodetection and bioremediation of heavy metals based on an engineered lead-specific operon. Environ Sci Technol 48(6):3363–3371. https://doi.org/10.1021/es4046567
Yin K, Lv M, Wang Q, Wu Y, Liao C, Zhang W, Chen L (2016) Simultaneous bioremediation and biodetection of mercury ion through surface display of carboxylesterase E2 from Pseudomonas aeruginosa PA1. Water Res 103:383–390. https://doi.org/10.1016/j.watres.2016.07.053
Zaman BT, Bakirdere EG, Kasa NA, Deniz S, Sel S, Chormey DS, Bakirdere S (2018) Development of an efficient and sensitive analytical method for the determination of copper at trace levels by slotted quartz tube atomic absorption spectrometry after vortex-assisted dispersive liquid-liquid microextraction in biota and water samples using a novel ligand. Environ Monit Assess 190(7):437. https://doi.org/10.1007/s10661-018-6735-y
Funding
This work was supported by the National High Technology Research and Development Program of China (2014AA06A514), the Key Discipline of Zhejiang Province in Medical Technology (First Class, Category A), and the Science and Technology Project of Wenzhou (S20170016).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflicts of interest
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 3542 kb)
Rights and permissions
About this article
Cite this article
Wang, W., Jiang, F., Wu, F. et al. Biodetection and bioremediation of copper ions in environmental water samples using a temperature-controlled, dual-functional Escherichia coli cell. Appl Microbiol Biotechnol 103, 6797–6807 (2019). https://doi.org/10.1007/s00253-019-09984-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-09984-9