Abstract
PCR primers targeting genes encoding the two proteins of anammox bacteria, hydrazine synthase and cytochrome c biogenesis protein, were designed and tested in this study. Three different ecotypes of samples, namely ocean sediments, coastal wetland sediments, and wastewater treatment plant (WWTP) samples, were used to assess the primer efficiency and the community structures of anammox bacteria retrieved by 16S ribosomal RNA (rRNA) and the functional genes. Abundances of hzsB gene of anammox bacteria in South China Sea (SCS) samples were significantly correlated with 16S rRNA gene by qPCR method. And hzsB and hzsC gene primer pair hzsB364f-hzsB640r and hzsC745f-hzsC862r in combination with anammox bacterial 16S rRNA gene primers were recommended for quantifying anammox bacteria. Congruent with 16S rRNA gene-based community study, functional gene hzsB could also delineate the coastal-ocean distributing pattern, and seawater depth was positively associated with the diversity and abundance of anammox bacteria from shallow- to deep-sea. Both hzsC and ccsA genes could differentiate marine samples between deep and shallow groups of the Scalindua sp. clades. As for WWTP samples, non-Scalindua anammox bacteria reflected by hzsB, hzsC, ccsA, and ccsB gene-based libraries showed a similar distribution pattern with that by 16S rRNA gene. NH4 + and NH4 +/Σ(NO3 − + NO2 −) positively correlated with anammox bacteria gene diversity, but organic matter contents correlated negatively with anammox bacteria gene diversity in SCS. Salinity was positively associated with diversity indices of hzsC and ccsB gene-harboring anammox bacteria communities and could potentially differentiate the distribution patterns between shallow- and deep-sea sediment samples. SCS surface sediments harbored considerably diverse community of Scalindua. A new Mai Po clade representing coastal estuary wetland anammox bacteria group based on 16S rRNA gene phylogeny is proposed. Existence of anammox bacteria within wider coverage of genera in Mai Po wetland indicates this unique niche is very complex, and species of anammox bacteria are niche-specific with different physiological properties towards substrates competing and chemical tolerance capability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The initially proposed missing lithotroph capable of oxidizing ammonium to nitrogen gas with nitrate/nitrite based on thermodynamical predictions (Broda 1977), the anaerobic ammonium-oxidizing (anammox) bacteria, was subsequently observed in a denitrifying fluidized bed reactor (Mulder et al. 1995). The lithotrophic bacteria capable of carrying out this biochemical process by combining ammonium and nitrite to generate dinitrogen gas (N2) were subsequently identified as deeply branched members within the order Planctomycetales (Jetten et al. 1998; Strous et al. 1999). Up to date, no pure culture of anammox bacteria is available, but only in enrichment cultures. Genomic/metagenomic analysis of them from enrichment cultures has been conducted for Ca. Kuenenia stuttgartiensis, Ca. Jettenia asiatica, Ca. Brocadia anammoxidans, Ca. Brocadia fulgida, Planctomycetaceae KSU-1 and Ca. Scalindua profunda (Gori et al. 2011; Hira et al. 2012; Hu et al. 2012; Strous et al. 2006; van de Vossenberg et al. 2012).
Since the first evidence of anammox bacteria found in the wastewater treatment plant, ubiquitous distribution of them has been reported widely, e.g., wetland and freshwater ecosystems (Dale et al. 2009; Moore et al. 2011; Wang et al. 2013), anoxic marine sediments (Brandsma et al. 2011; Hong et al. 2011; Yoshinaga et al. 2011), terrestrial ecosystems (Hu et al. 2011a; Sun et al. 2014; Wang and Gu 2013a; Wang and Gu 2013b; Zhu et al. 2011), coastal mangrove wetlands (Li et al. 2011a, c), animal intestinal tracts (Chan et al. 2016; Li and Gu 2016), and even subsurface ecosystem such as petroleum reservoirs (Li et al. 2010a) and hydrothermal vents (Byrne et al. 2009). They occur ubiquitously on a global basis, but the diversity pattern could be very limited, e.g., Scalindua sp. in marine environments (Schmid et al. 2007).
Anammox process may contribute significantly to the N2 evolved in selective ecosystem. Incubation with 15N-labeled nitrate or ammonium showed 24-67%, and 19-35% were due to anammox reaction in two continental shelf sites of Baltic-North Sea (Thamdrup and Dalsgaard 2002) and in anoxic water column of Golfo Dulce of N2 production (Dalsgaard et al. 2003), respectively. Further studies also indicated that about 50% of the dinitrogen gas loss in oxygen minimum zones from marine water columns was contributed by this process alone (Kuypers et al. 2005; Lam et al. 2009; Thamdrup et al. 2006). Moreover, anammox bacteria in lakes and agricultural soils were also reported to be as high as 9-40 and 4-37% of the total nitrogen loss in isolated cases (Hu et al. 2011b). Due to the lack of sufficient information on its diversity and abundance and relationship to ecological niches, discovery of new anammox bacteria is still a high priority in research on ecophysiology and application of them to improve applications in global N assessment and pollution control.
Techniques currently available for detecting the activity and distribution of anammox bacteria include isotope paring technique by 15N labeling compounds, ladderane lipids analysis and molecular detecting methods of PCR, quantitative PCR, RT-PCR, and FISH and CARD-FISH methods (Li and Gu 2011; Schmid et al. 2005). Due to the unique phylogeny of anammox bacteria in the phylum Planctomycetes, primers of 16S ribosomal RNA (rRNA) gene and several functional genes are used in detecting the existence and phylogeny of anammox bacteria and also their abundance (Han and Gu 2013; Li and Gu 2011).
The phylogeny of 16S rRNA gene sequences of anammox bacteria forms a monophyletic clade within Planctomycetes, which was named as a new order Brocadiales, showing less than 80% similarity with other genera in Planctomycetes (Schmid et al. 2005). PCR targeting 16S rRNA gene sequences to discover the phylogeny of new species is routinely used with universal primers for Planctomycetes, such as Pla46F and Amx368F, and specific primers for anammox bacteria, such as BS820R and Amx820R (Jetten et al. 2009; Li and Gu 2011). Wide divergence of 16S rRNA gene among different genera within Brocadiales (<87.1% identity) made it hard to compromise specificity and coverage simultaneously to achieve high coverage and also sensitive detection (Junier et al. 2010; Li and Gu 2011), but a pair of more universal and specific primers A438f-A684r was designed capable to cover 100 and 93.5% of anammox bacterial 16S rRNA gene sequences with one or no mismatch (Humbert et al. 2012). Functional gene encoding enzymes involved in specific metabolism pathways are also suitable biomarkers to detect anammox bacteria. Unlike 16S rRNA gene, functional genes as biomarkers are highly specific for the existence of catalyzing activity physiologically and ecologically related to the targeting microorganisms. In addition, functional genes of specific types of microorganisms serve better in detecting minor members within communities dominated by other species as interference (Junier et al. 2010; Kartal et al. 2011a; Li and Gu 2011). Hydroxylamine/hydrazine oxidoreductase (HAO/HZO) targeting primers were used to detect the corresponding hao/hzo genes in ammonium-oxidizing bacteria (AOB) and anammox bacteria. The primers targeting gene sequence of cluster 1 HZO of anammox bacteria are good biomarkers as verified for biochemical activity and existence in all anammox genera up to now (Hirsch et al. 2011; Li et al. 2010b; Quan et al. 2008; Schmid et al. 2008). Cytochrome cd 1-containing nitrite reductase (NIR) was testified with only one copy in Ca. Kuenenia stuttgartiensis, and its encoding gene nirS was largely different from nirS gene of the denitrifiers, making it a suitable functional biomarker for anammox bacteria (Li et al. 2011b; Strous et al. 2006). Two pairs of primers AnnirS and ScnirS targeting Scalindua and other genera of anammox bacterial nirS gene were tested on marine and coastal sediment samples (Lam et al. 2009; Li et al. 2011b). Hydrazine synthase encoded by hzsCBA gene cluster (kuste2859-kuste2861 in Kuenenia stuttgartiensis genome) is responsible for the formation of the N-N bond in hydrazine from ammonium and nitric oxide as the precursors (Fig. S1) (Kartal et al. 2011b; Kuenen 2008; Strous et al. 2006). This gene cluster is functionally specific on hydrazine synthesis in anammox bacteria, not detected in other bacteria species (Gori et al. 2011; Harhangi et al. 2012; Hira et al. 2012; Hu et al. 2012; Strous et al. 2006; van de Vossenberg et al. 2012). Among hydrazine synthase cluster encoding genes, hzsAB gene sequences were analyzed and targeting primers were subsequently developed and tested on sequencing batch reactor enrichment cultures, marine and freshwater sediment samples, paddy soil, and estuary sediment samples (Harhangi et al. 2012; Wang et al. 2012a; Wang et al. 2012b). However, genomes/metagenomes of newly defined anammox species are released and a comprehensive identification and verification of suitable primer sites on the hzsBC gene sequences is highly desirable.
More than 200 gene encoding proteins directly participated in the catabolism and respiration pathways in Ca. Kuenenia stuttgartiensis, indicating the existence of versatile branched respiring chain and emphasizing its important role in the unique anammox metabolism (Strous et al. 2006). Cytochrome c synthesis gene is abundant, and at least one full set of system II cytochrome c maturation pathway in all recovered anammox bacteria is evident (Ferousi et al. 2013). CcsAB proteins in system II form a channel-like structure, facilitating the heme transport to anammoxsome and maintaining the reducing state. Two sets of CcsAB complexes were recovered in four genera of anammox bacterial genomes up to date (Ferousi et al. 2013). Canonical CcsAB complex encoding genes with heme-binding motif phylogenetically different from the equivalents of other species were aligned and used to design anammox bacteria-specific primers in this study (Fig. S1). General PCR primers and qPCR primers were analyzed for efficiency; comparison between 16S rRNA and functional gene primers on delineating anammox bacterial community characteristics and relationships with physicochemical parameters were also investigated. Meanwhile, a further expanded survey and analysis of Scalindua sp. harbored in South China Sea (SCS) surface sediments based on newly designed anammox bacterial 16S rRNA gene primer pairs were also carried out in this study.
Materials and methods
Sampling and physicochemical parameters
Surface sediment samples of the South China Sea were collected on the SCS Open Cruise in July 2008, and their physicochemical parameters were measured as previously (Table S1) (Cao et al. 2011a; Cao et al. 2011b). Wetland sediment samples were collected from Mai Po Nature Reserve of Hong Kong in Match 2012; both surface and subsurface sediments from intertidal mudflats and mangrove forest were sampled. At time of sampling, temperature, pH, and redox potential were measured in situ as described previously (Han and Gu 2013). Concentrations of nitrate, nitrite, and ammonium in the pore water of these samples after centrifugation were measured by flow injection analyzer (FIA) with standard methods (Table S2) (Han and Gu 2013). Two wastewater treatment plant (WWTP) samples were obtained from granules and wastewater slurry for nitrogen removal by anammox reaction in Bali, Taiwan.
Sequence alignment and primer design
According to available genomic/metagenomic data of anammox bacteria enrichment cultures from NCBI and JGI genome databases, the hzsBC gene sequences from Ca. Kuenenia stuttgartiensis (NCBI:PRJNA16685), Ca. Jettenia asiatica (personal communication with Ziye Hu), Ca. Brocadia anammoxidans (JGI:2081372000), Ca. Brocadia fulgida (JGI:2225789020), Planctomycetaceae KSU-1 (NCBI:PRJDA163683 and PRJDB68, phylogenetically as Ca. Jettenia sp.), and Ca. Scalindua profunda (JGI:2017108002 and 2022004002) were obtained (Gori et al. 2011; Hira et al. 2012; Hu et al. 2012; Strous et al. 2006; van de Vossenberg et al. 2012). They were aligned by CLC Main Workbench 7 to identify potential primer designing sites with relatively high conservative values and moderately variable 3′ end, degenerate primers to cover the known species of anammox bacteria were developed.
Canonical cytochrome c synthesis protein CcsAB encoding gene sequences were retrieved from above genomic/metagenomic data, except for Ca. Jettenia asiatica, which did not show satisfactory BLAST results. Five sequences with full ccsAB gene length coverage were used to form alignments. By applying the same approach, the regions with high conservative values and moderately variable 3′ ends were chosen to develop degenerate primers. Sequence differences and percent identity values of 16S rRNA gene, hzsABC and cssAB gene alignments of known anammox bacteria species, were generated and illustrated by CLC Main Workbench 7 (Fig. S2).
DNA extraction, PCR amplification, and cloning library
Frozen sediment samples were thawed before genomic DNA extraction by the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Inc.). Approximately 0.25 g of wet weight were applied to carry out DNA extraction according to the manual. Moisture contents were measured by oven-drying the samples at 120 °C for 24 h to a constant weight to allow conversion of wet weight to dry weight in the final expression. For anammox bacterial 16S rRNA gene amplification, the same procedures by Han and Gu (Han and Gu 2013) were followed. In each 25 μl of total PCR mixture, it contained 5 μl of 5× GoTaq Flexi buffer (Promega), 5 nmol dNTPs (Invitrogen), 10 μg of BSA (Roche), 62.5 nmol Mg2+, 0.5 μl of each primer (A438f: 5′-GTCRGGAGTTADGAAATG-3′, A684r: 5′-ACCAGAAGTTCCACTCTC-3′) in 20 mM and 0.2 μl of GoTaq Flexi polymerase (5 U/ml, Promega) (Humbert et al. 2012), and 1 μl of DNA template (~25 ng). The PCR thermal cycling steps were set as following: an initial denaturation at 95 °C for 5 min, 33 cycles of 95 °C for 45 s, 54 °C for 30 s, and 72 °C for 50 s, followed by 10 min of final extension at 72 °C.
For the hzsB and hzsC, and cssA and cssB gene amplification, each 25 μl volume of total PCR mixture contained 5 μl of 5× GoTaq Flexi buffer (Promega), 5 nmol of dNTPs (Invitrogen), 62.5 nmol of Mg2+, 10 μg of BSA (Roche), 1 μl of each primer in 20 mM and 0.2 μl of GoTaq Flexi polymerase (5 U/ml, Promega), and 1 μl of DNA template (~25 ng). For each round of PCR thermal cycling, it involved an initial denaturation step at 95 °C for 5 min, and every cycle consisting of 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 50 s, and the final extension at 72 °C for 10 min. For one-step direct PCR procedure and two steps of nest PCR procedure, 40 and 35 cycles were selected to obtain the final PCR products, respectively. Double volumes of PCR reaction were employed to generate enough amplicons from those PCR products with poor amplification. PCR products were separated by 1× Tris-acetate-EDTA (TAE) electrophoresis in 1% agarose gels, stained with 1/40,000 GelRed staining solution (Biotium) and checked and photographed under Gel Doc (Bio-Rad).
PCR products with the correct sizes were checked and retrieved from TAE gel by clear surgical knives. Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) was employed to purify the PCR products according to the manufacturer’s instructions. Clone libraries were conducted by inserting PCR products into PMD-18T plasmid (TaKaRa, Japan) and transferred into DH5α Escherichia coli. The successful inserted clones with ampicillin (Amp) resistance gene could survive on the Amp-containing Luria-Bertani (LB) agar plates. Then, from each plate, the colonies were picked and transferred into one tube containing LB broth medium and Amp. After overnight incubation, plasmids from cultures were isolated and sequenced by using M13F primer. Sequences acquired from individual clones formed one clone library.
Quantitative PCR
By applying anammox bacterial 16S rRNA gene primers (A438f/A684r) and hzsB and hzsC qPCR primers designed in this study, the targeting gene abundance in each DNA sample was measured. Quantitative PCR measurement and statistical analysis were carried out by StepOnePlus Real-Time PCR System (Applied Biosystems). A randomly picked plasmid with successful targeted gene inserted was used to measure the anammox bacterial gene copy numbers by the equation: Abundance of gene copy number/μl = (amount/μl × 6.022 × 1023)/(length × 1 × 109 × 660). The successive ten-time dilution series were made for the corresponding plasmid to generate the standard curves respectively. The qPCR reaction mixture in total of 15 μl volume contained 7.5 μl of iTaq™ universal SYBR® Green Supermix (Bio-Rad), 1 μl of DNA template (~25 ng), and 0.75 μl of each forward and reverse primer in 20 mM and 12 μg of BSA (Roche). The annealing temperature for primer pair A438f/A684r was 54 °C and that for hzsB and hzsC gene qPCR primer pairs was 55 °C. The remaining thermocycle setting for the time and temperature of denaturation and extension were assigned according to the manufacturer’s instructions.
Phylogeny and community analysis
Short 16S rRNA gene sequences less than 200 bp were deleted, and potential chimeric sequences were screened out by using the DECIPHER online software (Wright et al. 2012). Phylogenetic analyses were conducted to check whether sequenced 16S rRNA genes belonged to anammox bacteria, and any non-specific Planctomycetes clones were screened out from the alignments. Qualified 16S rRNA gene sequences were aligned for each qualified sample by MEGA 5.05 (Tamura et al. 2011), and operational taxonomic units (OTUs) were divided by the mothur software with cutoff criterion of 0.03 (Schloss et al. 2009). Obtained richness and diversity indices, such as rarefaction curve, Shannon, and Chao1 diversity indices, were analyzed. OTU representative sequences were applied together with anammox bacteria reference sequences downloaded from NCBI database to build a comprehensive phylogenetic tree by Neighbor-Joining method at the following settings, Jukes-Cantor model, bootstrap 1000 times for resampling, and pairwise deletion treatment.
For hzsB and hzsC, and ccsA and ccsB sequences, OTUs were first divided by mothur with 0.05 cutoff according to nucleotide sequences, and also the richness and diversity indices were obtained; then, the representative sequences were translated to amino acid sequences with coding frame checked and pooled together to construct phylogenetic trees by Neighbor-Joining method at the following settings: p-distance model, 1000 times of bootstrap for resampling, and pairwise deletion treatment.
In order to avoid negative branch length value, recruited OTU representative sequences from 16S rRNA gene and functional gene alignments were submitted to RAxML-HPC BlackBox (8.0.24) in CIPRES Science Gateway to build maximum likelihood-based phylogenetic trees. Then, trees were uploaded to the Fast UniFrac online software, together with sample ID and category files, to visualize the dissimilar relationship among samples graphically by Principal Coordinate Analysis (PCoA) and Jackknife sample clustering method (Hamady et al. 2009). The weighted UniFrac matrix and normalized algorithms were chosen to draw PCoA diagram. For Jackknife sample clustering, 100 times of permutation were performed by choosing the same number of sequences in each sample. In order to further delineate how environmental factors shaped the microorganism community, CANOCO 4.5 was used to conduct the gradient analysis (Braak and Šmilauer 2002). Pearson correlation analyses were conducted by GraphPad Prism 5 to test whether there were statistically significant correlations between every two groups (Motulsky 1999).
Accession numbers of sequences retrieved
After checking the quality of sequences amplified and sequenced, those with length above 200 bp were uploaded to NCBI GenBank database: anammox bacterial 16S rRNA gene sequences with the accession nos. from KP003359 to KP003631; hzsB gene sequences with the accession nos. from KP002276 to KP002877; hzsC gene sequences with the accession nos. from KP002878 to KP003358; testified ccsA gene sequences which highly believed belonging to anammox bacteria with the accession nos. from KP001599 to KP001743, other ccsA gene sequences believed to be cytochrome c biogenesis ccsA (resC) with the accession nos. from KP001744 to KP001940; and ccsB (resB) gene sequences with the accession nos. from KP001941 to KP002275.
Results
Physicochemical properties
Three types of samples, including marine sediments, coastal wetland sediments, and wastewater treatment plant samples, were applied in this study. SCS samples included both deep- and shallow-sea sediments depending on seafloor depth. Salinity and temperature changed gradually along the increase in water depth, from the continental slope of the Pearl River Delta Estuary to pristine deep ocean (Table S1). The concentration of NO3 −, NO2 −,and NH4 +, together with pore water salinity, organic matters represented biogeochemical property in situ at each location as described previously (Cao et al. 2011a; Cao et al. 2011b). Mai Po Nature Reserve is characterized as an intertidal marsh with mangrove forest. It is a 1500-ha wetland designated as a Ramsar site since 1995, aiming to manage and protect local biodiversity and migratory birds. Facing the Inner Deep Bay, adjacent to Shenzhen and Kam Tin rivers, Mai Po wetland suffered from deteriorated conditions by the intrusion of anthropogenic and industrial wastes since the rapid economic development a few decades ago (Lau and Chu 1999; Liang and Wong 2003). Two sampling sites were chosen (sites 1 and 3), and near each site, we collected intertidal mudflats and mangrove field sediment samples from surface and subsurface layers. A regular pattern of NO3 − + NO2 − and NH4 + concentrations between two sites was obtained, which indicating representative sampling could generally reflect the nitrogen-associated physicochemical properties in Mai Po wetland (Table S2). Two WWTP samples were collected from reactors involving active anammox process.
Sequence alignments and primer pair design
Hydrazine synthase is involved in the metabolic pathways mediating the anammox energy conservation, and it helps to build N-N bond from catalyzing synthesis of nitric oxide and ammonium into hydrazine (Kartal et al. 2011b). For its high specificity in anammox bacteria, none of other organisms known up to now, it serves as an ideal functional gene-targeting biomarker for anammox bacteria. Based on the recently released genomic profiles of anammox bacteria enrichments, only a single copy has been identified in individual anammox bacterial genome, making it most suitable than other candidate functional genes in quantifying the abundance of anammox bacteria. Owing to the differences in evolutionary history, hydrazine synthase clusters in Scalindua species differ from other non-Scalindua species in that fused Hzsβγ protein instead of isolated ones (Fig. S1). Comparing with the sequence identities in 16S rRNA and hzsA gene alignments, both hzsB and hzsC genes have the same range of sequence similarity around 60-80% among all known anammox bacteria (Fig. S2). A total of six sets of hzsB and hzsC gene sequences from the sequenced genomes were used and aligned by CLC Main Workbench 7, and 15-25 nt covering sites with high conservation among all sequences were used to design the new PCR primers of this study (Figs. S3 and S4). Degeneracy degrees were constrained by deleting several tail nucleotides so that a list of short primers was made. A list of the primers assigned with forward and reserve directions is shown in Table S3, and three parts of combinations for hzsC gene primer pairs are also included.
CcsA-CssB complex, located on the membrane of anammoxosome, as a membrane-anchored protein complex, transfers heme from nucleoid to paryphoplasm and maintains its reduced state (Frawley and Kranz 2009) in the system II cytochrome c maturation system. Two distinct sets of CcsA-CssB complex encoding genes with functional motifs and essential residues in the up-to-date anammox bacterial genomes were discovered (Ferousi et al. 2013). One of them shared canonical heme-binding motifs in CcsA with published reference, while the other one acquired modified heme-binding CcsA motifs, indicating a biologically different binding configuration with heme (Ferousi et al. 2013). In the alignment with accessible ccsA and ccsB gene sequences to date, between 40 and 80% of similarity is shared among the known anammox bacteria (Fig. S2). The most conservative nucleotide sites were used to design degenerate primers in this study (Figs. S5 and S6), and a list of forward and reverse primers and the combination of those primers with additional information are shown in Table S4.
Primer combinations and PCR performance
In order to find the best combinations of primer pairs to amplify the targeted functional gene from marine and coastal wetland samples, firstly, a pair of forward and reverse primers, which could amplify proper PCR products of expected length, was chosen. And then, the combinations of primer pairs with short nucleotide length but high degeneracy (parts II and III of hzsB gene combinations) were also applied. For the forward primer hzsC496f_An&Sc, the concentration ratio of hzcC496f_An/hzcC496f_Sc as 4:1 was suggested. All PCR thermocycles were the same when comparing the effectiveness of primer pairs. The initial denaturation step was as 95 °C for 5 min, followed by 40 cycles at 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 60 s (expecting PCR products shorter than 1000 bp) or 90 s (expecting PCR products longer than 1000 bp); the final extension time was 72 °C for 10 min. Two DNA samples as templates, E702S/MP7 (1:1 ratio) and E702S alone, were used to compare the outcome with different DNA sources. The PCR reaction mix was the same as described in the “Materials and methods,” with each primer final concentration at 1 mM. After PCR amplification, all PCR products were separated in agarose gels with the same loading quantities. Finally, results from each combination of the designed primers were obtained in this study with lane numbers in the same order with the combination labels shown in Tables S3 and S4 (Figs. S7 and S8).
Generally, when amplifying DNA templates from deep-sea E702S sample, the PCR products were shown as clear and correct bands. And hzsB primer pairs showed great ability to successfully amplify the templates from both samples; most of the combinations showed good results with clear and single bands. However, for hzsC and ccsB primers, poor results were resulted from most of the combinations of these two functional gene primer pairs. Further optimization of PCR conditions should be tried to optimize the reaction systems to amplify the small amount of anammox functional genes from the large DNA template pools.
After the preexperimental test of primers on each sample from SCS and Mai Po wetland, the best combinations of primers with each functional gene were obtained (Table 1). For one-step PCR, 40 cycles were used while as for two-step nested PCR, 35 cycles for each step were used. qPCR primer pairs with the best performance were also chosen and applied (Table 1).
For WWTP samples from Taiwan, only one-step PCR was required to amplify 16S rRNA and functional genes: A438f-A684r for 16S rRNA gene, hzsB364f-hzsB790r for hzsB gene, hzsC559f-hzsC862r for hzsC gene, ccsA376f-ccsA668r for ccsA gene, and ccsB1116f-ccsB1589r for ccsB gene.
In total, 16 hzsB and hzsC gene clone libraries from 3 deep SCS samples (08CF7S, E425S, and E702S), 7 shallow SCS samples (E704S, E707S, E708S, E709S, E201S, E510S, and E706S), 4 Mai Po wetland samples (MP1-4), and 2 WWTP samples (2A and 2B) were successfully acquired. Additional 10 more 16S rRNA gene clone libraries (Han and Gu 2013) were incorporated into the analysis. Due to the poor primer efficiency of A438f/A684r on MP3 sample, clone library of MP3 sample showed biased community structure mainly from non-anammox bacteria so that only MP1, MP2, and MP4 clone libraries were available for analysis from Mai Po wetland. For ccsA and ccsB genes, 8 clone libraries from SCS samples (E425S, E510S, E706S, E702S, E704S, E707S, E708S, and E709S) were successfully obtained for the analysis.
Primer efficiency on different environmental samples
For anammox bacterial 16S rRNA gene primer pairs A438f-A684r, sequences from three types of samples combining with the data from Han and Gu (Han and Gu 2013) were used in the further analysis. After removing the non-anammox bacteria Planctomycetes 16S rRNA gene sequences, A438f-A684r displayed 54.3-100% efficiency with an average value of 88.1% in retrieving anammox bacterial 16S rRNA gene from SCS samples. For three out of the four Mai Po samples, this primer pair showed very good efficiency with an average value of 93.6%, but MP3 only achieved 3.8% of sequences belonging to anammox bacteria after excessive sequencing efforts on the clone library. For hzsB, hzsC, and ccsB gene primers, nearly 100% of successfully amplified sequences were obtained clustering with known anammox bacteria. Except for the ccsA gene results, 92.1-100.0% of SCS-derived sequences were cytochrome c biogenesis CcsA (ResC) encoding sequences. However, sequence identities of their amino acid sequences with those of anammox bacterial ccsA genes could not conclusively be differentiated from those with other non-anammox bacterial species (amino acid sequence identities are around 45-50% with KSU-1, Ca. Kuenenia stuttgartiensis, and other non-anammox bacteria). After removing those ambiguous sequences, ccsA gene primer efficiency in retrieving anammox bacterial sequences showed considerable differences among different clone libraries, from as low as 0 to the highest 93.9% (Table S5). When applying all primer pairs in this study on the two WWTP samples, all retrieved sequences belonged to anammox bacteria without any problem.
Gene abundance by quantitative PCR
After screening primer pairs targeting hzsB and hzsC genes with the best performance and moderately short PCR product length, hzsB364f-hzsB640r and hzsC745f-hzsC862r were chosen for hzsB gene and hzsC gene, respectively. The abundance of each gene from samples collected from SCS, coastal Mai Po wetland, and WWTP are presented in Fig. 1 and Table S6. The r 2 value and amplification efficiency of all qPCR reactions in this study covered the range of 0.972-0.998 and 88.3-110%. In SCS samples, 16S rRNA gene-based abundance was higher than hzsB and hzsC gene in E702S, E704S, and E425S samples, but a reverse trend was detected in the rest of SCS samples. In particular, the ratios of hzsB and hzsC gene abundance over 16S rRNA gene abundance were much higher at around 6-9 and 52-64 times, respectively, in two shallow-sea samples, E201S and E510S. In Mai Po wetland, hzsC gene abundance prevailed over hzsB gene, and hzsB and hzsC gene abundances were much higher (3 to 100 times) than the corresponding 16S rRNA gene abundance. While, in MP7 sample, the abundance level among the three genes was almost identical at 8-9 × 104 gene copies/g dry sediment. Among SCS samples, the abundances measured by the three genes in gene copies per gram dry sediment varied considerably, ranging from 103 to 106 for anammox bacterial 16S rRNA gene, from 105 to 106 for hzsB gene, and from 105 to 106 for hzsC gene. However, these three gene abundances were less variable in Mai Po samples except for MP7 sample at the level of 104 and 105 for anammox bacterial 16S rRNA gene and hzsB gene and 105 and 106 for hzsC gene. WWTP samples harbored the most abundant amount of these three genes than other environmental samples due to the enrichment of anammox in the treatment process. Anammox bacterial 16S rRNA gene abundance was around 8-9 × 108 gene copies per gram of dry sediment, while, hzsB and hzsC gene abundance were approximately 7-9 × 109 gene copies per gram of dry sediment in WWTP samples. The abundances of hzsB and hzsC gene were nearly 10 times higher than those of 16S rRNA gene in WWTP samples. Pearson correlation analysis showed that these three pairs of qPCR primers obtained in considerably correlated results on quantifying the gene abundance in SCS samples (Table S7). However, no detectable correlation among these three genes in Mai Po wetland samples was evident. The limited sample numbers did not support any statistical conclusions for the WWTP samples.
Clone library richness and diversity and phylogenetic analysis
Except for ccsA genes, clone library sizes obtained by anammox bacterial 16S rRNA and functional gene primers were reasonably large (coverage values were higher than 80% except for E707S site, which was 68.57%). Combining the data from a previous study, OTU number, Chao1, and Shannon diversity index based on anammox bacterial 16S rRNA gene from the three types of samples (except for MP3, which obtained inadequate sequences in its clone library because of the pool efficiency of primer pair) were obtained (Han and Gu 2013). For cssB gene clone libraries, two types of environmental samples (SCS and WWTP) were included because of the poor amplification efficiency from Mai Po wetland samples, while, for hzsB and hzsC genes, richness and diversity indices based on the corresponding clone libraries from all types of samples were obtained. All clone library richness and diversity indices were calculated by using the mothur software, and data are presented in Table S8.
The representative OTU sequences from each sampling site in this study and a previous one (Han and Gu 2013) together with additional sequences from others deposited in NCBI were retrieved to build the phylogenetic tree based on the anammox bacterial 16S rRNA gene (Fig. 2, Table S9). In the large clade of Scalindua spp., eight subclades were assigned: Scalindua pacifica, Scalindua arabica, Scalindua zhenghei-I, S. zhenghei-II, and S. zhenghei-III, Scalindua wagneri, Scalindua brodae/sorokinii/marina, and Scalindua nanhaiensis (Hong et al. 2011; Jetten et al. 2003; Schmid et al. 2003; Woebken et al. 2008). The mean evolutionary distances between these eight groups were ranged within 0.046-0.106 and all above the boundary of new species (Table S10). S. nanhaiensis group showed the nearest distance to S. zhenghei-I and S. zhenghei II with value of 0.054 and 0.046 among all subclades. Most of SCS samples harbored sequences in Scalindua clade only except for E201S with one clone affiliated into Brocadia clade. SCS samples covered most of the Scalindua clades with sequence fraction ranging within 11-24%, except for S. wagneri (two clones from E201S) and S. zhenghei-III (no clones from SCS samples, while all from Mai Po samples). Samples from Mai Po wetland harbored sequences covering most of Scalindua clades, except for S. arabica and S. zhenghei-I.
This tree was conducted using the Neighbor-Joining method with 1000 times bootstrap test supporting. Nucleotide sequences from this study, Han et al.’s study, and other anammox bacterial 16S rRNA gene referential sequences from database were involved (Han and Gu 2013). The supporting values of branches according to the frequency were displayed on each node. The evolutionary distances were analyzed using Jukes-Cantor method, and the branch lengths were drawn to scale according to the evolutionary distance with a scale ruler on the bottom of the figure. Detailed sequencing accession number and clone information were provided in Table S9
Clones from MP1 and MP3 of mangrove forest and intertidal mudflat samples (17 clones from Han and Gu 2013, 1 clone from this study) joined together to form a new clade named Mai Po clade, representing 0.088-0.150 evolutionary distance with other four defined non-Scalindua clades. Its best hits of anammox bacterial 16S rRNA gene in NCBI database were those from surface samples of intertidal marshes of Yangtze River Estuary with the similarity ranging from 95 to 100%, and other hits showed similarity lower than 96%, indicating that niche-specific other than geographic-specific distribution of Mai Po clade. Kuenenia sp. sequences were the most dominant in the two WWTP samples, and also the most ubiquitous in Mai Po wetland. Sequences from MP1 covered all defined clades of terrestrial non-Scalindua groups (Table S11).
Well-defined hzsB gene from anammox bacteria genome/metagenome data and all known indicative hzsB genes were pooled together to build the phylogenetic tree. Combining hzsB representative gene sequences from all OTUs retrieved in this study, 203 sequences were used to build the tree (Fig. 3) with 12 sequences from E708S, E510S, E709S, and E707S of shallow SCS affiliated into the same clade with Scalindua profunda, which was inoculated and enriched from 116-m deep sediment from Gullmar Fjord, Sweden. Its phylogenetic position belonged to Scalindua brodae/sorokinii clade according to its 16S rRNA gene relatedness to these two typical species with sequence identity ranging from 96.8 to 99.5% (van de Vossenberg et al. 2008). This clade-harboring hzsB gene of Scalindua profunda could be reasonably believed to represent Scalindua brodae/sorokinii group in hzsB gene tree. The majority of hzsB gene sequences from SCS belonged to Scalindua clade, while six sequences from E201S falling into Kuenenia clade and one sequence from E510S, two sequences from E706S, and three sequences from E704S forming one unique clade. There was also one unique sequence sharing the lowest similarity with defined anammox bacterial hzsB gene according to the phylogenetic construction. Majority of the sequences of two WWTP samples (30/34 from 2A, 13/23 from 2B) formed a neighboring clade to Kuenenia, and the remaining ones joined in a parallel clade close to Brocadia.
This phylogenetic tree was conducted using the Neighbor-Joining method based on the OTU representative hzsB gene sequences from this study and referential sequences from online database. The hzsB gene sequences were translated into amino acid sequences. The evolutionary distances between sequences were computed using the p-distance method, and 203 sequences in total were involved in this tree. The supporting values on each node were based on the frequency of 1000 times bootstrap testing. This tree was drawn to scale with scale ruler on the bottom of the figure. Taxon name with _X_Y meant this taxon was the X-th OTU with Y sequences in this OTU. Taxon name with only _Z at the end meant several OTUs from the same sampling site’s library were merged into one in this clade, and there were Z number of sequences from this sampling site’s library
After two rounds of nested PCR, 15 hzsC gene clone libraries were obtained from the three types of samples. OTU numbers were fewer, and Shannon and Chao1 index were lower comparing with the richness and diversity indices of hzsB clone libraries, respectively. The phylogenetic tree based on the translated OTU representative sequences and other hzsC gene amino acid sequences is shown in Fig. 4. Consistent with seawater depth of SCS samples, the Scalindua clade could be divided into two subclades: deep SCS and shallow SCS clades, although E704S with the water depth of 175 m fell into deep SCS clade. Only one MP3 hzsC gene sequence appeared as an out-group in Scalindua clade. All sequences from WWTP samples were found in Kuenenia clade, neighboring to a clade formed by 37/38 hzsC gene sequences from E707S sample. Other sequences from Mai Po samples formed a clade dissimilar to any defined non-Scalindua clade and parallel to Kuenenia clade.
This phylogenetic tree was conducted using Neighbor-Joining method with 1000 times bootstrap test supporting. The representative hzsC gene sequences from each OTU were firstly translated into amino acid and then used to build the tree together with referential anammox bacterial hzsC sequences from the database. The evolutionary distances were computed using p-distance method, and the tree was drawn to scale with scale ruler on the bottom of the tree. Taxon name with _X_Y meant this taxon was the X-th OTU with Y sequences in this OTU
For ccsA gene phylogenetic tree construction, due to the low specificity of designed primers in this study in retrieving anammox bacterial ccsA gene and existence of other ccsA gene homologs in the DNA pool of SCS samples, some of the clone libraries did not contain enough clones to meet the coverage requirement. All sequences were pooled together, and OTUs were defined at 0.05 nucleotide cutoff; finally, 55 OTUs were obtained. All ccsA gene sequences from this study, including canonical ccsA gene of defined anammox bacteria-type species, and reference sequences of other species were used to build the tree (Fig. 5). Only SCS and WWTP sequences were used in this study because of the poor performance of primers in amplifying ccsA gene from Mai Po samples with complex DNA background. Scalindua profunda ccsA gene was grouped with the majority of sequences from SCS, while other ccsA genes of defined anammox bacteria formed a separated paraphyletic clade and clustered with cssA genes of other species. There were two clades formed exclusively by deep SCS sequences and another one exclusively by shallow SCS sequences, indicating that ccsA gene groups from two categories of sediments with different seawater depth could be phylogenetically differentiated. In common with phylogenetic distribution pattern analyzed by 16S rRNA gene sequences, cssA gene sequences of WWTPs all fell into non-Scalindua clade. As for 16S rRNA gene sequences, 1/33 and 5/34 of 2A and 2B, respectively, were affiliated with Brocadia fulgida/caroliniensis and the rest were affiliated with Kuenenia. For ccsA gene sequences, 3/34 and 6/35 of 2A and 2B, respectively, were affiliated within Brocadia fulgida clade and the rests were affiliated to Kuenenia. The translated ccsA gene amino acid sequences were blasted, and non-anammox bacterial ccsA gene amino acid sequences shared less than 60% identity with sequences from this study. For the ccsB gene, representative sequences of each OTU from clone libraries of SCS and WWTP samples were used to build the phylogenetic tree. Two clades of marine Scalindua group and terrestrial non-Scalindua group are distinguished in Fig. 6. There were three large subclades in Scalindua clade, while Scalindua profunda was located in the second clade surrounded by deep SCS sequences from E425S and E702S. As for sequences from WWTP samples, similar to 16S rRNA and cssA gene, 1/34 and 11/34 of 2A and 2B were affiliated within Brocadia fulgida clade, and the remaining were affiliated to Kuenenia.
This phylogenetic tree was conducted using Neighbor-Joining method with 1000 times bootstraps test supporting. All clarified anammox bacteria-like ccsA genes were pooled as one library, after divided into OTUs with 0.05 cutoff criterion, then translated into amino acid sequences. The evolutionary distances were calculated using p-distance method, and the scale bar was given on the bottom of the figure. Other ccsA genes from non-anammox bacteria were also included. Taxon name with _X_Y meant this taxon was the X-th OTU with Y sequences in this OTU, and the taxon name with only _Z meant there were Z number of sequences in this OTU
This phylogenetic tree was conducted using Neighbor-Joining method with 1000 times bootstrap supporting. The ccsB genes retrieved in this study together with the anammox bacterial ccsB gene from database were translated into amino acid sequences and then used to build this tree. The node was tagged with bootstraps supporting value based on the frequency. The evolutionary distance was calculated using p-distance method. The scale bar was given on the bottom of the figure. Taxon name with _X_Y meant this taxon was the X-th OTU with Y sequences in this OTU
Community analysis and environmental factors
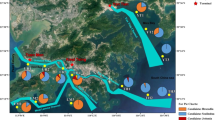
PCoA was used to delineate the relationship of library composition of samples based on UniFrac distance matrix (Fig. 7). The first two principal components are depicted as the axes x and y. The larger value of percentage of variation explanation of axis means the more fraction of variance that the axis could account for. PCoA analysis could clearly divide three types of environments according to the diagrams based on anammox bacterial 16S rRNA and hzsB gene. Deep and shallow SCS samples are positioned in the left upper corner of anammox bacterial 16S rRNA gene diagram; while, two WWTP samples are located in the right site and Mai Po wetland samples are dispersed in the lower part of the diagram (Fig. 7a). As for hzsB gene diagram, it is very similar to anammox bacterial 16S rRNA gene diagram with three divided groups according to three types of environmental samples (Fig. 7b). In hzsC and ccsB gene diagrams, two deep SCS samples are separated from each other, while, the shallow SCS sample E704S (175 m water depth) is located close to them in both diagrams (Fig. 7c, d). The two WWTP samples were closely located in both diagrams. In hzsC gene diagram, Mai Po samples are located closely together while MP3 sample was slightly apart; however, E707S sample was separated from the rest of the shallow SCS samples. As for ccsB gene diagram, shallow SCS samples grouped together in the lower left corner except for E704S sample. Jackkinfe clustering analyses based on the above four genes libraries also showed a similar clustering pattern as described by PCoA analysis (Fig. S9).
Pearson correlation relationships between the richness and diversity values of 16S rRNA, hzsB and hzsC genes, and environmental parameters from SCS samples were analyzed (Table 2). Anammox bacterial 16S rRNA was positively correlated with seawater depth with strong statistical support. Shannon index of hzsC gene was strongly correlated with salinity. Shannon index of ccsB gene was also correlated with salinity. Chao1 index of anammox bacterial 16S rRNA gene was strongly correlated with NH4 +, but Shannon index was negatively correlated with organic matter concentration. Chao1 index of hzsB gene was positively correlated with NH4 +/Σ(NO3 − + NO2 −). The rest of the correlation coefficient values with p values within 0.01-0.05 are shown in Table 2.
In the Canonical Correspondence Analysis (CCA) diagram depicted by CANOCO 4.5, the first two axes could account for 40.8% variance explanation of the species dataset, with the eigenvalues of 0.764 and 0.658. The first canonical axis showed its p value of 0.074, indicating it was a reliable statistically significant pattern. The length of individual environmental factor indicated its influence on the distribution of samples, and its angle with each axis indicated correlated level with that axis. The figure was based on the hzsB gene phylogenetic tree and SCS environmental factors (Fig. 8). Seawater depth and Σ(NO3 − + NO2 −) largely shaped the three SCS samples, namely E425S, E702S, and 08CF7S, and moderately influenced E704S. In particular, E201S was mostly shaped by organic matter concentration separated far from the others, while the length of this arrow indicated its limited influence on that direction. Temperature, pH value, salinity, and NH4 + nearly coinciding together with NH4 +/Σ(NO3 − + NO2 −) influenced the rest of the shallow SCS samples to some extent, respectively.
Canonical Correspondence Analysis (CCA) diagram representing the effect of environmental variations on anammox bacteria distribution pattern. Species units were defined on each clade from hzsB gene phylogenetic tree, and environmental elements were acquired from measured physicochemical parameter from South China Sea sediment samples. The angle direction and length indicated the correlation between environmental factor and axes, and the coordinates of samples showed the samples’ relationship driven by environmental factors. The eigenvalues of the first two axes were 0.764 and 0.658. Test of significance of first canonical axis showed that p value was 0.0740
Discussion
New functional gene primer application and efficiency
Degenerate PCR primers can be used for recovering all possible nucleotide combinations of the primary targeting sites to acquire gene fragments within the certain taxonomic level differences (Steffan and Atlas 1991). However, they could be so efficient to amplify non-specific gene fragments of anammox bacteria when the target group is only a very small portion (<1~10%) of the total DNA repository and background of the DNA samples is very complex (Kartal et al. 2011a; Schmid et al. 2007). In this study, primers based on hzsB gene showed good capability of retrieving targeted gene fragments with a number of combinations, and moreover, when applying on complex DNA samples of Mai Po wetland sediment, good efficiency was achieved with targeted PCR product length ranging 200-800 bp (Fig. S7a, e). In this study, one-step PCR with 40 cycles was sufficient to retrieve hzsB gene fragments from all three types of environmental samples. Different combinations of hzsC gene primers showed that a more “clear” DNA background from marine sediment samples allowed the primers to amplify the correct targeting genes compared to Mai Po samples. Several combinations of hzsC gene primers acquired unambiguous DNA bands from both marine and wetland samples (Fig. S7). Nested PCR method using two rounds of primer pair combinations designed in this study was successful in amplifying clear and bright DNA bands from nearly all samples of the above two environments, except for E201S (data not shown).
In the application of ccsA and ccsB gene primers on Mai Po and SCS samples, several combinations of ccsA gene primer pair showed good efficiency in retrieving targeting gene from both samples. Due to the few conservative sites on ccsB gene homologs (Fig. S2), one-step PCR was not capable of obtaining satisfactory results from both of the above environments (Kranz et al. 2009), but two-step nested PCR with long and short primer pair combinations successfully achieved ccsB gene libraries from all SCS samples without non-specific PCR products (data not shown).
When applying all the above four sets of designed primer pairs on WWTP samples, very clear and sharp bands from these samples were achieved, owing to the dominant anammox bacterial population and activity in the whole community. However, in order to discover new potential species and delineate the distribution pattern of anammox bacteria community, it is still important to develop PCR primers performing efficiently with various environments rather than enrichment cultures.
In terms of the capability of primers in distinguishing phylogenetic positions of anammox bacterial genera, results of functional genes were compared with those of 16S rRNA gene; the former performed better to reflect the diversity patterns of anammox bacteria in different ecotypes than the latter. Taking anammox bacteria community in WWTP samples as an example, 16S rRNA gene phylogeny indicated that most of the anammox bacteria belonged to Kuenenia while a small fraction belonged to Brocadia fulgida clade. When implemented with functional gene primers in this study, similar pattern appeared for hzsB clones (4/34, 10/23), for ccsA clones (3/34, 6/35), and for ccsB clones (1/34, 11/34) belonging to Brocadia, while the remaining belonged to Kuenenia. Furthermore, only one clone in E201S anammox bacteria clone library from SCS fell into non-Scalindua sp., as depicted by 16S rRNA gene-based phylogenetic tree, but hzsB gene phylogeny of marine sediment samples showed higher proportions of non-Scalindua. Both deep- and shallow-sea samples could be differentiated clearly, indicative of distinctive distribution of these two groups of anammox bacteria species towards marine sediment depth in the hzsC gene phylogenetic tree (Fig. 4). Phylogenetic tree of ccsA gene also displayed several clades with exclusive sequences from shallow and deep sea. Seawater depth might be a key factor shaping those two functional gene-containing communities.
Two pairs of functional gene primers targeting hzsB and hzsC genes were applied to quantitative detection of gene copies in the samples, and they were also compared with 16S rRNA gene qPCR primers. Pearson correlation analysis showed that anammox bacterial 16S rRNA gene abundance was positively correlated with hzsB gene; hzsB gene abundance was correlated with hzsC gene with high statistical support among SCS samples. In three SCS samples (E702S, E704, and E425S), anammox bacterial 16S rRNA gene abundance was higher than hzsB and hzsC gene abundance, which is in accordance with previous detection of hzsA and anammox bacterial 16S rRNA gene in anammox reactors, showing that hzsA gene abundances were 3-6 times lower than anammox bacterial 16S rRNA gene abundance in six out of eight reactors (Harhangi et al. 2012). Both hzsB and hzsC gene abundances were higher than the corresponding anammox bacterial 16S rRNA gene abundance in the remaining SCS samples. Newly sequenced “Candidatus Scalindua brodae” draft genome showed that fusion of hydrazine synthase operon hzsBC seemed to undergo a duplication in the genome so that there might also be two pairs of hzsBC operon in other unknown marine Scalindua species or even other unknown anammox bacteria groups (Speth et al. 2015). This could be a major reason for some of SCS marine samples, more hzsB and hzsC genes were detected than anammox bacterial 16S rRNA gene. Alternatively, hzsB and hzsC gene primer pairs of this study (hzsB364f-hzsB640r and hzsC745f-hzsC862r) were more efficient in detecting anammox bacteria abundance than anammox bacterial 16S rRNA gene due to the possibility that 16S rRNA gene primer pair A438f/A684r might be limited in its ability in amplifying 16S rRNA gene from new species of anammox bacteria (Humbert et al. 2012). In Mai Po and WWTP samples, similar abundance pattern among these three genes showed that hzsB and hzsC gene reflected higher anammox bacteria abundance than that in the 16S rRNA gene. Nearly identical gene abundance was detected with these three genes in MP7 from Mai Po wetland. Due to the different efficiency in retrieving genes from complex DNA background other than relatively simpler SCS samples, the correlation relationship of these three gene abundances was not so consistent when applying statistical analysis to Mai Po wetland samples. It is not known if non-specific amplification of hzsB and hzsC genes in Mai Po and WWTP samples or the primer bias and mismatch of 16S rRNA gene primer pair (Humbert et al. 2012) causes the differences in detecting anammox bacteria abundance. Functional gene hzsB and hzsC primer pairs are highly recommended in combination with anammox bacterial 16S rRNA gene primers to quantify anammox bacteria more accurately and reliably.
Anammox bacteria community patterns and influence of environmental factors
Scalindua spp. could be regarded as niche-specific species in marine environments and non-Scalindua spp. in freshwater/terrestrial environments (Schmid et al. 2007; Woebken et al. 2008). In this study, all retrieved 16S rRNA gene sequences from SCS belonged to Scalindua spp., with only one sequence from E201S fell into Brocadia fulgida/caroliniensis clade, indicating its alternative origin from terrestrial import. The Scalindua groups are divided into eight subclades, namely pacifica, arabica, wagneri, brodae/sorokinii/marina, zhenghei-I, zhenghei-II, zhenghei-III, and nanhaiensis in the phylogeny. The non-Scalindua subclades include Mai Po clade, Brocadia fulgida/caroliniensis, Brocadia anammoxdians/sinica, Anammoxoglobus, Jettenia, and Kuenenia. One new candidate species, S. nanhaiensis, is erected as a new group of Scalindua species which is discovered in this study and acquires average evolutionary distance of 0.039 within species, a suitable value for assigning a new species (nan.hai.en’sis. N.L. masc. adj. nanhaiensis stands for the SCS in Chinese). Each marine subclade was distributed rather evenly among SCS samples, with the composition changes from 11 to 23%, except for Scalindua zhenghei-III (no clone from SCS) and wagneri subclade (two clones from E201S). They could be reasonably missing due to primer bias and their very low representation in SCS samples, because other evidence of sequences affiliated to these above two subclades is available (Hong et al. 2011; Li et al. 2013b). Comparing with the previously estimated diversity pattern of marine anammox bacteria, SCS surface samples in this study harbored 10.6% sequence variance, greater than the maximum ∼5% 16S rRNA gene sequence variance observed by molecular investigation of four separated suboxic waters and ∼8% sequence variance proposed if all Scalindua species were recovered (Woebken et al. 2008). This finding largely expands the group of Scalindua marine species and also indicates the existence of considerably diversified groups of anammox bacteria inhabiting in SCS sediments.
In delineating the distribution of anammox bacteria from SCS samples by 16S rRNA and hzsB gene, their apparent separation from other environmental samples was evident in PCoA diagrams (Fig. 7). Community shift from the shallow SCS to deep SCS was detected before as a coastal-ocean distribution pattern depending on the ocean water depth not only reflected from anammox bacteria but also from aerobic ammonium-oxidizing prokaryotes (Cao et al. 2011a; Han and Gu 2013; Li et al. 2013a; Li et al. 2013b; Wang et al. 2012a). Functional gene hzsB is also capable of revealing the community composition shift as by 16S rRNA gene with better resolution. Based on the CCA analysis, this coastal-ocean pattern could be further substantiated that depth factor might be cooperated with ammonium, pH value, temperature, and salinity as an assemblage to shape the distinctive separation between shallow- and deep-sea samples. And organic matter concentration also shaped the anammox bacterial community of E201S sample. Pearson correlation analysis between the abundance and richness with environmental factors indicated that anammox bacterial abundance positively correlated by seawater depth, however is not consistent with the finding by others previously, stating that NO3 − concentration influenced most on their abundance (Han and Gu 2013) or abundance of anammox bacteria and nirS-encoding nitrite-reducing bacteria were strongly correlated with pH value, organic matter contents, and NH4 +/Σ(NO3 − + NO2 −) (Li et al. 2013b). Probably, congruence of the abundance of three genes (16S rRNA, hzsB, and hzsC gene) reflected the most accurate anammox bacteria abundance in this study, though inactive microbial cells could also be counted. Alternatively, the option of using different functional genes will lead to the emphasis on the distribution pattern of specific functional groups of anammox bacteria, while the different functional gene copy numbers in anammox bacteria might be different, and inactive genes may also cause the disparities. NH4 + positively correlated with Chao1 index while organic matters negatively with Shannon index of 16S rRNA gene strongly. NH4 +/Σ(NO3 − + NO2 −) positively correlated with Chao1 index of hzsB gene. Salinity positively correlated with Shannon index of hzsC and ccsB genes. Investigation of anammox bacteria community under anthropogenic influences in Cape Fear River Estuary stated that salinity strongly shaped the diversity and abundance of anammox bacteria. Variations of salinity improved diversity of anammox bacteria and correlated with the dominance of certain anammox bacteria species locally, and its abundance increased along the gradient of salinity in estuarine environments (Dale et al. 2009). The coastal-ocean pattern of this study suggested that the abundance of anammox bacteria increased along the slope from shallow- to deep-sea sediments, and NH4 + together with NH4 +/Σ(NO3 − + NO2 −) were positively correlated with the diversity indices of anammox bacterial communities derived from16S rRNA and hzsB genes. Furthermore, hzsC and ccsB gene-based community composition showed that two characteristic groups consisting of shallow- and deep-sea sediment samples were clearly differentiated (Fig. 7), and, according to hzsC gene-based phylogenetic tree, the Scalindua sp. clade could be divided into two large separated subclades comprising of exclusive shallow- or deep-sea sediment-derived sequences (Fig. 4). In terms of hzsC and ccsB gene-based community distribution patterns, distances among shallow-sea samples were smaller than those among deep-sea samples (Fig. 7). Unlike the variation tendency of community diversity indices reflected by anammox bacterial 16S rRNA and hzsB genes, Shannon indices of hzsC and ccsB genes were positively correlated with salinity. Given the aforementioned evidence, salinity might be the driving force for the differentiation of the distribution pattern of hzsC and ccsB gene-harboring anammox bacteria. Our research indicated that more detail influential factors responsible for the coastal-ocean distribution pattern might be undercover and needing for higher resolution investigation. Niche-specific influential factors along the coastal-ocean gradient, such as direct substrates and little organic compound amount, might be the primary force behind shaping specific anammox bacteria communities (Li et al. 2013a). The anthropogenic influence with terrestrial input from Pearl River Delta Estuary to pristine ocean could also be possibly responsible for imported exogenous terrestrial anammox bacteria groups in the ocean. Both of the explanations can be combined to explain the processes, and anammox bacteria composition could serve as a bioindicator for monitoring anthropogenic/terrestrial input to marine environments (Li et al. 2013a).
New Mai Po clade paralleling with other non-Scalindua groups was detected and then proposed in this study, and its relatedness with a group of Yangtze River Estuary sequences was shown. Its existence indicated that the distribution pattern of this clade was not geographically differentiated, but rather, certain groups with close evolutionary and functional properties may share similar niche-specific environments (Woebken et al. 2008). There is a paralleling clade to Kuenenia evident in both hzsB and hzsC gene phylogenetic trees formed by sequences from Mai Po wetland, representing a distinct group of terrestrial non-Scalindua clade (Figs. 3 and 4). The new Mai Po clade discovered in this study represents a new anammox bacteria species based on <97% similarity of 16S rRNA gene sequence. Mai Po wetland is influenced by terrestrial input and consists of a mosaic of different niches including coastal Mai Po forest wetland and intertidal mudflats. The in situ physicochemical properties are also imposed by the interactions among mangrove trees, macrofauna/microfauna, and microorganisms (Cao et al. 2011c; Dai et al. 2008). In addition, dynamics of seasonality and rhizosphere activity of the mangrove forest influence the sediment properties through root metabolism and leaf/debris absorption, shaping the microbial community composition (Wang et al. 2013). PCoA analysis showed that Mai Po sample coordinates, considering the depth and sediment properties, dispersed from each other more for community structures by both 16S rRNA and hzsB genes. However, communities represented by hzsC gene showed a tight grouping, possibly due to constraints of nested PCR method. In the diagram depicted by hzsB gene, surface samples grouped closer than subsurface samples, indicative of the depth rather than the sediment properties, imposing more influence on the anammox bacteria communities. Functional genes rather than the 16S rRNA gene should be better markers for physiological and metabolic information of anammox bacteria. Certain anammox bacteria are much more versatile due to utilization of propionate and acetate as energy sources simultaneously with anammox reaction (Güven et al. 2005). As an example, Anammoxoglobus propionicus outcompetes other anammox bacteria and heterotrophic denitrifiers in the presence of ammonium and propionate (Kartal et al. 2007). Additionally, genomic evidence of Kuenenia stuttgartiensis showed its versatile heterotrophic capability of iron respiration with formate as the elector donor and that of Scalindua profunda indicated its ability of reducing nitrate via nitrite to ammonium using organic matters and the assimilation of small organic compounds as alternative versatile lifestyle when the electron acceptor is limited (van de Vossenberg et al. 2012). There are significant differences in growth rate; influences by pH, temperature, dissolved oxygen (DO), phosphate and salinity on anammox activity; affinity for ammonium, nitrite, and tolerance of nitrite of three identified anammox bacteria enrichments (Oshiki et al. 2011). Different anammox bacteria species have differences in adapting to various environmental conditions, and it contributes to form the unique community composition in a specific microniche. Available organic compounds from the degradation of debris, root excretion, and external input may shape the distribution pattern of anammox bacteria when substrate nitrite is very limited. As a result, a wide coverage of different known anammox bacteria genera can be discovered in Mai Po wetland (Han and Gu 2013; Li et al. 2011c). Other influential factors, not only inorganic nitrogen chemicals but also ions, dipeptides/oligopeptides, and small organic compounds, should be further taken into consideration in the future research.
The consistency of 16S rRNA, hzsB, and hzsC gene abundances in SCS and WWTP samples and suitability of hzsB gene primers on quantifying gene abundance in complex Mai Po wetland suggested that qPCR primers based on hzsB gene in this study are more suitable as a biomarker to detect anammox bacterial abundance. Considerable diversity pattern of Scalindua species detected in SCS samples is confirmed again with the 16S rRNA gene phylogenetic analysis, and functional genes served well in unraveling anammox bacteria richness and diversity among SCS samples in this study.
Given the uniqueness and specificity of functional gene encoding central metabolic enzymes involved in anammox activity, newly discovered genes can serve well in phylogenetic reconstruction and also facilitate and support further detection and monitoring the dynamics and activities of anammox bacteria in natural and man-made environments. A coastal-ocean distribution pattern is evident based on data collected from SCS sediment samples. NH4 + and NH4 +/Σ(NO3 − + NO2 −) positively correlated with anammox bacterial gene diversity, and organic matter concentration negatively correlated with anammox bacterial gene diversity in SCS. Salinity was positively associated with diversity indices of hzsC and ccsB gene-harboring anammox bacteria communities and could potentially differentiate the distribution patterns between shallow- and deep-sea sediment samples. Because of complexity of ecological properties and nutrient dynamics, Mai Po wetland harbors highly complex anammox bacterial communities. Wider coverage of anammox bacterial genera at this site is probably niche-specific, possibly including hydrodynamics, nutrients, and redox conditions. Furthermore, comprehensive understanding on the distribution pattern of anammox bacteria in coastal and ocean environments will allow delineation of the microbial responses to human impacts on the natural ecosystems.
References
Braak Ct, Šmilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, New York
Brandsma J, van de Vossenberg J, Risgaard-Petersen N, Schmid MC, Engstrom P, Eurenius K, Hulth S, Jaeschke A, Abbas B, Hopmans EC, Strous M, Schouten S, Jetten MSM, Damste JSS (2011) A multi-proxy study of anaerobic ammonium oxidation in marine sediments of the Gullmar Fjord, Sweden. Environ Microbiol Rep 3(3):360–366. doi:10.1111/j.1758-2229.2010.00233.x
Broda E (1977) Two kinds of lithotrophs missing in nature. Z Allg Mikrobiol 17(6):491–493
Byrne N, Strous M, Crepeau V, Kartal B, Birrien J-L, Schmid M, Lesongeur F, Schouten S, Jaeschke A, Jetten M, Prieur D, Godfroy A (2009) Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J 3(1):117–123. doi:10.1038/ismej.2008.72
Cao H, Hong Y, Li M, Gu J-D (2011a) Diversity and abundance of ammonia-oxidizing prokaryotes in sediments from the coastal Pearl River estuary to the South China Sea. Antonie Van Leeuwenhoek 100(4):545–556
Cao H, Hong Y, Li M, Gu J-D (2011b) Phylogenetic diversity and ecological pattern of ammonia-oxidizing archaea in the surface sediments of the Western Pacific. Microb Ecol 62(4):813–823
Cao H, Li M, Hong Y, Gu J-D (2011c) Diversity and abundance of ammonia-oxidizing archaea and bacteria in polluted mangrove sediment. Syst Appl Microbiol 34(7):513–523
Chan HW, Meng H, Gu J-D (2016) Anammox bacteria detected in fish intestinal tract systems. Appl Environ Biotechnol 1(1):13-18 doi:10.18063/AEB.2016.01.010
Dai M, Wang L, Guo X, Zhai W, Li Q, He B, Kao S-J (2008) Nitrification and inorganic nitrogen distribution in a large perturbed river/estuarine system: the Pearl River Estuary, China. Biogeosci Disc 5(2):1545–1585
Dale OR, Tobias CR, Song B (2009) Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River Estuary. Environ Microbiol 11(5):1194–1207
Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuna-Gonzalez J (2003) N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422(6932):606–608. doi:10.1038/nature01526
Ferousi C, Speth DR, Reimann J, den Camp HJO, Allen JW, Keltjens JT, Jetten MS (2013) Identification of the type II cytochrome c maturation pathway in anammox bacteria by comparative genomics. BMC Microbiol 13(1):265
Frawley ER, Kranz RG (2009) CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A 106(25):10201–10206
Gori F, Tringe SG, Kartal B, Marchiori E, Machiori E, Jetten M (2011) The metagenomic basis of anammox metabolism in Candidatus ‘Brocadia fulgida’. Biochem Soc Trans 39(6):1799–1804
Güven D, Dapena A, Kartal B, Schmid MC, Maas B, van de Pas-Schoonen K, Sozen S, Mendez R, den Camp HJO, Jetten MS (2005) Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 71(2):1066–1071
Hamady M, Lozupone C, Knight R (2009) Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4(1):17–27
Han P, Gu J-D (2013) More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl Microbiol Biotechnol 97(8):3653–3663
Harhangi HR, Le Roy M, van Alen T, Hu B-l, Groen J, Kartal B, Tringe SG, Quan Z-X, Jetten MS, den Camp HJO (2012) Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl Environ Microbiol 78(3):752–758
Hira D, Toh H, Migita CT, Okubo H, Nishiyama T, Hattori M, Furukawa K, Fujii T (2012) Anammox organism KSU-1 expresses a NirK-type copper-containing nitrite reductase instead of a NirS-type with cytochrome cd 1. FEBS Lett 586(11):1658–1663
Hirsch MD, Long ZT, Song B (2011) Anammox bacterial diversity in various aquatic ecosystems based on the detection of hydrazine oxidase genes (hzoA/hzoB). Microb Ecol 61(2):264–276
Hong Y-G, Li M, Cao H, Gu J-D (2011) Residence of habitat-specific anammox bacteria in the deep-sea subsurface sediments of the South China Sea: analyses of marker gene abundance with physical chemical parameters. Microb Ecol 62(1):36–47
Hu B-l, Rush D, van der Biezen E, Zheng P, van Mullekom M, Schouten S, Damste JSS, Smolders AJP, Jetten MSM, Kartal B (2011a) New anaerobic, ammonium-oxidizing community enriched from peat soil. Appl Environ Microbiol 77(3):966–971. doi:10.1128/aem.02402-10
Hu B, Shen L, Xu X, Zheng P (2011b) Anaerobic ammonium oxidation (anammox) in different natural ecosystems. Biochem Soc Trans 39(6):1811
Hu Z, Speth DR, Francoijs K-J, Quan Z-X, Jetten M (2012) Metagenome analysis of a complex community reveals the metabolic blueprint of anammox bacterium “Candidatus Jettenia asiatica”. Front Microbio 3. doi:10.3389/fmicb.2012.00366
Humbert S, Zopfi J, Tarnawski SE (2012) Abundance of anammox bacteria in different wetland soils. Environ Microbiol Rep 4(5):484–490. doi:10.1111/j.1758-2229.2012.00347.x
Jetten MSM, Strous M, van de Pas-Schoonen KT, Schalk J, van Dongen U, van de Graaf AA, Logemann S, Muyzer G, van Loosdrecht MCM, Kuenen JG (1998) The anaerobic oxidation of ammonium. FEMS Microbiol Rev 22(5):421–437. doi:10.1016/s0168-6445(98)00023-0
Jetten MSM, Sliekers O, Kuypers M, Dalsgaard T, van Niftrik L, Cirpus I, van de Pas-Schoonen K, Lavik G, Thamdrup B, Le Paslier D, Op den Camp HJM, Hulth S, Nielsen LP, Abma W, Third K, Engstrom P, Kuenen JG, Jorgensen BB, Canfield DE, Damste JSS, Revsbech NP, Fuerst J, Weissenbach J, Wagner M, Schmidt I, Schmid M, Strous M (2003) Anaerobic ammonium oxidation by marine and freshwater planctomycete-like bacteria. Appl Microbiol Biotechnol 63(2):107–114. doi:10.1007/s00253-003-1422-4
Jetten MS, Lv N, Strous M, Kartal B, Keltjens JT, Op den Camp HJ (2009) Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol 44(2–3):65–84
Junier P, Molina V, Dorador C, Hadas O, Kim O-S, Junier T, Witzel K-P, Imhoff JF (2010) Phylogenetic and functional marker genes to study ammonia-oxidizing microorganisms (AOM) in the environment. Appl Microbiol Biotechnol 85(3):425–440
Kartal B, Rattray J, van Niftrik LA, van de Vossenberg J, Schmid MC, Webb RI, Schouten S, Fuerst JA, Damsté JS, Jetten MS (2007) Candidatus “Anammoxoglobus propionicus” a new propionate oxidizing species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 30(1):39–49
Kartal B, Geerts W, Jetten MS (2011a) Cultivation, detection, and ecophysiology of anaerobic ammonium-oxidizing bacteria. Methods Enzymol 486(Part A):89–108
Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, den Camp HJO, Harhangi HR, Janssen-Megens EM, Francoijs K-J (2011b) Molecular mechanism of anaerobic ammonium oxidation. Nature 479(7371):127–130
Kranz RG, Richard-Fogal C, Taylor J-S, Frawley ER (2009) Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev 73(3):510–528
Kuenen JG (2008) Anammox bacteria: from discovery to application. Nat Rev Microbiol 6(4):320–326
Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, Jorgensen BB, Jetten MSM (2005) Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci U S A 102(18):6478–6483. doi:10.1073/pnas.0502088102
Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D, Gutiérrez D, Amann R, Jetten MS, Kuypers MM (2009) Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci U S A 106(12):4752–4757
Lau S, Chu L (1999) Water quality degradation at the Mai Po Marshes Nature Reserve (Hong Kong) with reference to nutrient enrichment. Hydrobiologia 403:195–203. doi:10.1023/A:1003759215909
Li M, Gu J-D (2011) Advances in methods for detection of anaerobic ammonium oxidizing (anammox) bacteria. Appl Microbiol Biotechnol 90(4):1241–1252
Li M, Gu J-D (2016) Molecular evidence of the existence of anaerobic ammonia oxidation bacteria in the gut of polychaete (Neanthes glandicincta). Appl Environ Biotechnol 1(1):19–29 doi:10.1820063/AEB.2016.01.011
Li H, Chen S, Mu B-Z, Gu J-D (2010a) Molecular detection of anaerobic ammonium-oxidizing (anammox) bacteria in high-temperature petroleum reservoirs. Microb Ecol 60(4):771–783. doi:10.1007/s00248-010-9733-3
Li M, Hong Y, Klotz MG, Gu J-D (2010b) A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol 86(2):781–790
Li M, Cao H, Hong Y-G, Gu J-D (2011a) Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po nature reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) Genes. Microbes Environ 26(1):15–22. doi:10.1264/jsme2.ME10131
Li M, Ford T, Li X, Gu J-D (2011b) Cytochrome cd1-containing nitrite reductase encoding gene nirS as a new functional biomarker for detection of anaerobic ammonium oxidizing (Anammox) bacteria. Environ Sci Technol 45(8):3547–3553
Li M, Hong Y-G, Cao H-L, Gu J-D (2011c) Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) encoding gene analyses. Ecotoxicology 20(8):1780–1790
Li M, Cao H, Hong Y, Gu J-D (2013a) Using the variation of anammox bacteria community structures as a bio-indicator for anthropogenic/terrestrial nitrogen inputs in the Pearl River Delta (PRD). Appl Microbiol Biotechnol 97(22):9875–9883
Li M, Hong Y, Cao H, Gu J-D (2013b) Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea. Microb Ecol:1–16
Liang Y, Wong MH (2003) Spatial and temporal organic and heavy metal pollution at Mai Po Marshes Nature Reserve, Hong Kong. Chemosphere 52(9):1647–1658
Moore TA, Xing Y, Lazenby B, Lynch MDJ, Schiff S, Robertson WD, Timlin R, Lanza S, Ryan MC, Aravena R, Fortin D, Clark ID, Neufeld JD (2011) Prevalence of anaerobic ammonium-oxidizing bacteria in contaminated groundwater. Environ Sci Technol 45(17):7217–7225. doi:10.1021/es201243t
Motulsky H (1999) Analyzing data with GraphPad prism. GraphPad Software Incorporated
Mulder A, Graaf A, Robertson L, Kuenen J (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized bed reactor. FEMS Microbiol Ecol 16(3):177–184
Oshiki M, Shimokawa M, Fujii N, Satoh H, Okabe S (2011) Physiological characteristics of the anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia sinica’. Microbiology 157(6):1706–1713. doi:10.1099/mic.0.048595-0
Quan ZX, Rhee SK, Zuo JE, Yang Y, Bae JW, Park JR, Lee ST, Park YH (2008) Diversity of ammonium-oxidizing bacteria in a granular sludge anaerobic ammonium-oxidizing (anammox) reactor. Environ Microbiol 10(11):3130–3139
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schmid M, Walsh K, Webb R, Rijpstra WI, van de Pas-Schoonen K, Verbruggen MJ, Hill T, Moffett B, Fuerst J, Schouten S (2003) Candidatus “Scalindua brodae”, sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst Appl Microbiol 26(4):529–538
Schmid MC, Maas B, Dapena A, van de Pas-Schoonen K, van de Vossenberg J, Kartal B, Van Niftrik L, Schmidt I, Cirpus I, Kuenen JG (2005) Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl Environ Microbiol 71(4):1677–1684
Schmid MC, Risgaard-Petersen N, Van De Vossenberg J, Kuypers MM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9(6):1476–1484
Schmid MC, Hooper AB, Klotz MG, Woebken D, Lam P, Kuypers MM, Pommerening-Roeser A, Op Den Camp HJ, Jetten MS (2008) Environmental detection of octahaem cytochrome c hydroxylamine/hydrazine oxidoreductase genes of aerobic and anaerobic ammonium-oxidizing bacteria. Environ Microbiol 10(11):3140–3149
Speth DR, Russ L, Kartal B, Op den Camp HJ, Dutilh BE, Jetten MS (2015) Draft genome sequence of anammox bacterium “Candidatus Scalindua brodae”, obtained using differential coverage binning of sequencing data from two reactor enrichments. Genome Announc 3(1):e01415–e01414
Steffan RJ, Atlas RM (1991) Polymerase chain reaction: applications in environmental microbiology. Annu Rev Microbiol 45(1):137–161. doi:10.1146/annurev.mi.45.100191.001033
Strous M, Fuerst JA, Kramer EH, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MS (1999) Missing lithotroph identified as new planctomycete. Nature 400(6743):446–449
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440(7085):790–794
Sun W, Xu MY, Wu WM, Guo J, Xia CY, Sun GP, Wang AJ (2014) Molecular diversity and distribution of anammox community in sediments of the Dongjiang River, a drinking water source of Hong Kong. J Appl Microbiol 116(2):464–476. doi:10.1111/jam.12367
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Thamdrup B, Dalsgaard T (2002) Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol 68(3):1312–1318. doi:10.1128/aem.68.3.1312-1318.2002
Thamdrup B, Dalsgaard T, Jensen MM, Ulloa O, Farias L, Escribano R (2006) Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol Oceanogr 51(5):2145–2156
van de Vossenberg J, Rattray JE, Geerts W, Kartal B, van Niftrik L, van Donselaar EG, Damste JSS, Strous M, Jetten MSM (2008) Enrichment and characterization of marine anammox bacteria associated with global nitrogen gas production. Environ Microbiol 10(11):3120–3129. doi:10.1111/j.1462-2920.2008.01643.x
van de Vossenberg J, Woebken D, Maalcke WJ, Wessels HJ, Dutilh BE, Kartal B, Janssen-Megens EM, Roeselers G, Yan J, Speth D (2012) The metagenome of the marine anammox bacterium ‘Candidatus Scalindua profunda’ illustrates the versatility of this globally important nitrogen cycle bacterium. Environ Microbiol
Wang J, Gu J-D (2013a) Dominance of Candidatus Scalindua species in anammox community revealed in soils with different duration of rice paddy cultivation in Northeast China. Appl Microbiol Biotechnol 97(4):1785–1798. doi:10.1007/s00253-012-4036-x
Wang Y-F, Gu J-D (2013b) Higher diversity of ammonia/ammonium-oxidizing prokaryotes in constructed freshwater wetland than natural coastal marine wetland. Appl Microbiol Biotechnol 97(15):7015–7033. doi:10.1007/s00253-012-4430-4
Wang S, Zhu G, Peng Y, Jetten MS, Yin C (2012a) Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environ Sci Technol 46(16):8834–8842
Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MS, Yin C, Op den Camp HJ (2012b) Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett 336(2):79–88
Wang Y-F, Feng Y-Y, Ma X, Gu J-D (2013) Seasonal dynamics of ammonia/ammonium-oxidizing prokaryotes in oxic and anoxic wetland sediments of subtropical coastal mangrove. Appl Microbiol Biotechnol 97(17):7919–7934. doi:10.1007/s00253-012-4510-5
Woebken D, Lam P, Kuypers MM, Naqvi SW, Kartal B, Strous M, Jetten MS, Fuchs BM, Amann R (2008) A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol 10(11):3106–3119. doi:10.1111/j.1462-2920.2008.01640.x
Wright ES, Yilmaz LS, Noguera DR (2012) DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78(3):717–725
Yoshinaga I, Amano T, Yamagishi T, Okada K, Ueda S, Sako Y, Suwa Y (2011) Distribution and diversity of anaerobic ammonium oxidation (anammox) bacteria in the sediment of a eutrophic freshwater lake, Lake Kitaura, Japan. Microbes Environ 26(3):189–197. doi:10.1264/jsme2.ME10184
Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MS, Yin C (2011) Anaerobic ammonia oxidation in a fertilized paddy soil. ISME J 5(12):1905–1912
Acknowledgments
This research was supported by a postgraduate student studentship from The University of Hong Kong (ZZ) and fund from RGC GRF grant No. 701913 (J-DG). Kelly Lau was thanked for laboratory assistance and technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the RGC GRF grant no. 701913 (J-DG) and a Hong Kong PhD fellowship (ZZ).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(4.61 MB)
Rights and permissions
About this article
Cite this article
Zhou, Z., Chen, J., Meng, H. et al. New PCR primers targeting hydrazine synthase and cytochrome c biogenesis proteins in anammox bacteria. Appl Microbiol Biotechnol 101, 1267–1287 (2017). https://doi.org/10.1007/s00253-016-8013-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-016-8013-7