Abstract
The variation of anammox bacteria community composition was evaluated in sediments collected from the Pearl River Delta area with an anthropogenic/terrestrial input gradient. Results indicated that the community composition of anammox bacteria shifted from estuarine environment to the South China Sea deep ocean along with the anthropogenic/terrestrial input gradient, where Scalindua genus of anammox bacteria predominated in the area with less anthropogenic/terrestrial influences, such as in the open oceanic area, while genera of Kuenenia/Brocadia anammox bacteria have higher proportions in the area with higher anthropogenic/terrestrial impacts. The canonical correspondence analysis demonstrated that salinity, organic matter contents, and ratio of NH4 + to (NO2 −+NO3 −) strongly affected the shifting of anammox bacterial community compositions within the same gradients. The results obtained in this study, together with the similar variation of anammox bacteria community structures in other several estuaries in the world, indicated that anammox bacteria might have a habitat-specific distribution pattern according to their living habits, and their community composition could be served as a bio-indicator to monitor the anthropogenic/terrestrial N inputs in coastal environments.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Estuaries, connecting the freshwater and marine environment, are among the most dynamic and the productive ecosystems on Earth. However, they are under threat from anthropogenic N loading, resulting in various environmental problems. The Pearl River Delta (PRD), the low-lying area surrounding the Pearl River Estuary (PRE) where the Pearl River flows into the South China Sea (SCS), is one of the fast-growing economic regions and is a major manufacturing center of the world with a population of approximately 20 million. In the PRD, many important cities in China, such as Hong Kong, Macau, Guangzhou, Shenzhen, and Dongguan, are located in this area; thus, the long-term sustainability of the regional economy and environmental quality is the central focus in urban planning. However, as one of the most densely urbanized regions in the world, PRD is notoriously polluted by sewage and industrial wastewater as observed on the steadily deterioration of water quality in recent years (Yin and Harrison 2007). It has been reported that 4.7∼8.3 billion tons of sewage were discharged into the ocean off the coast of PRD area in each year (http://www.gdofa.gov.cn/index.php/Catagories/index/id/247). Pollutions are evident in the PRD, and among them, reactive N species (NO3 −, NO2 −, NO x , etc.) are a class of the most important pollutants in the PRE. In the water column, the inorganic N is high accounting for 85 % of total N, while NO3–N is 95 % of inorganic N, but both NO2–N and NH4–N are low. In contrast, the content of total N is relatively high in the surface sediment on average 1,649 mg kg−1, in which 83 % are organic N that averaged at 1,374 mg kg−1. NH4–N is the main form of inorganic N on average of 209 mg kg−1, with NO3–N and NO2–N together at 54 mg kg−1 (http://www.gdofa.gov.cn/index.php/Catagories/index/id/247). According to the available evidence, the enrichment of N in subtropical PRE coastal water and sediment has resulted in various symptoms of eutrophication and other many associated consequences, including the low diversity of benthic infauna (Shen et al. 2010) and the increasing frequency of algal blooms (Yin and Harrison 2007; Yin et al. 2004). Given that the demand for N in food production is continuing to increase, the intensity of pollution from N use may increase by N discharges associated with human activities, such as untreated domestic and industrial wastewaters, into estuarine and coastal environments (Galloway et al. 2008). The additional anthropogenic reactive nitrogen is expected to affect the chemistry of the atmosphere, and the composition and function of the terrestrial and aquatic ecosystems, with possible implications even for global climate change (Mulder et al. 1995; Vandegraaf et al. 1995). Although limited reports have been involved in the N cycle in the PRD (Dai et al. 2008; Wang et al. 2012; Yin et al. 2001; Zhang et al. 1999), no mechanism for the progressively worsening environmental conditions has been formulated.
Current knowledge on N cycle has not been fully accounted for the N flux in estuarine ecosystems, but the intensity and duration of estuarine eutrophication and the rate of estuarine recovery strongly depend on microbial N removal processes, including anammox and denitrification (Nicholls and Trimmer 2009). Denitrification has been widely studied in various estuaries, while anammox is much less studied in estuarine ecosystems. Existing studies indicate that anammox rates and its contribution to total N2 production (anammox significance) are system specific and controlled in part by reaction-scale substrate limitations and by environmental parameters (Dale et al. 2009). Furthermore, the community structure of anammox bacteria, linking to their activities, has shown a strong variation along with environmental gradients in natural ecosystems, such as the estuaries (Nicholls and Trimmer 2009; Rich et al. 2008; Risgaard-Petersen et al. 2004; Wang et al. 2012). Several recent reports also indicated that the structures of anammox bacterial communities might be strongly affected by anthropogenic pollution inputs (Amano et al. 2011; Dang et al. 2010; Li et al. 2010, 2011a), and our previous studies have evaluated the responses of aerobic and anaerobic ammonia/ammonium-oxidizing microorganisms to anthropogenic pollution in coastal environments, providing some hints that anammox bacteria may serve as bio-indicators for environmental quality (Cao et al. 2011; Li et al. 2011a); however, more evidence is needed to more comprehensively elucidate this hypothesis.
To better understand the relationship of anammox bacteria communities and the anthropogenic/terrestrial nitrogen inputs, this study will investigate the variation of anammox bacterial community structures from PRD coastal sediments to SCS open ocean surface and subsurface sediments, which is representing an anthropogenic or terrestrial input gradient. In addition, the results of this study will also be compared with several other estuarine coastal ecosystems, including Yodo River Estuary, Cape Fear River Estuary, and Colne Estuary. The results indicate that the anammox bacteria community structures shifted along with the anthropogenic/terrestrial input gradients, and the variation of their community composition could serve as a bio-indicator to monitor the anthropogenic or terrestrial inputs to marine coastal environments.
Materials and methods
Data collection and description
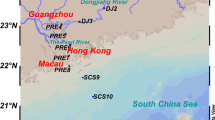
Three different ecosystems selected in this study are the estuary, the mangrove wetland, and the SCS, and the details of the sampling sites are shown in Fig. 1. Based on the previous studies, the anammox bacteria community structures have been independently described base on the 16S rRNA and hzo genes in three ecosystems (Hong et al. 2011; Li et al. 2010, 2011a, b, 2013a, b). Sequences obtained from these three ecosystems were divided into five groups, representing as summer estuarine sediments (MP-5), winter estuarine sediments (MP-11), mangrove sediments (mangrove), and surface (SCS-SS) and subsurface sediments (SCS-SB) of the SCS. Based on the basic physicochemical characteristics and their geological location, five group sediment samples form an anthropogenic/terrestrial input gradient with different influences (Table 1). In addition, variations of anammox bacteria community structures in several estuarine ecosystems, including Yodo River Estuary in Japan, Cape Fear River Estuary in USA, and Colne Estuary in UK, are also analyzed to parallel confirm the influences of anthropogenic or terrestrial inputs on the anammox bacterial community structures.
Phylogenetic and statistical analyses
All sequences of anammox bacterial 16S rRNA (nucleic acids) and hzo (amino acids) genes retrieved from the selected sampling sites of the present study are aligned using the ClustalW program (Thompson et al. 1994). Phylogenetic trees of 16S rRNA and hzo genes were constructed by MEGA 5.0 with the neighbor-joining method with 1,000 bootstrap replicate resampling method to estimate the confidence intervals of the tree nodes. Sequences of 16S rRNA and hzo genes were analyzed using the DOTUR program to compare their diversity and richness (Schloss and Handelsman 2005). Three percent cutoff for nucleotides (16S rRNA gene) and 5 % amino acid (hzo gene) sequence variations to define an operational taxonomic unit (OTU) (Schloss and Handelsman 2005). The anammox bacterial community compositions were analyzed based on the sequences of phylogenetic affiliation and their proportions in each sequence groups. Canonical correspondence analysis (CCA) was performed in CANOCO 4.5 for Windows to identify the relationships between anammox bacteria community structures and environmental parameters.
Variation of anammox bacterial community structures in other estuarine ecosystems
The variation of anammox bacteria community compositions in several other estuarine ecosystems was also calculated based on the available 16S rRNA gene sequences in the database. These estuarine ecosystems include the Yodo River Estuary in Japan (AB522738-AB522761), the Cape Fear River Estuary in USA (Dale et al. 2009), and the Colne Estuary in UK (Dong et al. 2009), and the proportions of different phylogenetic groups of anammox bacteria were calculated along with the anthropogenic or terrestrial input gradients in each ecosystem.
Nucleotide sequence accession numbers
The GenBank accession numbers of sequences reported in the PRD are the following: GQ427230 to GQ427485 and HM209472 to HM209609 for MP-5 and MP-11, HQ665558 to HQ66592 for SCS-SS, GQ331139 to GQ331201 and GQ1202 to GQ331244 for SCS-SB (16S rRNA gene), GQ427486 to GQ427673 and HM209610 to HM209725 for MP-5 and MP-11, GQ849414 to GQ849421 and HQ665927 to HQ666219 for SCS-SS, and GQ331245 to GQ331332 for SCS-SB (hzo gene).
Results
Phylogenetic diversity analysis
Diversity of anammox bacteria in the PRD was evaluated through the phylogenetic analyses of 16S rRNA and hzo genes, and two biomarkers of anammox bacteria obtained 41 and 136 OTUs from all selected sample groups, respectively. The OTU numbers showed similar variation trends with the values of Shannon, Chao, and Simpson diversity indices, indicating samples from the SCS-SS and MP-5 have a relatively higher diversity of anammox bacteria, while the lowest diversity of anammox bacteria was found in the SCS-SB samples (Table 2). From the phylogenetic relationship of anammox bacteria, 16S rRNA genes could be divided into six different phylogenetic groups, including Kuenenia cluster, Scalindua wagneri cluster, Scalindua zhenghei clusters I to III, and Scalindua arabica/brodae cluster, while the hzo genes were grouped into five different phylogenetic clusters representing as Kuenenia cluster and Scalindua clusters 1 to 4, respectively. Furthermore, different phylogenetic groups of anammox bacteria also showed that habitat-specific characteristics, such as S. zhenghei I of 16S rRNA and Scalindua cluster 2 of hzo, were only detected in the SCS surface and subsurface sediments (Figs. 2 and 3).
Community structure variation
After the phylogenetic diversity analysis, anammox bacteria in each phylogenetic group were calculated to clearly understand the community compositions and their variation in the five selected sample groups (Fig. 4). From the community compositions, it clearly showed that different phylogenetic groups of anammox bacteria in five sample groups have quite different proportions. In 16S rRNA gene database, the proportion of S. arabica/brodae cluster decreased from the SCS-SB (65.8 %) to the MP-5 (13.1 %) along with the increasing anthropogenic/terrestrial inputs, while S. zhenghei II group has relatively higher proportions in estuary and mangrove sediment samples than that in the SCS surface and subsurface sediments (Fig. 4a). However, the proportion of anammox bacterial hzo gene in Kuenenia cluster increased from under detection to 87.8 % from the SCS to the MP-5 sediment samples, and hzo Scalindua cluster 4 also has much higher proportions in the SCS samples than that in mangrove and estuary sediment samples (Fig. 4b).
Furthermore, the anammox community compositions in the Cape Fear River Estuary of USA (Dale et al. 2009), the Yodo River Estuary of Japan, and the Colne Estuary of UK (Dong et al. 2009) were also analyzed. Although no 16S rRNA gene sequences were found to be related to the S. zhenghei I–III in these estuaries, the relative proportion of S. wagneri cluster also decreased from the marine sites to the terrestrial sites along with the salinity decreasing gradients, while the Brocadia/Kuenenia clusters showed the opposite variation patterns at the same gradient (Fig. S1, results of Yodo River Estuary and the Cole Estuary were not shown in here). The variations of anammox bacteria community in these estuaries were consistent with that of our study.
Correlations between anammox bacterial community compositions and environmental factors
The CCA results showed that the CCA axes in 16S rRNA gene and hydrazine oxidoreductase (HZO) sequence-deduced schemes could explain more than 43.6 and 65.0 % of the cumulative variance of the correlation between the environmental factors and the anammox bacterial community distribution, respectively (Fig. 5). Furthermore, both 16S rRNA and HZO sequences indicated that all of the anammox bacterial assemblages fell into three groups, representing as MP-11 and mangrove, MP-5, and SCS (Fig. 5). From the diversity of anammox bacteria, it could be found that salinity negatively correlated with the anammox bacteria in Kuenenia, S. wagneri, and S. zhenghei II clusters and positively correlated with S. arabica/brodae and S. zhenghei I clusters in the 16S rRNA CCA plot, while in the HZO CCA plot, salinity negatively correlated with the anammox bacteria in Kuenenia and Scalindua cluster 1 and positively correlated with Scalindua clusters 2 and 4, indicating S. arabica/brodae and S. zhenghei I clusters of 16S rRNA gene as possible coordinate with Scalindua clusters 2 and 4 of HZO sequences. Furthermore, the ratio of NH4 + to (NO2 − + NO3 −) and organic matter also positively correlated with anammox bacteria in the Kuenenia cluster and S. zhenghei II and S. wagneri clusters, respectively (Fig. 5). From the correlations between the distribution of anammox bacteria and environmental factors, both 16S rRNA and HZO CCA plots also indicated the same results that salinity and pH are positively correlated with anammox bacteria distribution in the SCS samples and organic matter, concentrations of NH4+ and (NO2 − + NO3 −) positively correlated with the distribution in MP-11 and mangrove samples, and the ratio of NH4 + to (NO2 − + NO3 −) positively correlated with the distribution in MP-5 samples (Fig. 5).
CCA ordination plots for the physicochemical parameters. Anammox bacteria groups (triangle) represented by 16S rRNA (a) and HZO (b) sequences and their sampling locations (circle). Correlations between environmental variables and CCA axes are represented by the length and angle of arrows (environmental factors)
Discussion
In this study, the dynamics of anammox bacteria community structure in five group sediment samples collected from three ecosystems located in the PRD were evaluated, where these sediment samples form an anthropogenic/terrestrial input gradient. Through the evaluations of two molecular biomarkers (16S rRNA gene and HZO), we found that the proportion of anammox bacteria affiliated with genera of Kuenenia/Brocadia would increase along with the increasing anthropogenic or terrestrial inputs, while the proportions of S. arabica/brodae (or the hzo Scalindua cluster 4) decrease at the same gradient. Thus, it is clearly showed that the anammox bacteria community compositions shift from the high anthropogenic/terrestrial input environments (MP-5) to the low anthropogenic/terrestrial inputs and pristine environments (SCS-SS and SCS-SB). Furthermore, higher diversity of anammox bacteria (genus level) was also found in the high anthropogenic/terrestrial-influenced environments though anammox bacteria might also have a very high microdiversity within the same genus in the low anthropogenic/terrestrial inputs or pristine environments (SCS-SS). In addition, similar results found in the previous study (Wang et al. 2012) and several other estuarine ecosystems from USA (Dale et al. 2009), Japan, and UK were also provided with strong evidence to confirm the shifting of anammox bacteria community structures along with the anthropogenic/terrestrial input gradient (Fig. 4).
To explain how anammox bacteria community structures verify in these terrestrial and marine interacted zones, several possibilities should be considered. If we assume that all anammox bacteria retrieved from these sediment samples are endogenous, the variation of anammox bacteria community structures might be due to the different responses of different anammox bacteria to the environmental conditions. According to previous study, different species of anammox bacteria in different genera show different affinity for ammonium and nitrite and also have quite different tolerances on nitrite, dissolved oxygen, phosphate, and salinity (Oshiki et al. 2011). Thus, the rapid changing of physicochemical conditions in estuarine ecosystems would shape different anammox bacteria community structures, which is also discussed in several published reports. On the other hand, the anammox bacteria recovered from different sediment samples in the estuarine ecosystems might also be exogenous, such as through the wastewater or terrestrial inputs. Previous studies have demonstrated that Scalindua anammox bacteria are the dominant group in marine environments (Schmid et al. 2007; Woebken et al. 2008), while Brocadia and Kuenenia anammox bacteria are usually found in engineered and terrestrial systems (Jetten et al. 2005; Jetten et al. 2009). Therefore, higher proportion of Brocadia or Kuenenia anammox bacteria would be detected in the sediment samples with higher anthropogenic/terrestrial inputs but lower proportions or even absence of Brocadia or Kuenenia anammox bacteria could be detected in marine ecosystems with less anthropogenic/terrestrial inputs. Furthermore, it might be more reasonable to explain the variation of anammox bacteria in estuarine ecosystems by combing these two processes, where some Brocadia and Kuenenia anammox bacteria discovered in estuaries might originate from wastewater or terrestrial inputs, and these exogenous anammox bacteria, together with the endogenous anammox bacteria, show different responses on the changing environmental factors. However, no matter which process really causes the changing of anammox bacteria in estuaries, it is still reasonable to use the variation of anammox bacteria community structures as a bio-indicator to monitor the anthropogenic/terrestrial inputs of marine environments.
To further confirm our hypothesis, the correlations between anammox bacteria diversity distribution and environmental factors are also analyzed in the PRD sediment samples (Fig. 5). It is clearly showed that salinity strongly affects the community structures and distribution of anammox bacteria in the PRD sediments, especially the group of S. arabica/brodae. However, the significant influences of organic matters, concentrations of NH4 + and (NO2 − + NO3 −), and their ratio on the anammox bacteria community and distribution further provide evidence on the hypothesis by using the variation of anammox bacteria community structures as a bio-indicator to monitor the anthropogenic/terrestrial inputs of marine environments.
In summary, we have evaluated the community structure and their variation in sediment samples collected from the PRD area, and the correlations between environmental factors and the community composition and distribution were also analyzed in this study. Combining the results in the PRD area and that in several estuarine ecosystems, it clearly demonstrated that the variation of anammox bacteria community structures could serve as a bio-indicator to monitor the anthropogenic/terrestrial inputs of marine environments.
References
Amano T, Yoshinaga I, Yamagishi T, Chu VT, Pham TT, Ueda S, Kato K, Sako Y, Suwa Y (2011) Contribution of anammox bacteria to benthic nitrogen cycling in a mangrove forest and shrimp ponds, Haiphong, Vietnam. Microbes Environ 26(1):1–6
Cao HL, Li M, Dang HY, Gu JD (2011) Responses of aerobic and anaerobic ammonia/ammonium-oxidizing microorganisms to anthropogenic pollution in coastal marine environments. Method Enzymol 496:35–62
Dai M, Wang L, Guo X, Zhai W, Li Q, He B, Kao SJ (2008) Nitrification and inorganic nitrogen distribution in a large perturbed river/estuarine system: the Pearl River Estuary, China. Biogeosciences 5(5):1227–1244
Dale OR, Tobias CR, Song BK (2009) Biogeographical distribution of diverse anaerobic ammonium oxidizing (anammox) bacteria in Cape Fear River Estuary. Environ Microbiol 11(5):1194–1207
Dang HY, Chen RP, Wang L, Guo LZ, Chen PP, Tang ZW, Tian F, Li SZ, Klotz MG (2010) Environmental factors shape sediment anammox bacterial communities in hypernutrified Jiaozhou Bay, China. Appl Environ Microb 76(21):7036–7047
Dong LF, Smith CJ, Papaspyrou S, Stott A, Osborn AM, Nedwell DB (2009) Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne Estuary, United Kingdom). Appl Environ Microb 75(10):3171–3179
Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai ZC, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA (2008) Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320(5878):889–892
Hong YG, Li M, Cao HL, Gu JD (2011) Residence of habitat-specific anammox bacteria in the deep-sea subsurface sediments of the South China Sea: analyses of marker gene abundance with physical chemical parameters. Microb Ecol 62(1):36–47
Jetten MSM, Cirpus I, Kartal B, van Niftrik L, van de Pas-Schoonen KT, Sliekers O, Haaijer S, van der Star W, Schmid M, van de Vossenberg J, Schmidt I, Harhangi H, van Loosdrecht M, Kuenen JG, den Camp HO, Strous M (2005) 1994–2004: 10 years of research on the anaerobic oxidation of ammonium. Biochem Soc Trans 33:119–123
Jetten MSM, van Niftrik L, Strous M, Kartal B, Keltjens JT, Op den Camp HJM (2009) Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol 44(2–3):65–84
Li M, Hong YG, Klotz MG, Gu JD (2010) A comparison of primer sets for detecting 16S rRNA and hydrazine oxidoreductase genes of anaerobic ammonium-oxidizing bacteria in marine sediments. Appl Microbiol Biotechnol 86(2):781–790
Li M, Cao HL, Hong YG, Gu JD (2011a) Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po nature reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes Environ 26(1):15–22
Li M, Hong YG, Cao HL, Gu JD (2011b) Mangrove trees affect the community structure and distribution of anammox bacteria at an anthropogenic-polluted mangrove in the Pearl River Delta reflected by 16S rRNA and hydrazine oxidoreductase (HZO) encoding gene analyses. Ecotoxicology 20(8):1780–1790
Li M, Hong Y, Cao H, Gu JD (2013a) Community structures and distribution of anaerobic ammonium oxidizing and nirS-encoding nitrite-reducing bacteria in surface sediments of the South China Sea. Microb Ecol doi:10.1007/s00248-012-0175-y
Li M, Hong Y, Cao H, Klotz MG, Gu JD (2013b) Diversity, abundance, and distribution of NO-forming nitrite reductase-encoding genes in deep-sea subsurface sediments of the South China Sea. Geobiology 11(2):170–179
Mulder A, Vandegraaf AA, Robertson LA, Kuenen JG (1995) Anaerobic ammonium oxidation discovered in a denitrifying fluidized-bed reactor. FEMS Microbiol Ecol 16(3):177–183
Nicholls JC, Trimmer M (2009) Widespread occurrence of the anammox reaction in estuarine sediments. Aquat Microb Ecol 55(2):105–113
Oshiki M, Shimokawa M, Fujii N, Satohl H, Okabe S (2011) Physiological characteristics of the anaerobic ammonium-oxidizing bacterium 'Candidatus Brocadia sinica'. Microbiology 157:1706–1713
Rich JJ, Dale OR, Song B, Ward BB (2008) Anaerobic ammonium oxidation (anammox) in Chesapeake Bay sediments. Microb Ecol 55(2):311–320
Risgaard-Petersen N, Meyer RL, Schmid M, Jetten MSM, Enrich-Prast A, Rysgaard S, Revsbech NP (2004) Anaerobic ammonium oxidation in an estuarine sediment. Aquat Microb Ecol 36(3):293–304
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microb 71(3):1501–1506
Schmid MC, Risgaard-Petersen N, van de Vossenberg J, Kuypers MMM, Lavik G, Petersen J, Hulth S, Thamdrup B, Canfield D, Dalsgaard T, Rysgaard S, Sejr MK, Strous M, den Camp HJMO, Jetten MSM (2007) Anaerobic ammonium-oxidizing bacteria in marine environments: widespread occurrence but low diversity. Environ Microbiol 9(6):1476–1484
Shen PP, Zhou H, Gu JD (2010) Patterns of polychaete communities in relation to environmental perturbations in a subtropical wetland of Hong Kong. J Mar Biol Assoc UK 90(5):923–932
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Vandegraaf AA, Mulder A, Debruijn P, Jetten MSM, Robertson LA, Kuenen JG (1995) Anaerobic oxidation of ammonium is a biologically mediated process. Appl Environ Microb 61(4):1246–1251
Wang S, Zhu G, Peng Y, Jetten MS, Yin C (2012) Anammox bacterial abundance, activity, and contribution in riparian sediments of the Pearl River estuary. Environ Sci Technol 46(16):8834–8842
Woebken D, Lam P, Kuypers MMM, Naqvi SWA, Kartal B, Strous M, Jetten MSM, Fuchs BM, Amann R (2008) A microdiversity study of anammox bacteria reveals a novel Candidatus Scalindua phylotype in marine oxygen minimum zones. Environ Microbiol 10(11):3106–3119
Yin K, Harrison PJ (2007) Influence of the Pearl River estuary and vertical mixing in Victoria Harbor on water quality in relation to eutrophication impacts in Hong Kong waters. Mar Pollut Bull 54(6):646–656
Yin KD, Qian PY, Wu MCS, Chen JC, Huang LM, Song XY, Jian WJ (2001) Shift from P to N limitation of phytoplankton growth across the Pearl River estuarine plume during summer. Mar Ecol Prog Ser 221:17–28
Yin KD, Zhang JL, Qian PY, Jian WJ, Huang LM, Chen JF, Wu MCS (2004) Effect of wind events on phytoplankton blooms in the Pearl River estuary during summer. Cont Shelf Res 24(16):1909–1923
Zhang J, Yu ZG, Wang JT, Ren JL, Chen HT, Xiong H, Dong LX, Xu WY (1999) The subtropical Zhujiang (Pearl River) estuary: nutrient, trace species and their relationship to photosynthesis. Estuar Coast Shelf Sci 49(3):385–400
Acknowledgments
This research was supported by a PhD studentship (M.L.), Environmental and Conservation Fund grant no. 15/2011(J-DG), South China Sea Open Cruise by R/V Shiyan 3, and South China Sea Institute of Oceanology, CAS. We thank Jessie Lai for general laboratory support at The University of Hong Kong during this study.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 105 kb)
Rights and permissions
About this article
Cite this article
Li, M., Cao, H., Hong, Y. et al. Using the variation of anammox bacteria community structures as a bio-indicator for anthropogenic/terrestrial nitrogen inputs in the Pearl River Delta (PRD). Appl Microbiol Biotechnol 97, 9875–9883 (2013). https://doi.org/10.1007/s00253-013-4990-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-013-4990-y