Abstract
Metals are essential to all organisms; accordingly, cells employ numerous genes to maintain metal homeostasis as high levels can be toxic. In the present study, the gene operons responsive to metal(loid)s were employed to generate bacterial cell-based biosensors to detect target metal(loid)s. The cluster of genes related to copper transport known as the cop-operon is regulated by the interaction between the copA promoter region (copAp) and CueR, turning on and off gene expression upon copper ion binding. Therefore, the detection of copper ions could be achieved by inserting a plasmid harboring the fusion of copAp and reporter genes, such as enzymes and fluorescent genes. However, copAp is not as strong a promoter as other metal-inducible promoters, such as znt-, mer-, and ars-operons; thereby, its sensitivity toward copper ions was not sufficient for quantification. To overcome this problem, we engineered Escherichia coli with a deletion of copA to interfere with copper export from cells. The engineered E. coli whole-cell bioreporter was able to detect copper ions at 0 to 10 μM in an aqueous solution. Most importantly, it was specific to copper among several tested heavy metal(loid)s. Therefore, it will likely be useful to detect copper in diverse environmental systems. Although additional improvements are still required to optimize the E. coli-based copper-sensing whole-cell bioreporters presented in this study, our results suggest that there is huge potential to generate whole-cell bioreporters for additional targets by molecular engineering.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Industrial development has led to explosive increases in the use of metals and the release of large quantities of metal(loid)s, including Cd, Fe, Hg, Ag, and As, into the environment. The adverse effects of metals were initially ignored, resulting in the common occurrence of disastrous consequences to the environment and human health related to metal exposure worldwide. Eventually, extensive studies clarified the toxic effects of metals and their severe threats to human health. As metal toxicity is not acute, it is difficult to detect pollution. To evaluate metal pollution in diverse environmental systems, instrumental analyses are typically applied to measure the concentrations of certain metals. However, it is difficult to characterize the adverse effects on organisms owing to the complexity of environmental systems. The biologically effective portion of metals, i.e., the bioavailability, not only is considered a critical factor to evaluate the adverse effects of metals, but is also difficult to determine and varies among systems (Alexander 2000; Juhasz et al. 2008; Reid et al. 2000). The adverse effects of toxic materials are generally assessed using model systems including worms, fishes, plants, and microorganisms (Foy et al. 1978; Javed and Usmani 2015; Rathnayake et al. 2013; Van Gestel and Ma 1988). Notably, previous analyses of risk based on concentration-dependent studies have only considered the total amount of toxic materials. Furthermore, these studies typically exclude the effects of environmental factors. Furthermore, toxic chemical compounds and metals can adopt a biologically inactive conformation depending on environmental factors. Thus, it is necessary to analyze the biological available portion to evaluate risk and pollution levels.
To assess the bioavailability of toxic materials, bacterial cell-based biosensors, also termed whole-cell bioreporters, have been developed based on intrinsic cellular defense mechanisms. The regulatory mechanisms that maintain homeostasis are initiated by external stimuli including toxic materials, resulting in the expression of a series of genes. The expression of these genes is suppressed by the binding of regulatory proteins to promoter regions; conversely, the specific recognition of toxic materials releases regulatory proteins from promoter regions, resulting in the expression of genes. Thus, it is feasible to generate specific toxic material sensors by genetic engineering; i.e., by the fusion of toxic material-specific promoters and reporter genes, such as enzymes or fluorescence proteins (Belkin 2003; Harms et al. 2006; Nivens et al. 2004). The presence of toxic materials can then be determined by measuring reporter protein levels. Among whole-cell bioreporters, extensive studies have focused on the development and application of metal-sensing bioreporters. In particular, since the initial identification of metal-responsive operons in microorganisms, these have been widely applied to assess the bioavailability of metals in environmental systems. The promoter region of each operon, which is turned on or off depending on the presence of target metal(loid)s, is fused to a reporter gene, enabling detection upon exposure to metal(loid)s. As signal levels correspond to the amount of metal(loid)s, it is possible to quantify the biologically active portion of metal(loid)s to cells. Of course, bioavailability is specific to host organisms and is not necessarily generalizable across species. However, bioavailability provides a more accurate assessment of the risk than the total amount of metal(loid)s.
Several studies have evaluated copper sensors using the cop-operon, known as the copper-responsive operon (Hakkila et al. 2004; Riether et al. 2001; Tom-Petersen et al. 2001). However, the response of eGFP fused to the copA promoter from Escherichia coli was not sufficient to quantify copper ions in the environment, and the correlation between the eGFP signal and copper concentration was weak. To develop a copper-sensing whole-cell bioreporter with improved sensitivity toward copper ions for quantification, in the present study, we generated genetically engineered E. coli strains by deleting copA and cueR to disrupt copper exportation from cells and to control the amount of regulatory protein, respectively. In this manner, the sensitivity of the whole-cell bioreporter was improved, enabling the quantification of bioavailable copper ions in diverse environmental systems.
Materials and methods
Plasmid construction
To amplify genes related to the copper (Cu) export system in E. coli, the genomic DNA of E. coli BL21 was extracted and used as the template for PCR. The upstream region of copA, − 300 to − 13 bp (including the promoter region of the cop-operon), and egfp, encoding enhanced green fluorescent protein, as the reporter gene was amplified by PCR using the primer sets listed in Table S1 in the Supplementary material. The PCR products for copAp and egfp were digested with NdeI/XbaI and BamHI/XhoI, respectively, and inserted into the pET-21(a) plasmid sequentially to generate the copAp::egfp fusion (pCopAp-eGFP). The gene encoding CueR, a copper (Cu)-binding regulatory protein, was amplified by PCR and inserted into pCDF-Duet with BamHI/XhoI to generate pCDF-Duet-CueR. The plasmids were confirmed by DNA sequencing.
Engineering Escherichia coli
To generate genetically engineered E. coli, copA (EcoGene accession number: EG13249) and cueR (EcoGene accession number: EG13256) were deleted using the Quick & Easy E. coli Gene Deletion Kit (Gene Bridges, Heidelberg, Germany). The primers used to delete copA and cueR were synthesized by Macrogen (Daejeon, Korea) and gene deletion was performed following the manufacturer’s protocol. Briefly, the pRedET plasmid carrying the Red/ET recombinase gene was transformed into BL21, followed by the introduction of the FRT-flanked PGK-gb2-neo cassette prepared by PCR with primers 5/6 and 7/8 targeting copA and cueR, respectively (Fig. S1 in the Supplementary material). Upon the addition of 10% arabinose solution, the target gene in the chromosomal DNA was replaced by a kanamycin resistance gene (kan). The detailed procedure used to generate E. coli mutants is described in the Supplementary methods (Online Resource 1). In this study, three mutant strains of E. coli, copA::kan, cueR::kan, and ΔcopA/cueR::kan, were generated and gene deletions in these strains were confirmed by PCR with a set of primers for each step (Fig. S1c). In the case of the double mutant, the kanamycin resistance gene replacing copA was first deleted by expressing FLP-recombinase using the 708-FLPe plasmid, and then cueR was replaced with kan. All mutant and wild-type E. coli BL21 strains were used as hosts for the biosensor plasmid to generate whole-cell bioreporters. The genetic information of engineered E. coli strains and plasmids used in this study is summarized in Table 1.
Whole-cell bioreporter assay
The whole-cell bioreporters generated by introducing a plasmid harboring pCopAp-eGFP into wild-type and mutant E. coli BL21 were grown in LB broth overnight. The culture was transferred to fresh LB and grown at 37 °C until the optical density at 600 nm (OD600) reached 0.4. Aliquots of 5 ml were added to test tubes and 0- to 10-μM metal(loid) salt solution prepared as 1-mM stock solutions by dissolving AsCl3, CdCl2, CrSO4, NiCl2, HgCl2, PbSO4, ZnCl2, and CuSO4 in distilled water was added. After 1 h of incubation, E. coli cells were harvested and the cell pellet was resuspended in Tris buffer (50-mM Tris-HCl, 160-mM KCl, pH 7.4) before measuring the intensity of eGFP. The eGFP signal was measured using a fluorescence spectrophotometer (Scinco, Seoul, Korea) at an emission wavelength from 500 to 600 nm with 480-nm excitation wavelength. The intensity of eGFP induced by exposure to copper was estimated as the induction coefficient, defined as the ratio of the eGFP intensity from exposure to copper exposure to eGFP intensity without copper exposure.
CueR recovery assay
The double gene deletion E. coli mutant strain BL21-copA/cueR was used as a host strain to generate WCB3 harboring pCopAp-eGFP only and WCB4 harboring both pCopAp-eGFP and pCDF-Duet-CueR. As the host strain lacked cueR, the endogenous cueR gene in the plasmid needed to be expressed to verify the role of CueR. To test the sensitivity toward copper ions, the assay was performed following the same procedures described above, except for the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) 1 h before copper exposure. Then, subsequent experiments were carried out at 30 °C to express the recombinant protein CueR. Copper at concentrations of 0 to 25 μM was used to treat WCB2, WCB3, and WCB4, and the intensity of eGFP was measured after 2 h.
Quantification of copper using WCB
To evaluate the capability of WCBs to quantify bioavailable copper, a WCB assay was performed using artificially contaminated and field water samples from the upper stream of Nak-Dong River in South Korea. The artificially contaminated samples were prepared by dissolving CuSO4 in distilled water at various concentrations. For the assay, 0.5 ml of water was applied to 4.5 ml of WCB cells, and the intensity of eGFP was measured after 1 h. Copper ions in artificial samples were quantified based on standard curves obtained for 0- to 10-μM copper ions. The accuracy of the WCB assay for copper quantification was determined by comparing the expected and measured concentrations. Then, the assay was applied to field samples collected from the Nak-Dong River to assess copper contamination.
Results
Copper sensing capability of E. coli BL21 harboring copAp::egfp
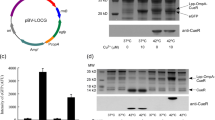
A plasmid able to sense copper ions was constructed by fusing the copA promoter from E. coli to egfp (Fig. 1), then transformed into E. coli BL 21 as a host strain to generate WCB (referred to as WCB1). WCB1 was grown overnight and inoculated into LB broth. When OD600 reached 0.3–0.4, diverse metal(loid)s were added to 5 ml of the culture to obtain a final concentration of 10 μM. However, there was no response to metal ions after 2 h of incubation (Fig. 2a), despite previous reports of copper sensing bioreporters using the same WCB1 system (Hakkila et al. 2004; Riether et al. 2001; Shetty et al. 2004; Tom-Petersen et al. 2001).
Summary of plasmid construction for copper-sensing E. coli-based WCBs. a Diagram of the cop-operon in E. coli and sequence information for the promoter region used to construct a copper-sensing plasmid. The red/underlined letters indicate the CueR binding site. b Diagram of the copper-sensing plasmid pCopAp-eGFP
Heavy metal(loid) selectivity and sensitivity toward copper of WCBs based on wild-type E. coli BL21 (DE3) and BL21-copA harboring pCopAp-eGFP. a WCB1 based on the E. coli BL21 (DE3) strain showed no response to the tested metal(loid)s and no selectivity. Unlike WCB1, WCB2 based on BL21-copA showed a response and selectivity toward copper. The metal(loid) solutions were prepared as 1-mM stock by dissolving AsCl3, CdCl2, CrSO4, NiCl2, HgCl2, PbSO4, ZnCl2, and CuSO4 in distilled water. b Comparison of eGFP induction coefficients from WCB1 and WCB2 treated with 0–10-μM copper ions
Enhancing copper sensing capability of the WCB based on engineered E. coli
To improve copper sensitivity, the copA gene, which encodes a critical component of the copper transport system in E. coli, was deleted to interfere with copper ion homeostasis. The gene was replaced with kan, a kanamycin resistance gene, and the deletion was confirmed by PCR (Fig. S1 in the Supplementary material). The engineered E. coli strain lacking copA was used as a host to receive copAp::egfp and was termed WCB2 (Table 2). To elucidate the effects of the copA deletion on E. coli-based WCBs, metal selectivity was tested using WCB2. As shown in Fig. 2a, the strain showed a strong response to copper with high specificity. These results indicated that loss of the CopA, copper-exporting channel, results in the accumulation of copper in cells. Subsequent comparison of the responses of WCB1 and WCB2 to copper at 0–10 μM clearly showed the induction of eGFP in WCB2, whereas eGFP was not induced in WCB1 (Fig. 2b and Fig. S2 in the Supplementary materials). These findings suggested that sensitivity and metal selectivity were modulated by genetic engineering targeting genes related to metal homeostasis.
Effect of CueR overexpression on copper sensitivity
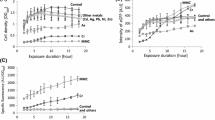
The genetic properties of all strains used in present study are summarized in Table 2. A plasmid harboring copAp::egfp, pCopAp-eGFP, was transformed into each strain and pCDF-Duet-CueR harboring recombinant cueR was inserted into double deletion mutant strains to recover CueR. Consequently, four types of WCBs were generated: WCB1 (wild type), WCB2 (copA deletion), WCB3 (copA and cueR deletion), and WCB4 (copA and cueR deletion with pCDFDuet-CueR). A copper sensitivity assay was performed following the same protocol described above, except 1-mM IPTG was added to express recombinant CueR. For the assay, WCB2, WCB3, and WCB4 were tested with 0- to 25-μM copper (Fig. 3). In concordance with previous results, the intensity of eGFP from WCB2 increased in a copper concentration-dependent manner with high background signals (Fig. 3a). There was no increase in eGFP levels in WCB3 lacking cueR, regardless of the presence of copper, and the response to copper was recovered in WCB4, which expressed endogenous cueR. These results revealed that the transcription of genes regulated by copAp was controlled by CueR. Additionally, it was noted that the background signal from WCB4 was much less (approximately 2-fold) than that from WCB2. However, it was still insufficient for quantification because the response of WCB4 toward copper was less than that of WCB2 based on the induction coefficient values (Fig. 3b). As the goal of the WCB assay was to quantify the amount of target materials, it was critical to analyze the relationship between the eGFP level and metal concentration. Accordingly, in Fig. 3c, the induction coefficients of WCB2 and WCB4 were plotted against the copper concentration. The WCB2 data were fit to a rectangular two-parameter hyperbola with R 2 = 0.988, and WCB4 data were fit to a linear regression model with R 2 = 0.983. It might therefore be concluded that both WCB2 and WCB4 could be used to measure copper in environmental samples based on the high R 2 values.
Effects of cueR deletion in WCBs on detection. a eGFP intensity in response to copper treatment for WCB2 (BL21-copA), WCB3 (BL21-copA/cueR), and WCB4 (BL21-copA/cueR + pCDF-Duet-CueR). b The intensity of eGFP was converted to an induction coefficient. c The correlation between induction coefficients and copper concentration of WCB2 and WCB4. R 2 for WCB2 was 0.988 based on a non-linear regression fit; R 2 for WCB4 was 0.983 based on a linear regression fit
Optimization of copper sensing WCBs by modulating CueR
As shown above, it was revealed that the background signals from the copA-deleted WCB (WCB2) were related to the amount of the regulatory protein, CueR, and that overexpression of CueR reduced the background signals. However, it also reduced the copper sensitivity. This may be explained by the transcription of genes under copAp being regulated too tightly by increasing the amount of CueR. Thus, it was proposed that a reduction in background signal without decreasing the actual response to copper may be achieved by modulating the amount of CueR. To verify this notion, we investigated the copper sensitivity and specificity of WCBs by regulating the amount of CueR in an IPTG concentration-dependent manner following the introduction of pCDF-Duet-CueR (Fig. 4). The ranges of 0–10-μM copper and 0–1-mM IPTG were applied to WCB2 and WCB5, and the induction coefficient of eGFP was measured after 1-h exposure. As shown in Fig. 4a, the intensity of WCB5 without IPTG treatment showed similar level of background signal as WCB2 because the amount of CueR in both WCBs was predicted to be similar although WCB5 carried endogenous cueR. Following IPTG treatment, the background eGFP signals clearly decreased as IPTG concentration was increased. Moreover, the response to copper of WCB5 was increased with 0.1-mM but not 1-mM IPTG induction. Although the mechanism by which 0.1-mM IPTG induction decreased background signals and increased the actual response to copper remained unclear, this result was consistent with our expectation. Specifically, the native amount of endogenous CueR was not considered sufficient to tightly regulate the cop-operon, whereas an excess amount of CueR would tightly suppress the transcription of genes. Thus, the copper sensing capability of WCB5 was achieved by stimulating the induction with a low concentration of IPTG. The enhanced copper sensing capability of WCB5 was clearly demonstrated in Fig. 4b by converting eGFP intensity to induction coefficient values. Moreover, the correlation between the induction coefficient values from WCB5 and copper concentration was linearly proportional with an R 2 value of 0.992 (Fig. 4c, indicating the suitability of WCB5 for application as a biosensor to quantify copper in environmental systems. Consequently, it could be concluded that the properties of bacterial cell-based biosensors could be modulated and improved by genetic and biochemical engineering.
Effects of modulating CueR expression on the copper sensitivity. a eGFP intensity of WCB2 and WCB5 with different amounts of IPTG treatment upon copper concentration. b The eGFP intensity was converted to an induction coefficient. c The relationship between the induction coefficient of WCB5 with 0.1-mM IPTG treatment against copper concentration ranged from 0 to 10 μM. The plot was fit to a linear regression curve with an R 2 value of 0.9923
Application of WCB to quantify copper in aqueous samples
To verify the applicability of WCBs to environmental systems, we quantified copper ions in artificially contaminated and field samples. Quantification was performed using WCB5, with 0.5-ml contaminated water samples being added to 4.5-ml WCBs. The standard curve for quantification was obtained from the plot of the eGFP induction coefficient against the concentration of copper ions ranging from 0 to 10 μM (Fig. 4c). Based on the standard curves, the concentrations of copper in contaminated samples were determined (Table 2). As the concentration of copper ions in the artificial samples was known, it was possible to determine the accuracy of the WCB assay. As shown in Table 2, the accuracy of copper quantification using WCB5 was 95% to 114%. Notably, even for a low expected concentration of 0.08 μM, the value obtained in the WCB assay was accurate (0.086 μM). Thus, these results revealed that the genetically engineered WCB exhibited enhanced sensitivity to copper and was capable of quantifying copper ions in water samples. The assay was then applied to field samples collected from Nak-Dong River to assess copper contamination and was performed following the same procedures, with the induction coefficient of eGFP as shown in the Supplementary information (Fig. S3). The positive control treated with 1-μM copper ions exhibited an induction coefficient of approximately 2.0, whereas there was no significant increase in signal levels in field samples, indicating that the contamination level of each field sample was not significant. In particular, the field samples labeled 5A and 10A exhibited an induction coefficient > 1; therefore, the amount of copper in these samples should be further compared with instrumental analysis. However, this was considered beyond the scope of the present study, the purpose of which was the development of a copper sensing bioreporter based on E. coli. Toward this goal, our findings support the possibility of generating WCBs with enhanced copper-sensing capabilities via genetic and molecular biological engineering approaches.
Discussion
There are diverse bacterial cell-based WCBs available to measure the bioavailability of toxic materials including heavy metal(loid)s. The WCBs targeted copper in the environments were reported in previous studies, and they were all based on the same molecular mechanism; i.e., the fusion of a copper-inducible promoter and reporter genes, although the host strains and reporter genes were different. Some studies have employed the promoter regions from Pseudomonas fluorescens and Saccharomyces cerevisiae with luxAB (Vibrio fischeri) and gfp (Aequoria victoria), respectively, and others have used E. coli as host cells with luxCDABE (V. fischeri) and firefly lucFF as reporter genes. In these studies, the concentration range yielding a linear response was 0.3–5000-μM copper (Hynninen and Virta 2009; Rensing and Grass 2003). Unlike other metal-sensing WCBs, the WCB using the cop-operon in E. coli developed here was not sensitive enough to quantify copper ions, even if we employed the promoter of cop-operon in E. coli as sensing domain and egfp as reporter domain. Thus, improved sensitivity was necessary to use our WCB as a copper sensor. To address this issue, we genetically engineered the host strain E. coli BL21 (DE3) to disrupt natural metal homeostasis, as suggested by another research group enhancing target metal sensitivity (Hynninen et al. 2010).
The copA known to encode CopA, a key protein to export copper to outside of E. coli cells, was the first target for deletion to disrupt copper homeostasis, and this mutant E. coli strain was used as host for WCB (WCB2). As described in previous reports, the sensitivity toward copper ions would be enhanced by the interruption of copper homeostasis with disruption of copper ion exportation following copA deletion expected to increase copper ion accumulation in E. coli cells (Cooksey 1994; Rensing and Grass 2003). As a result, the deletion of copA indeed increased copper sensitivity as shown in Fig. 2, and it was consistent with our expectations, in part, although some issues remained. Although the sensitivity was enhanced by genetic engineering, it remained insufficient for quantification because of increasing background signal from WCB2 without copper exposure (Fig. S2). The actual eGFP intensity from WCB2 was increased over 100 times more than WCB1. However, it was not significant when the eGFP intensity was converted to induction coefficient values ranged 1–3 with 0–10 μM of copper treatment (Fig. 2). It might be used for the quantification, but the background signal issue should be resolved to achieve accurate quantification.
It is not clear why the intensity of eGFP increased, although it is possible that the amount of endogenous CueR produced by E. coli was not sufficient to fully suppress copAp::egfp plasmids. To verify this hypothesis, we generated more WCBs based on an additional mutant E. coli strain with deletions of both copA and cueR. This shortcoming from WCB2 may be explained by the interaction between CueR and copAp (Outten et al. 2000; Stoyanov et al. 2001). It was speculated that deletion of copA increased the concentration of endogenous copper ions in cells, and then endogenous CueR was released from copAp by conformational changes upon copper binding; thereby resulting expression of egfp. Therefore, it was proposed that this shortcoming from would be solved by increasing the amount CueR. To verify this notion, CueR was overexpressed in WCB4 from the exogenously introduced plasmid pCDF-Duet-CueR by IPTG induction. As a result, it was observed the background signals were decreased in WCB4 (Fig. 3a). By increasing the amount of CueR, the role of regulatory protein as a repressor of cop-operon would be functioned properly against increased concentration of copper. The same is expected for the copAp::egfp plasmid, resulting in enhanced sensitivity in response to copper ions. However, the deletion of copA also affected background signal levels from WCBs not exposed to copper (Fig. S2), limiting the ability to quantify copper in environmental systems. In particular, the increase in background signal was not attributed to the copA deletion and was resolved by the overexpression of CueR. Thus, we consider that our proposed mechanism was at least partially accurate.
Nonetheless, the response to copper ions from WCB4 was decreased although the background signal was diminished. Also, the induction coefficient values of WCB4 against copper concentration was lower than that of WCB2 (Fig. 3b). Since the sensitivity toward copper was increased by deleting copA from WCB2 and it was decreased from WCB4, it might be improved by modulating the total amount of CueR in WCBs. If the amount of CueR was matched to the copper ions in E. coli cells without copA, the optimum responses toward copper might be obtained. To verify this notion, the WCB5 was generated by inserting exogenous cueR into WCB2 possessing endogenous cueR gene. And then the amount of CueR was modulated by different concentration of IPTG induction. As shown in Fig. 4, it was clearly shown that the specificity toward copper was varied upon IPTG treatment, and the WCB5 with 0.1-mM IPTG treatment showed enhanced sensitivity with low background signals. The induction coefficient values were ranged from 0 to 7 with 0–10-μM ranges of copper. Moreover, the relationship between induction coefficient and copper concentration was fit to a linear regression curve. And it would be practically applied to quantify the copper bioavailability as a biosensor.
Conclusively, we developed copper-sensing WCBs using the promoter regions of a copper-induced operon in E. coli and egfp as a reporter gene. Under the native state of E. coli, the system did not properly respond to copper ions. The reason for the lack of a response may have been that the strength of the copA promoter was not sufficient or that the system to maintain copper homeostasis in E. coli-reduced copper levels in the cells. To resolve these detection issues, we used genetic engineering techniques to interrupt the copper homeostasis system in E. coli by deleting key genes, resulting in the enhancement of copper sensitivity. Moreover, the WCBs for copper sensing were optimized by further genetic and molecular biological engineering. In this manner, we achieved a WCB system for the quantification of the amount of copper ions in the field. Of course, further studies are needed to improve and to optimize the detection ability of WCBs. Nonetheless, we believe that our study provides valuable information for the development of WCBs for the detection of various targets.
References
Alexander M (2000) Aging, bioavailability, and overestimation of risk from environmental pollutants. Environ Sci Technol 34(20):4259–4265. https://doi.org/10.1021/es001069+
Belkin S (2003) Microbial whole-cell sensing systems of environmental pollutants. Curr Opin Microbiol 6(3):206–212. https://doi.org/10.1016/S1369-5274(03)00059-6
Cooksey DA (1994) Molecular mechanisms of copper resistance and accumulation in bacteria. FEMS Microbiol Rev 14(4):381–386. https://doi.org/10.1111/j.1574-6976.1994.tb00112.x
Foy C, Chaney R, White M (1978) The physiology of metal toxicity in plants. Annu Rev Plant Physiol 29(1):511–566. https://doi.org/10.1146/annurev.pp.29.060178.002455
Hakkila K, Green T, Leskinen P, Ivask A, Marks R, Virta M (2004) Detection of bioavailable heavy metals in EILATox-Oregon samples using whole-cell luminescent bacterial sensors in suspension or immobilized onto fibre-optic tips. J Appl Toxicol 24(5):333–342. https://doi.org/10.1002/jat.1020
Harms H, Wells MC, van der Meer JR (2006) Whole-cell living biosensors—are they ready for environmental application? Appl Microbiol Biotechnol 70(3):273–280. https://doi.org/10.1007/s00253-006-0319-4
Hynninen A, Virta M (2009) Whole-cell bioreporters for the detection of bioavailable metals. Whole Cell Sensing System II Springer, pp 31–63
Hynninen A, Tõnismann K, Virta M (2010) Improving the sensitivity of bacterial bioreporters for heavy metals. Bioeng Bugs 1(2):132–138. https://doi.org/10.4161/bbug.1.2.10902
Javed M, Usmani N (2015) Impact of heavy metal toxicity on hematology and glycogen status of fish: a review. Proc Natl Acad Sci India Section B Biol Sci 85(4):889–900
Juhasz AL, Smith E, Weber J, Naidu R, Rees M, Rofe A, Kuchel T, Sansom L (2008) Effect of soil ageing on in vivo arsenic bioavailability in two dissimilar soils. Chemosphere 71(11):2180–2186. https://doi.org/10.1016/j.chemosphere.2007.12.022
Nivens DE, McKnight T, Moser S, Osbourn S, Simpson M, Sayler G (2004) Bioluminescent bioreporter integrated circuits: potentially small, rugged and inexpensive whole-cell biosensors for remote environmental monitoring. J Appl Microbiol 96(1):33–46. https://doi.org/10.1046/j.1365-2672.2003.02114.x
Outten FW, Outten CE, Hale J, O'Halloran TV (2000) Transcriptional activation of an Escherichia coli copper efflux regulon by the chromosomal MerR homologue, CueR. J Biol Chem 275(40):31024–31029. https://doi.org/10.1074/jbc.M006508200
Rathnayake I, Megharaj M, Krishnamurti G, Bolan NS, Naidu R (2013) Heavy metal toxicity to bacteria—are the existing growth media accurate enough to determine heavy metal toxicity? Chemosphere 90(3):1195–1200. https://doi.org/10.1016/j.chemosphere.2012.09.036
Reid BJ, Jones KC, Semple KT (2000) Bioavailability of persistent organic pollutants in soils and sediments—a perspective on mechanisms, consequences and assessment. Environ Pollut 108(1):103–112. https://doi.org/10.1016/S0269-7491(99)00206-7
Rensing C, Grass G (2003) Escherichia coli mechanisms of copper homeostasis in a changing environment. FEMS Microbiol Rev 27(2–3):197–213. https://doi.org/10.1016/S0168-6445(03)00049-4
Riether K, Dollard M-A, Billard P (2001) Assessment of heavy metal bioavailability using Escherichia coli zntAp::lux and copAp::lux-based biosensors. App Microbiol Biotechnol 57(5):712–716. https://doi.org/10.1007/s00253-001-0852-0
Shetty RS, Deo SK, Liu Y, Daunert S (2004) Fluorescence-based sensing system for copper using genetically engineered living yeast cells. Biotechnol Bioeng 88(5):664–670. https://doi.org/10.1002/bit.20331
Stoyanov JV, Hobman JL, Brown NL (2001) CueR (YbbI) of Escherichia coli is a MerR family regulator controlling expression of the copper exporter CopA. Mol Microbiol 39(2):502–512. https://doi.org/10.1046/j.1365-2958.2001.02264.x
Tom-Petersen A, Hosbond C, Nybroe O (2001) Identification of copper-induced genes in Pseudomonas fluorescens and use of a reporter strain to monitor bioavailable copper in soil. FEMS Microbiol Ecol 38(1):59–67. https://doi.org/10.1111/j.1574-6941.2001.tb00882.x
Van Gestel C, Ma W-C (1988) Toxicity and bioaccumulation of chlorophenols in earthworms, in relation to bioavailability in soil. Ecotoxicol Environ Saf 15(3):289–297. https://doi.org/10.1016/0147-6513(88)90084-X
Acknowledgements
This study was supported by Konkuk University in 2016 (to Y.Y.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
ESM 1
(PDF 193 kb)
Rights and permissions
About this article
Cite this article
Kang, Y., Lee, W., Kim, S. et al. Enhancing the copper-sensing capability of Escherichia coli-based whole-cell bioreporters by genetic engineering. Appl Microbiol Biotechnol 102, 1513–1521 (2018). https://doi.org/10.1007/s00253-017-8677-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-017-8677-7